Abstract
Chinese Wuchuan Black cattle is an important indigenous breed in Guizhou province, China. However, little is known about the genetic characterization and origin of this breed. In the present study, the polymorphism and variation of complete mitochondrial DNA D-loop sequences were analyzed. Fifty-one nucleotide polymorphic sites were detected; the nucleotide diversity (π) was 5.475%. Eleven haplotypes were determined (accession numbers: HM106460–HM106470) by biological software with a haplotype diversity (h) of 0.909. This suggests that plenty of genetic diversity exists in mtDNA D-loop region in Chinese Wuchuan Black cattle. A phylogenetic tree based on mtDNA D-loop sequences suggested that the origin of Chinese Wuchuan Black cattle was affected by Bos taurus and Bos indicus. In addition, coding sequence (CDS) regions of Mc1R gene for this breed were sequenced. Six polymorphic sites were detected with out insertions and deletions, only one transversion was observed. A novel single nucleotide polymorphism (SNP;[T/C]) at the position of 663 bp of Mc1R gene was detected in this cattle breed. This suggests that the novel SNP (T/C) might be used as a specific molecular marker to aid for breed identification.
1. Introduction
It has been reported that 28 Chinese native cattle breeds and many local cattle populations have been divided into three groups: northern, central, and southern groups (Qiu Citation1988). Genetic variation of mitochondrial D-loop region and evolution analysis for majority of Chinese cattle breeds have been studied (Chen et al. Citation1990). A mitochondrial DNA restriction fragment length polymorphism has been used to demonstrate that the southern Chinese cattle may have three origins: Bos taurus, Bos indicus, and Bos grunniens, of which B. taurus and B. indicus have the major influence (Yu et al. Citation1999). Microsatellite markers have been applied to identify the multiple origins of yellow cattle of China from B. taurus and B. indicus (Zhang et al. Citation2007). Among these studied, four native cattle breeds from Guizhou province of China including Liping, Sinan, Guanling, and Weining have been involved. However, little is known about phylogenetic information for Chinese Wuchuan Black cattle, a unique native cattle breed lived in Wuchuan county, Guizhou province of the southern China many years ago. In fact, Wuchuan Black cattle were reared in 150 years ago in The Qing Dynasty according to relevant records (Animal resources planning in Zunyi region of Guizhou Province, China in 2008, unpublished). It has not been registered until animal genetic resources have been resurveyed by Guizhou government in 2006 (Complementary surveying report of animal genetic diversity in four provinces of the southern region in China in 2006, unpublished).
Compared with the other indigenous cattle breeds in Guizhou province, Wuchuan Black cattle possess special characteristics with black coat color and larger body conformation. In China, black cattle such as Bohai Black cattle were listed as a native cattle breed (Mao et al. Citation2007). In the past, breed's identification was mostly based on the body conformation, physiological distribution, and productive performances. In Europe, coat color has been usually considered as an aid to breed identification. Registration of the animals to the herd-books usually required a specific coat color and color distribution pattern typical of that particular breed (Vincenzo et al. Citation2007).
Recently, studies have been conducted on coat color inheritance in a variety of mammalian species (Searle Citation1968; Adalsteinsson et al. Citation1995). In cattle, the Mc1R gene (melanocortin 1 receptor) has been the subject of several studies with the aim to elucidate the biology of coat color. In addition, (Mc1R) gene has been found to have a major function in the regulation of black versus red pigment synthesis within melanocytes (Jackson Citation1993). Three alleles of Mc1R gene have found in cattle, a point mutation in the dominant allele ED gives black coat color, allele e gives red color, whereas a frame shift mutation, producing a prematurely terminated receptor, in homozygous e/e animals, produces red color. The wild-type allele E+ produces a variety of colors. Then, another allele (E1), determined by a duplication of 12 bp that subsequently causes a duplication of four amino acids in the third intracellular loop of the Mc1R protein, was reported (Rouzaud et al. Citation2000; Kriegesmann et al. Citation2001). Its effect on coat color has not yet been completely clarified. Two other alleles, indicated as ED1 and ef, have been identified and their activity partially characterized in vitro (Maudet & Taberlet Citation2002; Graphodatskaya et al. Citation2002).
In this study, we sequenced the complete mtDNA control regions of Chinese Wuchuan Black cattle breed, polymorphic loci were detected and haplotypes were determined using biological software. Phylogenetic tree was constructed based on D-loop haplotypes to reveal the possible maternal origin of the breed. In addition, sequence variants and single nucleotide polymorphism (SNP) of the Mc1R gene were analyzed to see if the specific SNPs could be found and used as an aid to breed identification.
2. Materials and methods
2.1. Animals and DNA extraction
A total of 56 blood samples were collected from Wuchuan county, a central area of Chinese Wuchuan Black cattle population, located in northern Guizhou province. The sampling process was relatively difficult because no intensive farm available for this breed, all samples were taken from farmer's backyards in this county, and carried back to Guizhou University.
Genomic DNA was extracted from blood samples using DNA extraction kit from TaKaRa in China.
2.2. Mitochondrial D-loop region amplification and sequencing
The D-loop region was amplified using the following primers previously published in Loftus et al. (Citation1994) and Troy et al. (Citation2001):
5′-CTGCAGTCTCACCATCAACC-3′
5′-GGGGTGTAGATGCTTGC-3′
The primers were synthesized by TaKaRa Company in Dalian. Polymerase chain reaction (PCR) was performed using 1 µL DNA in a 25-µL volume reaction containing 2.5 µL of 10 × buffer, 2.5 µL of dNTPs (deoxy-ribonucleoside triphosphate including dATP, dGTP, dTTP, dCTP, dUTP), and 0.1 µL of each 100 mM primer. PCR conditions had an initial denaturing temperature of 94°C for 5 min followed by 35 cycles of a three-step process of 30 s at 94°C, 60 s at 60°C, and for 90 s at 72°C followed by a final step of 10 min at 72°C. PCR products were checked by electrophoresis in a 1% agarose 1 × Tis-Borat-EDTA buffer (TBE) gel and visualized by ethidium bromide staining and UV light. After purification, the PCR products were sequenced by Sanggon Biotech (Shanghai) Co., Ltd.
2.3. SNPs of the Mc1R gene
The coding sequence (CDS) region of Mc1R gene was amplified in this study. Primers were designed according to the sequence of Mc1R gene of Angus cattle published in GenBank (accession number: AF445641), the forward primer 5′ GGGCAACCGCACATCCAG 3′, and reverse primer 5′ GGTCTAGCCGATCCTCTTTG 3′; PCR was performed using a 50 µL of volume reaction, containing 2.5 µL of 2× Taq PCR MasterMix, 4 µL of DNA template, 2 µL of primers, and 17 µL of ddH2O. PCR conditions had an initial denaturing at 95°C for 5 min followed by 35 cycles of denaturation at 94°C for 40 s, annealing at optimum temperature 61°C for 60 s and extension at 72°C for 90 s with a final extension at 72°C for 10 min. store at 4°C. PCR products were checked, purified using TaKaRa Agarose Gel DNA Purification, and then sequenced by Sanggon Biotech (Shanghai) Co., Ltd.
2.4. Statistical analysis
For complete mtDNA sequences, program SeqMan from DNAstar (www.dnastar.com) was applied to compare with the D-loop sequences of B. taurus (accession number: V00654); Haplotype and nucleotide diversity were computed using DNASP4.50 (Librado & Rozas Citation2009). We constructed an unrooted neighbor-joining (NJ) tree of all sequences under the Tajima and Nei model using MEGA 5.2 software (Tamura et al. Citation2007). Eight referenced D-loop sequences from Chinese yellow cattle breeds such as Wannan (AY521122), Bashan (AY902385), Sanjiang (AY302393), Bohai black(DQ166070), Nanyang (DQ166103), Minnan (DQ166118), EF524121 (representing B. indicus), and Japanese black cattle (U87650) were included to better illustrate the genetic relations among Chinese cattle. The cattle mtDNA newly sequenced in this study have been deposited in GenBank under accession numbers HM106460–HM106470. For Mc1R sequences analysis, the consensus sequences were acquired using program SeqMan software of DNAstar; Clustalx (Thompson et al. Citation1997) was used to calibrate all Mc1R sequences.
3. Results
3.1. Nucleotide contents and variation of complete mtDNA D-loop
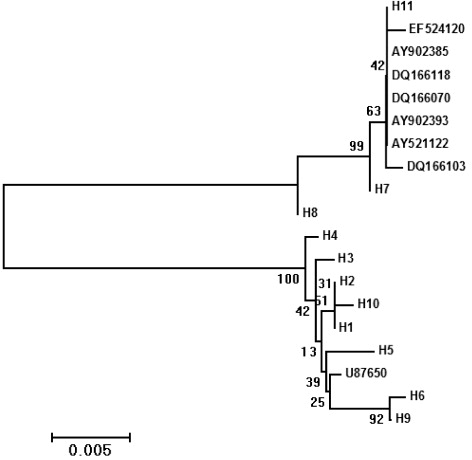
A total of 56 mtDNA D-loop sequences of Wuchuan Black cattle had been acquired, the length of whole D-loop region was 910 bp among 21 sequences, but one of them was 911 bp. This is probably because there is an insert of base C in 216–221 bp. The contents of A, C, G, and T were 33.1%, 25.0%, 13.5%, and 28.3%, respectively; Content for A + T was 61.4%, whereas G + C was 38.5%, indicating that a rich AT (A: adenine, T: thymus pyrimidine) contents in D-loop sequence. Comparison of all D-loop sequences revealed that 51 polymorphic sites were observed. Of the polymorphic sites, there were 45 transitions, 2 transversions, 3 insertions or deletions, and 1 site had both a transition and transversion (). Of 11 haplotypes observed in this study (accession numbers: HM106460–HM106470), 7 belonged to the haplotype of B. taurus and 4 belonged to the haplotypes of B. indicus. The haplotype diversity (h) was 0.909 ± 0.044, and nucleotide diversity (π) was 5.475%. Phylogenetic tree based on Kimura2-parameter genetic distances was constructed using NJ methods. Chinese yellow cattle mentioned above and EF524120 representing B. indicus were added to analyze. Confidence levels for NJ tree were assessed by bootstrapping from 1000 replications. The result indicated that all cattle were divided into two groups. H7 and H11 were grouped into the same cluster with the most of reference sequences, mainly affected by B. indicus. While the other haplotypes H1–H6, H8–H10, and H11 were clustered together with Japanese black cattle (U87650), largely influenced by B. taurus ().
3.2. Sequences variation of Mc1R gene
The whole length of Mc1R gene was 1238 bp including 954 bp of CDS region, encoding 317 amino acids. No insertions and deletions were detected. Average contents of T, C, A, and G were 22.9%, 35.9%, 14.9%, and 26.3%, respectively. The content for A + T was less than that of G + C.
Polymorphic sites for Mc1R gene of all individuals were analyzed using the sequence of Mc1R gene as a control downloaded from GenBank (accession number:AF445641). Six polymorphic sites were observed, only one transversion was observed whereas five were transition sites. Of six nucleotide mutation, four of which were missense mutation. These variable sites lead to produce different amino acids ().
Table 1. Polymorphic sites of Mc1R gene sequences in Wuchuan Black cattle.
Partial sequencing maps at position 296 showed the existence of heterozygote of C/T. Two genotypes of Mc1R gene: ED and E+ genotypes were classified according to the variable types at position 296. Of 56 Chinese Wuchuan Black cattle, 33 of which were E+/E+, while 23 belonged to ED/E+.
Here, it is worth mentioning that there was a T/C mutation at position 663 in coding region of Mc1R gene in Chinese Wuchuan Black cattle compared to that submitted in GenBank (accession numbers: AF445642, AF445642, and GU982927).
4. Discussion
It has been reported that frequency of transition is higher than that of transversion in mtDNA D-loop sequences during evolutionary process (Liu et al. Citation2006; Lai et al. Citation2006). In the present study, the nucleotide transition rate of all sequences in Chinese Wuchuan Black cattle was 88.24%, being consistent with the report. These results are in line with the rules of mtDNA evolution in mammals (Chen et al. Citation1993). The higher haplotype diversity value (h = 0.909) and higher nucleotide diversity value (π = 5.475%) were detected in this study. These two values are higher than that (av. h = 0.567, av. π = 2.37%) of the other 4 native cattle breeds in Guizhou province in Liu's report, and also similar to that (av. h = 0.932, av. π = 2.27%) of 16 Chinese indigenous cattle breeds (Zhang et al. Citation2009). This suggests that a plenty of polymorphism exists in mtDNA D-loop region in Chinese Wuchuan Black cattle.
The maternal origin of Chinese yellow cattle has been of concern all over the world. Western scholars think it originated simultaneously from B. taurus and India zebu B. indicus. Study on polymorphism of Y-mitochondrial DNA in Chinese cattle suggested that the northern cattle were largely influenced by B. indicus, the southern cattle were mainly affected by B. indicus, whereas the central ones were influenced by B. taurus and B. indicus at the same time (Lei et al. Citation2004; Zhang et al. Citation2009). Study on phylogenetic analysis of mtDNA D-loop region from four native cattle breeds (Liping, Sinan, Guanling, and Weining) in Guizhou province suggested that they were affected evenly by B. taurus and B. indicus (Liu et al. Citation2006). Report from Songjia Lai believed that the proportion of B. indicus in cattle breeds from Yungui plateau was larger than that of cattle breeds from Sichuan flatland (Lai et al. Citation2005). In theses research, black cattle breeds have been classified into the indigenous Chinese yellow cattle breeds. Actually, only several black cattle breeds such as Bohai, Tiandi, and Wuchuan have been distributed in China, the classification was conducted by local government due to no big differentiation on body conformation compared to yellow breeds. Phylogenetic analysis based on mtDNA D-loop sequences showed that Bohai and Wuchuan black cattle breeds, together with other Chinese native yellow cattle breeds, were influenced by B. taurus and B. indicus. (Mao et al. Citation2007; Zhang et al. Citation2009). For this reason, we conducted phylogenetic analysis using all D-loop haplotypes of Chinese Wuchuan Black cattle, the European cattle (AF492351), and Indian zebu (AF492351) were added to analyze. The result indicated that 33 haplotypes belonged to lineage of zebu (58.9%), 23 of which were lineage of B. taurus (41.1%), meaning that Chinese Wuchuan Black cattle had not been separated from B. taurus and B. indicus. This result supports the point of view that Chinese Wuchuan Black cattle was influenced by B. taurus and B. indicus, simultaneously (Mao et al. Citation2007; Zhang et al. Citation2009).
Mc1R gene of cattle with a length of 954 bp was mapped in chromosome 18, encoding 317 amino acids (Suzuki et al. Citation1996). It plays an important role in coat color determination because of its major function in the regulation of black versus red pigment synthesis within melanocytes (Jackson Citation1993). Previous genetic study in cattle revealed that Mc1R gene could be considered as assistance to breed identification (Jackson et al. Citation1995; Joerg et al. Citation1996). Recently, Mc1R gene polymorphisms in some Italian cattle breeds have been considered as specific DNA markers that can be used for a breed traceability strategy for some mono-breed products (Vincenzo et al. Citation2007). Therefore, we think that it is possible to find some specific SNPs markers of Mc1R gene to be used for an aid to breed identification in black cattle. In the present study, we have found a new SNP (T/C) at the position of 663 bp detected only in Chinese Wuchuan Black cattle compared with Mc1R gene sequences published on GenBank. Two genotypes, ED and E+, of Mc1R gene were classified according to the variable types at position 296. Of 56 Chinese Wuchuan Black cattle, 33 of which were E+/E+ while 23 belonged to ED/E+. The dominant allele ED caused by a T > C missense mutation in the Mc1R coding region determining an activation of the encoded receptor gives black coat color, whereas the wild-type allele E+ produces a variety of colors, depending on the Agouti locus. This also explains why all individuals of this breed are in black color. Based on this result, the new SNP (T/C) might be used as a specific molecular marker to identify the cattle breed. Considering that a small number of sample size in this study, the further study on Mc1R gene using the more sample size and the other indigenous yellow cattle breeds are necessary to confirm the new SNP (T/C).
Acknowledgments
This work was supported by Foundation for Overseas Returnee of Chinese Educational Ministry [(2009)1590], Foundation for Excellent Researchers Introduction of Guizhou University (2008030). We would like to thank Zhonghui Shi from Zhunyi Animal Husbandry Bureau, Xiong Tian from Wuchuan Animal Husbandry Bureau of Guizhou province, China, for their assistances in sample collection of Chinese Wuchuan Black cattle.
References
- Adalsteinsson S, Bjarnadottir S, Vage DI, Jonmundsson JV. 1995. Brown coat color in Icelandic cattle produced by the loci Extension and Agouti. J Heredity. 86:395–398.
- Chen H, Qiu H, Zhang TS, Jia JX. 1993. Studies on sex chromosome polymorphism of four local cattle (Bos taurus) breeds in China. Hereditas. 15:14–17.
- Chen YC, Wang YY, Cao HH. 1990. Characteristic of Chinese yellow cattle ecospecies and their course of utilization. Beijing (China): Agricultural Publishing House. Chinese.
- Graphodatskaya D, Joerg H, Stranzinge G. 2002. Molecular and pharmacological characterization of the MSH-R alleles in Swiss cattle breeds. J Receptor Signal Transd Res. 22:421–430. 10.1081/RRS-120014611
- Jackson IJ. 1993. Color-coated switches. Nature. 362:587–588. 10.1038/362587a0
- Jackson H, Vage DI, Gomez-Raya L, Adalsteinsson S, Lien S. 1995. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm Genome. 6:636–639. 10.1007/BF00352371
- Joerg H, Fries HR, Meijerink E, Stranzinger GF. 1996. Red coat color in Holstein cattle is associated with a deletion in the MSHR gene. Mamm Genome. 7:317–318. 10.1007/s003359900090
- Kriegesmann B, Dierkes B, Leeb T, Jansen S, Brenig B. 2001. Two breed-specific bovine MC1R alleles in Brown Swiss and Saler breeds. J Dairy Sci. 84:1768–1771. 10.3168/jds.S0022-0302(01)74612-7
- Lai SJ, Liu YX, Li XW, Zhang XL, Liu YP. 2005. Study on mitochondrial DNA genetic polymorphism of cattle breeds in Sichuan province. Acta Veterinaria et Zootechnica Sinica. 36:887–889. Chinese.
- Lai SJ, Liu YP, Liu YX, Li XW, Zhang XL, Liu YP. 2006. Genetic diversity and origin of Chinese cattle revealed by mtDNA D-loop sequence variation. Mol Phyl Evol. 38:146–154. 10.1016/j.ympev.2005.06.013
- Lei CC, Chen H, Yang GS, Song LS, Lei XQ, Sun WB, Li RB, Liu XL. 2004. Study on mitochondrial DNA genetic diversity of some cattle breeds in China. Acta Genetica Sinica. 31:51–62. Chinese.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. 10.1093/bioinformatics/btp187
- Liu RY, Xia XL, Lei CZ, Zhang MZ, Chen H, Yang GS. 2006. Genetic diversity of mitochondrial DNA D-loop sequences in cattle breeds in Guizhou. Hereditas. 28:279–284.
- Loftus RT, Machugh DE, Bradley DG, Sharp PM, Gunningham P. 1994. Evidence for two independent domestications of cattle. Proc Natl Acad Sci USA. 91:2757–2761. 10.1073/pnas.91.7.2757
- Mao YJ, Chang H, Yang ZP, Zhang L, Xu M, Sun W, Chang GB, Song GM, Henner S. 2007. Genetic structure and differentiation of three Chinese indigenous cattle populations. Acta Veterinaria et Zootechnica Sinic. 38:125–132. Chinese.
- Maudet C, Taberlet P. 2002. Holstein's milk detection in cheeses inferred from melanocortin receptor 1 (MC1R) gene polymorphism. J Dairy Sci. 85:707–715. 10.3168/jds.S0022-0302(02)74127-1
- Qiu H. 1988. Bovine breeds in China. Shanghai (China): Shanghai Scientific and Technical Publishing House. Chinese.
- Rouzaud F, Martin J, Gallet P, Delourme D, Goulemont-Leger V, Amigues Y, Ménissier F, Levéziel H, Julien R, Oulmouden A. 2000. A first genotyping assay of French cattle breeds based on a new allele of the extension gene encoding the melanocortin-1 receptor (Mc1r). Genet Sel Evol. 32:511–520. 10.1186/1297-9686-32-5-511
- Searle AG. 1968. Comparative genetics of coat color in mammals. London: Logos Press.
- Suzuki I, Cone RD, Im S, Nordlund J, Abdel-Malek ZA. 1996. Binding of melanotropic hormones to the melanocortin receptor MC1R on human melanocytes stimulates proliferation and melanogenesis. Endocrinology. 137:1627–1633. 10.1210/en.137.5.1627
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. 10.1093/molbev/msm092
- Thompson JD, Gibson TJ, Pelewniak F. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. 10.1093/nar/25.24.4876
- Troy CS, MacHugh DE, Bailey JF, Magee DA, Loftus RT, Cunningham P, Chamberlain AT, Sykes BC, Bradley DG. 2001. Genetic evidence for Near-Eastern origins of European cattle. Nature. 410:1088–1091. 10.1038/35074088
- Vincenzo R, Luca F, Emilio S, Marco T, Stefania D, Roberta D. 2007. Analysis of melanocortin 1 receptor (MC1R) gene polymorphisms in some cattle breeds: their usefulness and application for breed traceability and authentication of Parmigiano Reggiano cheese. Ital J Anim Sci. 6:257–272.
- Yu Y, Nie L, He ZQ. 1999. Mitochondrial DNA variation in cattle of South China: origin and introgression. Anim Genet. 30:245–250. 10.1046/j.1365-2052.1999.00483.x
- Zhang GX, Wang ZG, Chen WS, Wu CX. 2007. Genetic diversity and population structure of indigenous cattle breeds of China using 30 microsatellite markers. Anim Genet. 38:550–559. 10.1111/j.1365-2052.2007.01644.x
- Zhang GX, Zheng YM, Wang ZG, Han X, Jia SG, Ceng H. 2009. Genetic diversity and origin of mitochondria DNA D-loop region of some Chinese indigenous cattle breeds. Heredita. 31:160–168. Chinese.