Abstract
In the present study, mitochondrial 16S rRNA gene sequence analysis was used for identification of Cattle (Bos taurus), buffalo (Bubalus bubalis), sheep (Ovis aries), goat (Capra hircus) and pig (Sus scrofa) species in fresh and processed meat. The DNA was extracted from fresh and processed meat including autoclaved meat and meat emulsion followed by polymerase chain reaction (PCR) amplification of about 497 bp DNA fragments of mitochondrial 16S rRNA gene. Then the amplified PCR fragments were sequenced and analysed to differentiate the species. No adverse effect of ingredients and processing conditions was observed on PCR amplification of DNA extracted from heat-treated meat and meat emulsion. The closely related species such as cattle and buffalo, sheep and goat were differentiated from each other by sequence analysis. Thus, PCR amplification and sequence analysis of mitochondrial 16S rRNA gene can be used as a tool for authentic identification of meat species.
Introduction
Misrepresentation of meat and meat product is a common fraudulence practice prevalent in the meat industry. Various methods were developed and employed for detection of falsification of meat and meat products. However, DNA-based methods particularly polymerase chain reaction (PCR) amplification and DNA sequencing were proved to be more precise and authentic tools for detection of species origin of meat. Both genomic and mitochondrial genes were targeted to identify species origin of meat by PCR amplification (Mane et al. Citation2007, Citation2012) followed by sequencing. Use of mitochondrial gene has various advantages such as the maternal inheritance of mitochondrial gene, the mitochondrial gene is present in thousands of copies per cell, the mutation rate of mitochondrial genes is ~10-fold higher compared to nuclear genes in vertebrates so that point mutations accumulate quickly enough to allow the discrimination of even closely related species, which ultimately increases the chances of positive result even in highly fragmented DNA due to severe processing conditions (Unseld et al. Citation1995; Greenwood and Paboo Citation1999; Bellagamba et al. Citation2001; Mane et al. Citation2011).
Different mitochondrial genes were targeted for identification of meat species by PCR amplification and DNA sequencing. The mitochondrial 12S rRNA gene for PCR amplification and DNA sequencing was targeted to identify various meat species (Girish et al. Citation2004). In another study, mitochondrial 12S rRNA gene was amplified and sequenced in a forensic case to prove unambiguously that skin sample, which was claimed to be of tiger, was from bovine (Prakash et al. Citation2000). Identification of buffalo, emu and crocodile species was done by PCR amplification and sequencing of cytochrome b gene (Forrest and Carnegie Citation1994). Recently, Girish et al. (Citation2007) employed partial sequence analysis of 402 bp PCR amplified fragments of cytochrome b gene for identification of animal species in fresh and processed samples of unknown origin. These workers identified the species in unknown samples by the sequence analysis of PCR amplified DNA fragments. Therefore, the present study was planned to developed the sequence database of 16S rRNA gene of various meat species and evaluate their applicability in identification/differentiation of meat species for solving the forensic/veterolegal cases.
Materials and methods
DNA extraction from meat samples
The fresh meat samples of cattle (ox), buffalo, sheep, goat and pig were collected from local slaughterhouses and experimental abattoir of the institute. After collection, samples were kept at −20°C till further processing.
DNeasy® Blood and Tissue Kit (Qiagen, USA) was used for extraction of DNA from meat samples as per the instructions given by manufacturer. The same kit was also used for extraction of DNA from heat-treated meat and meat products.
PCR amplification
In the present study, primer pairs for PCR amplification were self-designed based on available gene sequences of mitochondrial 16S rRNA in National Center for Biotechnology Information (NCBI) database using primer designing software (DNA-STAR Inc., USA). Initially, primer pair was designed based on 16S rRNA gene sequence of cattle (accession code: AF492351) followed by comparison of designed primer pair with 16S rRNA gene sequence of buffalo (accession code: AF547270), sheep (accession code: AF010406), goat (accession code: AF533441) and pig (accession code: AF034253). The other 16S rRNA gene sequence of above species is also checked for consistency of 3′ and 5′ ends of primer pair. The following primer pair was designed and used in the study:
Forward: 5′ ACA TGC CTA ACG AGC CTG GTG ATA 3′ | |||||
Reverse: 5′ TTG TGT TTG CCG AGT TCC TTT TAC 3′ | |||||
Each PCR reaction was performed in a total volume of 50 µl containing 5 µl of 10× PCR buffer, 200 µM each of dNTP, 1–2 units of Taq DNA polymerase (Qiagen, USA), 10–20 pmol each of forward and reverse primer, 1 µl of DNA template (20–30 ng) and remaining nuclease-free water (Fermentas, USA) to make up the reaction volume. All the ingredients were taken using filter tips to avoid any cross-contamination. Every-time negative control (without template DNA) was put to make sure that there was no contamination in PCR system. The PCR cycling conditions were: 2 min at 94°C for initial denaturation, followed by 35 cycles of denaturation at 94°C for 0.5 min, annealing at 60°C for 0.5 min and extension at 72°C for 1 min. The final extension was done at 72°C for 5 min.
Analysis of PCR products by electrophoresis
The submarine horizontal agarose gel electrophoresis was used for analysis of PCR products. Two per cent agarose was used for preparation of gel. For that 0.4 g of agarose (Ambion, USA) was put in 20 ml of 1× TBE solution (Fermentas, USA) and heated to completely dissolve the agarose. Subsequently, 1 µl (5%) of ethidium bromide solution was added as gel visualising agent and mix thoroughly. After gel setting, the electrophoresis was done for 90 min at 80 V. The PCR product was finally analysed using UV transilluminator and documented by Gel documentation system (Alpha Imager, USA). The ready to use 100 bp ladders (Fermentas, USA) were used in the present study.
Sequencing and analysis of PCR amplified DNA fragments
PCR products were sequenced using ABI Prism 377 DNA sequencer at DNA sequencing facility, University of Delhi, South Campus, New Delhi. The obtained sequences were analysed using Edit Seq of Laser gene software (DNA-STAR Inc., USA). The mitochondrial 16S rRNA gene sequences of cattle, buffalo, sheep, goat and pig were sequenced in this study and their sequence comparison was made for meat species identification. The comparison of sequence was done by Clustal method using MegalignTM software package (DNA-STAR Inc., USA). The nucleotide sequences of 16S rRNA gene of cattle, buffalo, sheep, goat and pig were submitted to NCBI nucleotide sequence database. GenBank accession numbers for these sequences are given in .
Table 1. Details of mitochondrial 16S rRNA gene sequences used for analysis.
Heat treatments given to meat and meat emulsion
The standard procedures were followed for the preparation of meat emulsion. In the present study, meat and meat emulsion were cooked at different temperatures under different conditions to evaluate applicability of standardised PCR assay. The details of heat treatments, temperature–time combination used for heat treatments of meat and meat emulsions were dry heat in oven at 180°C for 30 min; steam cooking at 10°C for 45 min; and autoclaving at 121°C, 15 psi for 15–20 min.
Results and discussion
The present study was planned to develop the sequence database by PCR amplification and sequence analysis of mitochondrial 16S rRNA gene for their use in differentiation/identification and subsequently evaluating their application in solving the forensic/veterolegal cases. The developed primer pair successfully amplified the desired DNA fragments from all the species targeted in this work, even in heat-treated meat and meat products. The PCR assay was successfully standardised for amplification of about 497 bp DNA fragment with primer pair designed based on mitochondrial 16S rRNA sequence of cattle, buffalo, sheep, goat and pig (). The amplification was observed within wide range of annealing temperatures (54–62°C); however, 60°C was found to be most suitable and chosen for further study. Earlier workers have also exploited mitochondrial 12S rRNA gene (Prakash et al. Citation2000; Girish et al. Citation2004; Rastogi et al. Citation2004) and mitochondrial cytochrome b gene (Meyer et al. Citation1995; Unseld et al. Citation1995; Hsieh et al. Citation2005) for meat species identification, and it has also been reported that the PCR amplification of mitochondrial 16S rRNA gene has been used for identification of snail species (Abdulmawjood and Buelte, Citation2002). Earlier also reported that the PCR amplification followed by DNA sequencing can be the best way to identify the species origin of meat from all meat animals (Girish et al. Citation2007).
Figure 1. PCR amplification pattern of mitochondrial 16S rRNA gene in various meat species.
Note: Lane M: 100 bp ladder, Lane 1: cattle, Lane 2: buffalo, Lane 3: sheep, Lane 4: goat, Lane 5: pig and Lane 6: negative control.
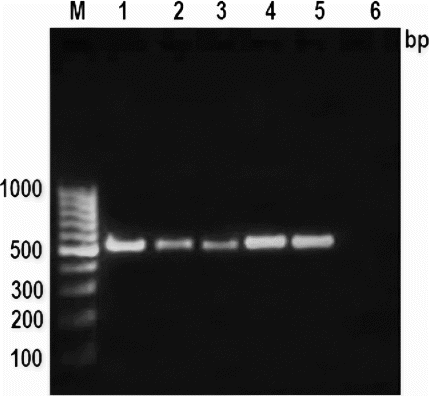
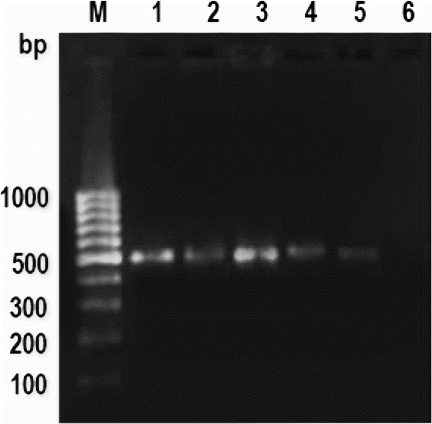
The successful amplification was observed in fresh and processed meat, i.e., cooked meat and meat emulsion () and autoclaved meat and meat emulsion (). No adverse effect of heat treatments, processing conditions and ingredients used for emulsion preparation was observed on PCR amplification. However, intensity was slightly weak in cooked and autoclaved meat emulsion in some species but this amplification is sufficient for their differentiation. This result was also similar to earlier reports of Matsunaga et al. (Citation1998) and Girish et al. (Citation2004), who observed weak amplification in autoclaved meat processed at 120°C for 30 min. In spite of variable effect of heat treatments, it is not surprising for amplification of about 497 bp DNA fragments of mitochondrial 16S rRNA gene in cooked and autoclaved meat and meat emulsion. This is due to large numbers of mitochondria are present in each cell, so that at least few copies survive even under intense processed meat, which are sufficient to give detectable amplification. The earlier workers also observed consistent amplification in heat-treated meat and meat products (Abdulmawjood and Buelte Citation2002; Mane et al. Citation2009, Citation2011, Citation2012), even up to 981 bp size PCR products (Zimmermann et al. Citation1998). The given meat sample can be simply subjected to following steps: DNA extraction, PCR amplification followed by sequencing and blast search in NCBI nucleotide sequence database (http://www.ncbi.nlm.nih.gov/blast). This will give a list of sequences in the order of the highest percentage of similarity, which can be used to identify the species in unknown meat sample (Girish et al. Citation2004).
Figure 2. PCR amplification pattern of mitochondrial 16S rRNA gene in cooked meat emulsion.
Note: Lane M: 100 bp Ladder, Lane 1: cattle, Lane 2: buffalo, Lane 3: sheep, Lane 4: goat, Lane 5: pig and Lane 6: negative control.
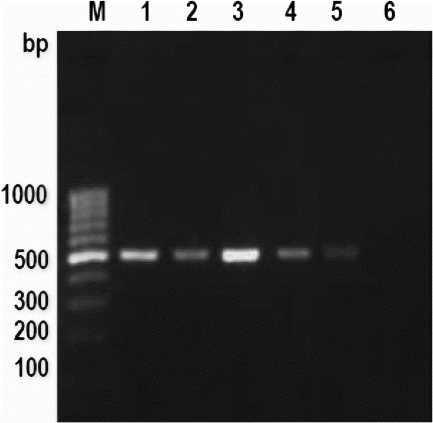
Figure 3. PCR amplification pattern of mitochondrial 16S rRNA gene in autoclaved meat emulsion.
Note: Lane M: 100 bp Ladder, Lane 1: cattle, Lane 2: buffalo, Lane 3: sheep, Lane 4: goat, Lane 5: pig and Lane 6: negative control.
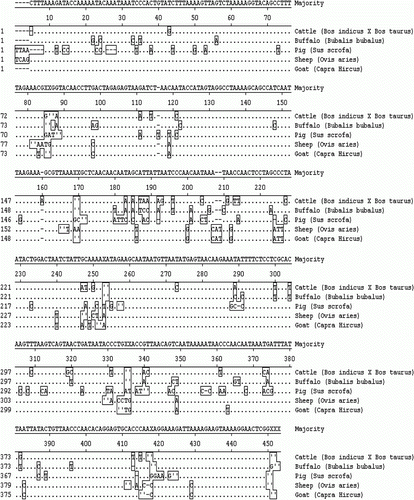
The sequence alignment reports in this study revealed that even the closely related species, such as cattle–buffalo and sheep–goat could also be differentiated from each other on the basis of their mitochondrial 16S rRNA gene sequences. The partial mitochondrial 16S rRNA gene sequences of cattle, buffalo, sheep, goat and pig in this work revealed that the per cent identity was higher in closely related species in comparison to other species, while divergence scores were lowest. These similarity and divergence scores showed that meat species can be precisely identify as difference is sufficient enough to identify the species and minor mutations that may happen will not affect the divergence values. Sequence alignment of about 410 bp mitochondrial 16S rRNA gene of cattle (Bos taurus), buffalo (Bubalus bubalis), sheep (Ovis aries), goat (Capra hircus) and pig (Sus scrofa) is given in . For sequence alignments Megalign™ programme of Laser gene software (DNA-STAR Inc.) was used. The availability of DNA sequence of mitochondrial 16S rRNA gene of large number of species makes this approach useful for identification of even unknown meat species by comparative analysis of the results obtained.
Figure 4. Nucleotide sequence alignment reports of mitochondrial 16S rRNA gene of cattle, buffalo, sheep, goat and pig.
Note: A dot indicates identity with the majority at the given position.
In conclusion, the PCR amplifications of mitochondrial 16S rRNA followed by sequencing and analysis proved to be very efficient for identification of species origin of meat. The technique is applicable in processed and heat-treated meat and meat products. This is due to high number copies of mitochondrial DNA in each cell and is sufficient for identification of meat species. However, this technique is not applicable to mixed or adulterated meat due to amplification of large number of different DNA fragments from each species make results interpretation more complex. This method is comparatively costly and time consuming, although high precise and authenticity suggest that the methods can be used in identification of species origin of meat from unknown samples.
Acknowledgements
The authors are thankful to Director, Indian Veterinary Research Institute, Izatnagar-243 122 (India) for providing necessary facilities for the present work.
References
- Abdulmawjood , A and Buelte , M. 2002 . Identification of ostrich meat by restriction fragment length polymorphism (RFLP) analysis of cytochrome b gene . Journal of Food Science , 67 : 1688 – 1691 . doi: 10.1111/j.1365-2621.2002.tb08706.x
- Bellagamba , F , Moreti , VM , Cominicini , S and Valfre , F. 2001 . Identification of species in animal feedstuffs by polymerase chain reaction–restriction fragment length polymorphism analysis of mitochondrial DNA . Journal of Agriculture and Food Chemistry , 49 : 3775 – 3781 . doi: 10.1021/jf0010329
- Forrest , ARR and Carnegie , PR. 1994 . Identification of gour meat using FINS (Forensically informative nucleotide sequencing) . Biotechnology , 17 : 24 – 26 .
- Girish , PS , Anjaneyulu , ASR , Viswas , KN , Anand , M , Rajkumar , N , Shivakumar , BM and Sharma , B. 2004 . Sequence analysis of mitochondrial 12S rRNA gene can identify meat species . Meat Science , 66 : 551 – 556 . doi: 10.1016/S0309-1740(03)00158-X
- Girish , PS , Anjaneyulu , ASR , Viswas , KN , Santhosh , FH , Bhilegaonkar , KN , Agarwal , RK , Kondaiah , N and Nagappa , K. 2007 . Polymerase chain reaction-restriction fragment length polymorphism of mitochondrial 12S rRNA gene: a simple method for identification of poultry meat species . Veterinary Research Communication , 31 : 447 – 455 . doi: 10.1007/s11259-006-3390-5
- Greenwood , A and Paboo , S. 1999 . Nuclear insertion sequences of mitochondrial DNA predominate in hair but not in blood of elephants . Molecular Ecology , 8 : 133 – 137 . doi: 10.1046/j.1365-294X.1999.00507.x
- Hsieh , HM , Tsai , CC , Tsai , LC , Chiang , HL , Huang , NE , Shih , RTP , Linacre , A and Lee , JC-I. 2005 . Species identification of meat products using the cytochrome b gene . Forensic Science Journal , 4 : 29 – 36 .
- Mane , BG , Mendiratta , SK and Tiwari , AK. 2009 . PCR assay for specific identification of chicken species in meat and meat products . Food Chemistry , 116 : 806 – 810 . doi: 10.1016/j.foodchem.2009.03.030
- Mane BG , Mendiratta SK , Tiwari AK , Bhilegaokar KN . 2012 . Development and evaluation of polymerase chain reaction assay for identification of buffalo meat . Food Analytical Methods 5 : 296 – 300 . doi: 10.1007/s12161-011-9237-x
- Mane , BG , Mendiratta , SK , Tiwari , AK , Bhilegaokar , KN , Ravindra , PV and Raut , AA. 2007 . Precise and rapid identification of ovine-caprine origin of cooked meat by specific PCR assay . Journal of Veterinary Public Health , 5 : 107 – 111 .
- Mane , BG , Mendiratta , SK , Tiwari , AK , Sharma , BD , Bhilegaokar , KN and Anjaneyulu , ASR. 2011 . Detection of pork in admixed meat and meat products by species-specific PCR technique . Indian Journal of Animal Science , 81 : 1178 – 1181 .
- Matsunaga , T , Shibata , K , Yamada , J , Shinmura , Y and Chikuni , K. 1998 . Identification of meat species based on the difference of 18S ribosomal RNA-genes . Journal of Japanese Society of Food Science and Technology , 45 : 719 – 723 . doi: 10.3136/nskkk.45.719
- Meyer , R , Hofelein , C , Luthy , J and Candrian , U. 1995 . Polymerase chain reaction-restriction fragment length polymorphism analysis: a simple method for species identification in food . Journal of AOAC International , 78 : 1542 – 1551 .
- Prakash , S , Patole , MS , Ghumatkar , SV , Nandode , SK and Shouche , YS. 2000 . Mitochondrial 12S rRNA sequence analysis in wild life forensics . Current Science , 8 : 1239 – 1241 .
- Rastogi , G , Dharne , M , Bharde , A , Pandav , VS , Ghumatkar , SV , Krishnamurthy , R , Patole , MS and Shouche , YS. 2004 . Species determination and authentication of meat samples by mitochondrial 12S rRNA gene sequence analysis and conformation-sensitive gel electrophoresis . Current Science , 12 : 1278 – 1281 .
- Unseld , M , Beyermann , B , Brandt , P and Hiesel , R. 1995 . Identification of the species origin of highly processed meat products by mitochondrial DNA sequences . PCR Methods and Applications , 4 : 241 – 243 . doi: 10.1101/gr.4.4.241
- Zimmermann , S , Zehmer , R and Mebs , D. 1998 . Identification of animal species in meat samples by DNA-analysis . Fleischwirtschaft , 78 : 530 – 533 .