Abstract
mRNA expression and DNA polymorphism of prolactin receptor (PRLR) gene were investigated and used to study their effects on litter size in goats. Real-time polymerase chain reaction (PCR) analysis revealed that PRLR mRNA levels in pituitary and endometrium of prolific Lezhi black goats were 39.0-fold and 29.9-fold higher than those in non-prolific Tibetan goats (P < 0.05), but there was no difference in ovary and liver between the two breeds. To further explore the possible association between variations in PRLR gene with litter size in Lezhi black goat, mutation screening of exon 10 of PRLR was performed by polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP). In P2 locus, the four genotypes AA, AB, AC, and AD had the genotypic frequencies of 0.136, 0.110, 0.162, and 0.594, respectively, but no polymorphism was detected in P1 locus. There were four mutations at C89T, A94C, C146G, and G157C of this fragment in all of the four genotypes, which led to a missense mutation Pro to Leu, Asn to His, Ala to Gly, and Ala to Pro, respectively. In addition, a mutation of C61T in genotype AA, A175G in genotype AD, T24C and C96T in genotype CD were detected. The Lezhi black does with genotype AD had 0.35 (P < 0.05) more kids than those with genotype AB, but there was no significant difference among other genotypes. It is concluded that PRLR gene expression and mutations in exon 10 of PRLR gene may be associated with the reproductive effects of the Lezhi black goat.
1. Introduction
Prolactin (PRL) is a polypeptide hormone, which is mainly synthesized and secreted by the anterior pituitary. It regulates more than 300 separate actions in vertebrates, including effects on water and salt balance, growth and development, endocrinology and metabolism, brain and behavior, reproduction, and immune regulation and protection (Bole-Feysot et al. Citation1998; Binart et al. Citation2010). The diverse biological actions of PRL are mediated through its receptor, the transmembrane receptor (PRLR), a member of the cytokine receptor superfamily (Binart et al. Citation2000). The PRLR gene has been detected in various tissues including brain (Muccioli et al. Citation1988), ovary (Bramley & Menzies Citation1987; Slomczynska et al. Citation2001), pituitary (Shao et al. Citation2008), liver (Schuler et al. Citation1997; Shao et al. Citation2008), and uterus (Young & Bazer Citation1989; Schuler et al. Citation1997) in several mammalian species. Mice homozygous for null mutations in PRLR are sterile due to a failure of embryonic implantation, irregular cycles, reduced fertilization rate, and defective embryonic development (Ormandy et al. Citation1997). Screening of several commercial pig lines indicated the existence of an association between the litter size and AluI polymorphism (Vincent et al. Citation1998). Recently, associations of PRLR polymorphisms with prolificacy in some sheep breeds (Chu et al. Citation2007) and goat breeds (Zhang et al. Citation2007; Chu et al. Citation2008; Di et al. Citation2011) have also been reported. All these characteristics make PRLR a strong candidate gene for reproductive traits. However, detailed analyses of PRLR mRNA expression in the goat breeds with different fecundity during the estrous cycle have not been reported.
The present study, for the first time, investigated the mRNA expression levels of PRLR gene in the Lezhi black goats, a local Chinese breed famous for its high fecundity (producing up to seven kids per kidding), and Tibetan goats (Capra hircus), a single-birth breed characterized by adapting to cold, hypoxic ecological conditions in the Qinghai-Tibet Plateau. The genetic variability of PRLR gene and its association with litter size in Lezhi black goat were also evaluated by polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) method. This intends to examine whether base mutations or expression levels of mRNA of these genes could be associated with the reproductive difference in goats.
2. Materials and methods
2.1. Animals and sample collection
Lezhi black goats were supplied by the Lezhi black goat breeding farm in Lezhi County (30°30′N, 105°02′E and 596.3 m altitude), China, and Tibetan goats were purchased from Li County (31°42′N, 103°16′E and 2800 m altitude) of Qinghai-Tibet Plateau in China during the breeding season (October). Lezhi black goats were kept in shelters, and Tibetan goats were grazed on the pasture. Animals aged 3–5 yr, with a history of multiple births (twin or triplet births) for Lezhi black goats (n = 6) and single birth for Tibetan goats (n = 6) were selected to investigate the nucleotide sequences and mRNA expression levels of PRLR gene. Estrus was detected twice a day, and a doe was considered in estrus only when she allowed the male to mount. Goats were slaughtered at 12–24 h after onset of estrus (in the middle of estrus) for collection of pituitaries, livers, ovaries, and endometria. Part of the removed tissue samples was snap-frozen in liquid nitrogen and then stored at 80 °C to be used for RNA extraction.
Blood samples were collected from 155 does of Lezhi black goats along with data on litter size for investigating polymorphism of PRLR gene and their relation to prolificacy in Lezhi black goats. Venous blood (5–6 mL) was collected using ethylenediaminetetraacetic acid (EDTA) as an anticoagulant. DNA extraction was performed within 24 h of blood collection, according to Sambrook and Russell (Citation2001). After checking the quality and quantity, DNA was diluted to a final concentration of 50 ng/µL in water and stored at 4 °C for immediate use while for long term kept at 20 °C.
2.2. DNA and RNA isolation and RT-PCR
DNA samples were extracted from blood samples according to specification of TIANamp Blood DNA Kit (Tiangen, China) and kept at 20 °C. Total RNA was extracted with RNAprep Pure Tissue Kit (Tiangen, China) following the manufacturer's instructions. cDNA was reverse transcribed from total RNA using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, EU) in a total volume of 20 µL reaction mixture, which contains 1 µg RNA, oligo(dT)18 primer 1 µL, random hexamer primer 1 µL, 5 × reaction buffer 4 µL, RiboLock RNase inhibitor 1 µL, 10 mM dNTP Mix 2 µL, RevertAid M-MuLV Reverse Transcriptase 1 µL, and 9 µL RNase free dH2O, at 42 °C for 30 min, 25 °C for 5 min, and 70 °C for 5 min.
2.3. Analysis of PRLR mRNA expression levels in different tissues
The PRLR mRNA expressions in tissues (ovary, liver, pituitary, and endometrium) were analyzed by quantitative RT-PCR. The primers were designed with Beacon Designer 7.0 software (Premier Biosoft Internationa l, Palo Alto, CA USA) based on the GenBank sequence (PRLR: NCBI Accession No. AC_000177.1; β-actin: NCBI Accession No. JX046106.1) and synthesized by Invitrogen (Shanghai, China). Primers for PRLR were 5′-AGAGGAAGGAGCCAACAT−3′ (forward) and 5′-GGAGACCGACATTTAATAAGC−3′ (reverse). Primers used for β-actin were 5′-GGAATCGTCCGTGACATCAAG−3′ (forward) and 5′-GGAAGGAAGGCTGGAAGAGA−3′ (reverse). Real-time PCR amplification mixture contained 5 µL SsoAdvanced™ SYBR Green Supermix (Bio-Rad), 1 µL of each of primers, 1 µL cDNA, and 2.5 µL ddH2O. Reactions were run on the CFX96TM real-time PCR detection system (Bio-Rad). The PCR conditions were as follows 3 min at 95 °C, 45 cycles of 10 s at 95 °C, 20 s at 58 °C. The samples in the RT-PCR were run in triplicate. The PCR efficiencies PRLR and β-actin were 84.3% and 95.2%, respectively. The threshold cycle (Ct) resulting from RT-PCR was analyzed by Pfaffl (Citation2001) method. The Ct of target gene PRLR was compared with the internal reference gene β-actin.
2.4. PCR amplification and SSCP assay
The specific PCR primers were designed based on the GenBank sequence (NCBI Accession Nos. AF091870 and AF041257) for PRLR gene () and synthesized by Invitrogen (Shanghai, China). PCR was performed in a 25 µL total volume reaction mixture containing 0.5 µL genomic DNA, 1 µL of each of primers, 10 µL ddH2O, and 12.5 µL Taq DNA polymerase (Tiangen, China). The conditions of PCR were as follows: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C/53.6 °C, and extension at 72 °C for 30 s and final extension at 72 °C for 10 min in thermal cycler T-gradient (Bio-Rad). The PCR products were run on 1.5% agarose gel and analyzed under the UV illuminator.
Table 1. Primers of goat PRLR gene designed for PCR-SSCP analysis.
The PCR products (5 µL) were mixed with 5 µL denaturing solution (95% formamide, 25 mM EDTA, 0.025% xylene–cyanole, and 0.025% bromophenol blue), heated at 98 °C for 10 min, and then chilled on ice for 7 min. The denatured DNA samples (2 µL) were then subjected to 8–10% PAGE (polyacrylamide gel electrophoresis) in 1 × TBE (Tris-borate EDTA) buffer and at a constant temperature (4 °C) under voltage of 180 V for 4 min, 120 V for 2.5 h. The gel (29:1 acrylamide:bis) was stained with 0.1% silver nitrate (Ji & Cao Citation2007). After the polymorphisms were detected, each of the DNA bands on the SSCP gel was extracted and the PCR products of the different electrophoresis patterns were sent for sequencing in both directions (repeated three times) in an ABI 377 DNA analyzer (Applied Biosystems) and the sequences were determined with DNAstar software (version 7.1) and blast in NCBI (National Center for Biotechnology Information).
2.5. Statistical analysis
Data on litter size in the first and the second parities from 155 does born in two successive years were used in the present study. Least square variance analysis was conducted for primer pairs, respectively. Therefore, the following statistical model was fitted to compare difference of litter size between genotypes. Yijkl = µ + Li + Pj + Gk + eijkl, where Yijkl is phenotypic value of litter size; µ is the population mean; Li is the fixed effect of the ith (i = 1,2) kidding year; Pj is the fixed effect of the jth (j = 1,2) parity; Gk is the fixed effect of the kth (k = 1,2) genotype for gene; and eijkl is random error effect of each observation. Calculations were achieved using the general linear model procedure of SAS (Ver 8.1) (SAS Institute Inc., Cary, NC, USA).
3. Results
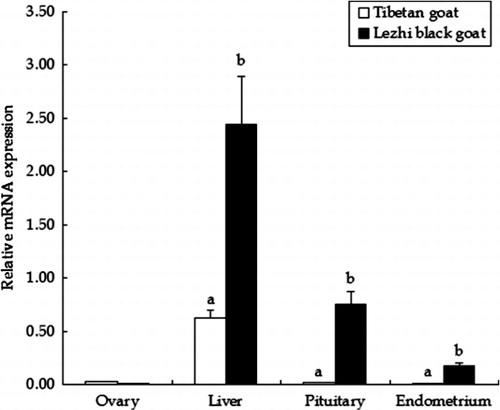
3.1. Tissue expression of PRLR gene in the Tibetan goats and Lezhi black goat
We used a quantitatively approach based on reverse transcription followed by real-time PCR amplification to investigate the expression of PRLR gene in ovary, liver, pituitary, and endometrium. The results showed that PRLR mRNA levels in pituitary and endometrium of prolific Lezhi black goats were 39.0-fold and 29.9-fold higher than those in non-prolific Tibetan goats (P < 0.05), but there was no difference in ovary and liver between the two breeds (P > 0.05). PRLR mRNA was higher in liver than in ovary, pituitary, and endometrium within the same breed ().
3.2. PCR-SSCP analysis of the PRLR gene in Lezhi black goat
According to international practice and other sources (Lan et al. Citation2007; Hu et al. Citation2009; Bhattacharya et al. Citation2011) regarding the naming of SSCP patterns, in primer 1 locus (P1 locus), no polymorphism was detected after electrophoresis (figure not shown). In primer 2 (P2 locus), four band patterns were detected (), named AA, AB, AC, and AD genotypes. The four genotypes had the genotypic frequencies of 0.136, 0.110, 0.162, and 0.594, respectively ().
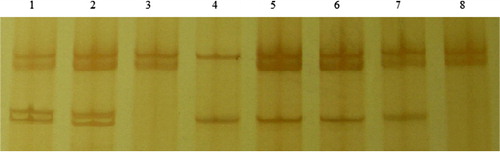
Table 2. Genotype and allele frequencies based on PCR-SSCP analysis of the exon 10 of PRLR gene at the P2 locus and litter size of different genotypes in Lezhi black goats.
3.3. DNA sequence analysis
In the P2 locus, the four amplified fragments with different PCR-SSCP patterns were sequenced. Compared with the known sequence (GenBank Accession No. AF041257), there were four mutations at C89T, A94C, C146G, and G157C of this fragment in all of the four genotypes, which led to a missense mutation Pro to Leu, Asn to His, Ala to Gly, and Ala to Pro, respectively. In addition, a mutation of C61T was detected in genotype AA, which led to Leu to Phe. A175G was detected in genotype AD, which led to Thr to Ala. T24C and C96T were detected in genotypes CD, which did not lead to the change of the amino acid.
3.4. Associations of polymorphisms of PRLR with litter size
The least squares mean and standard error for litter size of different PRLR genotypes in Lezhi black goats were given in . The Lezhi black does with genotype AD had 0.35 (P < 0.05) more kids than those with genotype AB, but there was no significant difference among other genotypes.
4. Discussion
In this study, we investigated the association of the PRLR gene with prolificacy in goat by comparing the expression levels of PRLR gene between highly prolific Lezhi black goat and non-prolific Tibetan goat, and exploring the polymorphism of exon 10 of PRLR gene in Lezhi black goat.
In agreement with the previous studies in other species (Bramley & Menzies Citation1987; Muccioli et al. Citation1988; Young & Bazer Citation1989; Schuler et al. Citation1997; Slomczynska et al. Citation2001; Shao et al. Citation2008), the q-PCR results of the present study showed that the expression of PRLR gene has been detected in all tissue samples of endometrium, liver, pituitary, and ovary of the two breeds. The previous studies showed that PRLR–/– female mice showed an absence of pseudopregnancy and an arrest of egg development immediately after fertilization, with only a few reaching the stage of blastocysts (Ormandy et al. Citation1997). The outcome is a complete sterility. The ovulation rate is not different between PRLR+/+ and PRLR–/– mice, and the corpus luteum is formed but an elevated level of apoptosis and extensive inhibition of angiogenesis occur during the luteal transition in the absence of PRL signaling (Bachelot & Binart Citation2007). In cyclic ewes, PRLR mRNA levels increased after Day 11. PRLR mRNA levels were not different between cyclic and pregnant ewes on Days 11–15, but PRLR mRNA levels increased (P < 0.01) between Days 11 and 19 in pregnant ewes (Stewart et al. Citation2000). However, comparative PRLR mRNA expression in high and low prolific goat breeds has not been reported. The present study, for the first time, reported that PRLR mRNA levels in pituitary and endometrium of prolific Lezhi black goats were greater than those in non-prolific Tibetan goats (P < 0.05), but there was no difference in ovary and liver between the two breeds (P > 0.05). PRLR mRNA was higher in liver than in ovary, pituitary, and endometrium within the same breed.
The PRLR gene was significantly associated with total number born and/or number born alive in several pig lines tested. The magnitude of the effects varied by line with effects exceeding 1 pig per litter increase between homozygous genotypes in one line (Rothschild et al. Citation1998; Vincent et al. Citation1998; Drogemuller et al. Citation2001; Putnova et al. Citation2002; van Rens & van der Lende Citation2002). For exon 10 of PRLR, the Small Tail Han ewes with genotype BB and AB had 0.66 and 0.54 more lambs than those with genotype AA, respectively (Chu et al. Citation2007). In goats, polymorphisms in intron 1 and intron 2 of PRLR gene were detected in high prolificacy (Jining Grey) and low prolificacy (Boer, Wendeng dairy, Liaoning Cashmere, and Beijing) native goats using PCR-SSCP. For intron 1, five genotypes (AA, AH, AK, HH, and HK) were identified in Jining Grey goats, and two (AA and AK) in the other four breeds. The Jining Grey does of genotype HH, HK, AH, and AK delivered by 0.65, 0.62, 0.59, and 0.57 more kids (P < 0.01) than those of genotype AA, respectively. For intron 2, three genotypes (CC, CD, and DD) were detected in Boer goats, and two (CC and CD) in the other four breeds. The Jining Grey does of genotype CD delivered by 0.55 (P < 0.01) more kids than those of genotype CC (Di et al. Citation2011). For exon 10 of PRLR, the Jining Grey does with genotype FG had 0.76 (P < 0.05) kids more than those with genotype FF (Zhang et al. Citation2007). In the present study, the Lezhi black does with genotype AD in exon 10 of PRLR had 0.35 (P < 0.05) more kids than those with genotype AB, but there was no significant difference among other genotypes. The Jining Grey does with genotype FG (Zhang et al. Citation2007) and Lezhi black does with genotype AD were found in different strains. Therefore, it seems likely that PRLR gene is a major gene of dominating prolificacy or is closely with genetic linkage in goat, but the single-nucleotide polymorphism (SNP) locus alone does not account for all the breed variance.
5. Conclusion
In conclusion, this study has demonstrated that the PRLR gene associated with prolificacy in the Lezhi black goats. However, further research will be necessary to more thoroughly understand the mechanisms behind fecundity in the Lezhi black goats.
Funding
This work was supported by Southwest University for Nationalities (12ZYXS72; 12NZYTH07; 2011XW D-S0905) and National Key Technology R&D Program (No. 2008BADC3B01).
Additional information
Funding
References
- Bachelot A, Binart N. 2007. Reproductive role of prolactin. Reproduction. 133:361–369. 10.1530/REP-06-0299
- Bhattacharya TK, Chatterjee RN, Sharm RP, Rajkumar U, Niranjan M. 2011. Genetic polymorphism at 5′ flanking region of the prolactin gene and its effect on egg quality traits in naked neck chickens. J Appl Anim Res. 39: 72–76. 10.1080/09712119.2011.565224
- Binart N, Bachelot A, Bouilly J. 2010. Impact of prolactin receptor isoforms on reproduction. Cell. 21:362–368.
- Binart N, Ormandy CJ, Kelly PA. 2000. Mammary gland development and the prolactin receptor. Adv Exp Med Biol. 480:85–92.
- Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. 1998. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in prolactin receptor knockout mice. Endocr Rev. 19:225–268. 10.1210/er.19.3.225
- Bramley TA, Menzies GS. 1987. Receptors for lactogenic hormones in the porcine corpus luteum: properties and luteal phase concentrations. J Endocrinol. 113:355–364. 10.1677/joe.0.1130355
- Chu MX, Mu YL, Fang L, Ye SC, Sun SH. 2007. Prolactin receptor as a candidate gene for prolificacy of Small Tail Han Sheep. Anim Biotechnol. 18:65–73. 10.1080/10495390601090950
- Chu MX, Zhang GX, Wang JY, Fang L, Ye SC. 2008. Polymorphism of prolactin receptor gene and its relationship with litter size of some goat breeds. J Agric Biotechnol. 16:725–726.
- Di R, Yin J, Chu MX, Cao GL, Feng T, Fang L, Zhou ZX. 2011. DNA polymorphism of introns 1 and 2 of prolactin receptor gene and its association with litter size in goats. Anim Sci Pap Rep. 29:343–350.
- Drogemuller C, Hamann H, Distl O. 2001. Candidate gene markers for litter size in different German pig lines. J Anim Sci. 79:2565–2570.
- Hu XC, Lu AJ, Chen H, Gao XY, Xu HX, Zhang CL, Fang XT, Lei CH. 2009. Preliminary evidence for association of prolactin and prolactin receptor genes with milk production traits in Chinese Holsteins. J Appl Anim Res. 36: 213–217. 10.1080/09712119.2009.9707062
- Ji YT, Cao BY. 2007. An optimal method of DNA silver staining in polyacrylamide gels. Electrophoresis. 28:1173–1175. 10.1002/elps.200600557
- Lan XY, Chen H, Pan CY, Ming LJ, Lei CZ, Hua LS, Zhang CL, Hu SR. 2007. Polymorphism in growth hormone gene and its association with production traits in goats. J Appl Anim Res. 32: 55–60. 10.1080/09712119.2007.9706846
- Muccioli G, Bellussi G, Ghe C, Pagnini G, Di Carlo R. 1988. Regional distribution and species variation of prolactin binding sites in the brain. Gen Comp Endocrinol. 69:399–405. 10.1016/0016-6480(88)90031-7
- Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, Edery M, Brousse N, Babinet C, Binart N, Kelly PA. 1997. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 11:167–178. 10.1101/gad.11.2.167
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2002–2007. 10.1093/nar/29.9.e45
- Putnova L, Knoll A, Dvorak J, Cepica S. 2002. A new Hpa II PCR-RFLP within the porcine prolactin receptor (PRLR) gene and study of its effect on litter size and number of teats. J Anim Breed Genet. 119:57–63. 10.1046/j.1439-0388.2002.00316.x
- Rothschild MF, Vincent AL, Tuggle CK, Evans G, Short TH, Southwood OI, Wales R, Plastow GS. 1998. A mutation in the prolactin receptor gene is associated with increased litter size in pigs. Anim Genet. 29;Suppl 1:69.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
- Schuler LA, Nagel RJ, Gao J, Horseman ND, Kessler MA. 1997. Prolactin receptor heterogeneity in bovine fetal and maternal tissues. Endocrinology. 138:3187–3194. 10.1210/en.138.8.3187
- Shao RJ, Nutu M, Weijdegard B, Egecioglu E, Fernandez-rodriguez J, Tallet E, Goffin V, Ling C, Billig H. 2008. Differences in prolactin receptor (PRLR) in mouse and human fallopian tubes: evidence for multiple regulatory mechanisms controlling PRLR isoform expression in mice. Biol Reprod. 79:748–757. 10.1095/biolreprod.108.070003
- Slomczynska M, Gregoraszczuk E, Kochman K, Stoklosowa S. 2001. Prolactin binding analysis and immunohistochemical localization of prolactin receptor in porcine ovarian cells. Endocr J. 48:71–80. 10.1507/endocrj.48.71
- Stewart MD, Johnson GA, Gray CA, Burghardt RC, Schuler LA, Joyce MM, Bazer FW, Spencer TE. 2000. Prolactin receptor and uterine milk protein expression in the ovine endometrium during the estrous cycle and pregnancy. Biol Reprod. 62:1779–1789. 10.1095/biolreprod62.6.1779
- van Rens BT, van der Lende T. 2002. Litter size and piglet traits of gilts with different prolactin receptor genotypes. Theriogenology. 57:883–893. 10.1016/S0093-691X(01)00693-8
- Vincent AL, Evans G, Short TH, Southwood OI, Plastow GS, Tuggle CK, Rothschild MF. 1998. The prolactin receptor gene is associated with increased litter size in pigs. Proceedings of the 6th World Congress on Genetics Applied to Livestock Production. 27:15–18.
- Young KH, Bazer FW. 1989. Porcine endometrial prolactin receptors detected by homologous radio receptor assay. Mol Cell Endocrinol. 64:145–154. 10.1016/0303-7207(89)90140-8
- Zhang GX, Chu MX, Wang JY, Fang L, Ye SC. 2007. Polymorphism of exon 10 of prolactin receptor gene and its relationship with prolificacy of Jining grey goats. Hereditas. 29:329–336.
