Abstract
Rapid and correct diagnosis of rotavirus infection is very important to offer immediate treatment. In the present study, ribonucleic acid-polyacrylamide gel electrophoresis (RNA-PAGE) and agarose gel electrophoresis (AGE) methods were standardised and compared for the detection of rotaviruses. Double-stranded RNA (dsRNA) of rotavirus was extracted from five positive faecal samples by QIAamp Viral RNA Mini Kit and subjected to RNA-PAGE where the concentration of separating gel was varying, i.e. 7.0%, 8.5% and 12.5%. The bands of the RNA segments were clearly separated on 7.0% and 8.5% separating gel. Extracted dsRNA was also subjected to AGE where the concentration of gel was varying at 0.8%, 1.2%, 1.6% and 2.0% containing ethidium bromide. All the 11 bands of 11 segments of dsRNA were clearly visualised in all the concentrations of gel while 1.2% gel showed clear and separated bands of ladder as well as samples. Different concentrations of rotavirus, i.e. 100–1000 ng, were also loaded on standardised RNA-PAGE and AGE, separately. However, both the methods showed same detection level up to 100 ng particles of dsRNA. Further, the time required for extraction of dsRNA from five samples, their PAGE running and staining by silver stain took around 6–7 h which was comparatively more than the AGE separation, took only 2–3 h for complete procedure. Extracted dsRNA of rotavirus from 100 stool samples of children and 42 faecal samples of piglets were subjected to standardised RNA-PAGE and AGE separately for detection of rotavirus. Interestingly, 38 (38%) samples from children and 3 (7.14%) from piglet were found to be positive for rotavirus by AGE. However, RNA-PAGE could detect only 34 (34%) samples from children and 3 (7.14%) from piglets. Out of 38 positive samples from children, 32 showed long electrophoretic patterns and remaining 6 showed short electrophoretic pattern while that for 3 piglet samples, all showed long electrophoretic pattern. Thus, from our results, it can be concluded that the AGE is highly sensitive (91.17%), marginally less specific (90.90%), reproducible, superior, efficient, less laborious, cost-effective and time-saving than the RNA-PAGE for rapid diagnosis of rotavirus from faecal samples of humans as well as animals.
1. Introduction
Rotavirus infections have been recognised as a common cause of acute gastroenteritis in humans and animals and are the most important cause of severe dehydrating diarrhoea in young children in both developed and developing countries (Kapikian et al. Citation2001; Temu et al. Citation2012). Rotaviruses cause an estimated 130 million cases of gastroenteritis and 800,000 deaths in children between the ages of 6 month and 2 year in developing countries (Das et al. Citation2003). Rotaviruses have a genome composed of 11 segments of dsRNA that encodes 6 structural (VP) and 5 or 6 non-structural proteins (NSPs; Estes & Kapikian Citation2006). Rotaviruses are classified into seven different groups (A–G) based on the antigenic specificity of the VP6 capsid proteins. Thus, groups A–G can be differentiated with polyclonal and monoclonal antibodies by immunofluorescence, ELISA and immunoelectron microscopy. However, group A rotaviruses are considered as an important viral diarrhoeal agents which can be typed as G (VP7 gene) and P (VP4 gene) genotypes based on sequence comparison or G serotypes and P serotypes based on the proteins of the outer capsid that elicit neutralising antibodies. To date, 23 distinct G genotypes and 32 P genotypes have been identified (Mukherjee et al. Citation2010; Adlhoch et al. Citation2011).
Owing to the poor adaptability of rotaviruses to serial propagation in cell cultures, laborious and sophisticated procedures are necessary and therefore rapid methods of direct and indirect detection and identification have been introduced into laboratory diagnostics. Initially, direct visualisation of stool material by electron microscopy was employed for rotavirus detection. However, availability of an electron microscope is a constraint in most of the laboratories. Many methods like reverse passive haemagglutination assay, enzyme immunoassay, complement fixation test and immunoelectrophoresis have been developed but are less sensitive. The most widely used methods for detection of rotavirus include latex agglutination, enzyme-linked immunosorbent assay (ELISA), lateral flow test and RNA-PAGE (de Góes et al. Citation2008). Reverse transcriptase-polymerase chain reaction (RT-PCR) is now being extensively used as a confirmatory method for detection of rotavirus (Dewar et al. Citation2005; Roman & Martinez Citation2005). RNA-PAGE following silver staining was found to be highly specific technique for detection of rotavirus from faecal samples, but it is tedious and time-consuming.
In India, one of every 250 children or about 90,000–153,000 children die of rotavirus diarrhoea each year (Jain et al. Citation2001; Broor et al. Citation2003; Parashar et al. Citation2009; Tate et al. Citation2009). Considering the background of morbidity and mortality caused by human rotavirus, detection methods have been employed routinely in clinical diagnostic and epidemiological studies. Therefore, the purpose of the present study was to find out the efficient method for detection of rotavirus from faecal samples at a lowest possible time and at a very low concentration of the virus. The two methods, RNA-PAGE and agarose gel electrophoresis (AGE), were standardised and compared for the evaluation of best suitable rapid method for detection of rotavirus from faecal samples.
2. Materials and methods
2.1. Extraction and purification of dsRNA
Five stool samples which were positive for rotavirus by RT-PCR in earlier studies carried out in 2010 in the Division of Veterinary Public Health, Indian Veterinary Research Institute, Izatnagar, Bareilly, India were used for standardisation of RNA-PAGE and AGE methods. Viral total RNA was extracted by using a QIAamp Viral RNA Mini Kit, Cat. No. 52904 (QIAGEN, Leusden, The Netherlands) containing carrier RNA, extraction buffer AW1 and AW2 and elution buffer as AVE. The extraction was carried out according to the manufacturer's instructions from 150 µl of each stool suspension in phosphate buffer solution (0.01 M phosphate buffer pH 7.2). The RNA pellet was suspended in 20 µl of nuclease-free water and stored at −20°C for further use. Similar protocol was also utilised for the extraction of dsRNA of rotavirus from children stool samples collected from hospitals as well as for piglet faecal samples.
2.2. RNA-PAGE
RNA-PAGE was performed as per the procedure described by Laemmli (Citation1970) and Herring et al. (Citation1982) with little modifications followed by silver staining to determine the presence of rotavirus. Briefly, the reagents of resolving gel [containing Tris HCL (pH 8.8), acry/bisacrylamide solution, 10% sodium dodecyl sulfate (SDS), ammonium persulphate and tetramethylene diamine] were mixed sequentially, and the solution was poured in the gel casting plates fixed in the gel casting apparatus. One ml of distilled water was overlaid on the upper surface of the gel and allowed the gel for polymerization. The reagents of stacking gel [5%; containing Tris HCL (pH 6.8), acrylamide/bisacrylamide solution, 10% SDS, ammonium persulphate and tetramethylene diamine] were mixed sequentially and carefully overlaid on the top of resolving gel after removing the water layer. Subsequently, the comb was put in the stacking gel solution and allowed to polymerize, which took approximately 30 min. After complete polymerization of the gel, the comb was removed, and the electrophoresis buffer (containing Tris base, glycine and distilled water) was loaded in upper and lower containers. The microfuge tubes containing the RNA samples were heated at 56°C for 5–10 min to dissolve the pellet. Subsequently, 10 µl of the sample was mixed with 5 µl of 6× loading dye on paraffin wax paper and loaded into the wells. The gel was run at constant voltage of 100 V till the dye reached the bottom of the gel (normally 1–1.5 h). The staining of the PAGE gel was carried out as per the protocol standardised by Kumar (Citation2006) where the gels were kept in a fixative solution (0.5% glacial acetic acid in 10% ethanol) for 30 min followed by staining in silver stain (0.185 g silver nitrate in 100 ml distilled water) for 30 min. After this, the gels were kept in a developer solution (3 g NaOH pellets in 100 ml distilled water and 0.75 ml formaldehyde) for 10–15 min followed by stop solution (5% glacial acetic acid).
The extracted dsRNA from five positive faecal samples was subjected to RNA-PAGE where the concentration of separating gel was varying, i.e. 7.0%, 8.5% and 12.5%. However, the stacking gel concentration was same as 5.0% in all the gels. The time required for the extraction of dsRNA, PAGE and staining was noted.
2.3. AGE
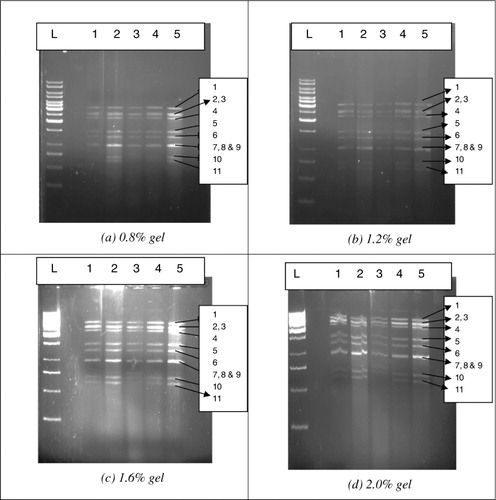
Electrophoresis was performed as per the procedure described by Chudzio et al. (Citation1989) to determine the presence of rotavirus. Ready-to-use ethidium bromide (2–3 µl) for staining was added directly into the agarose (Sigma) during its preparation. The agarose gel was prepared in 0.09-M Tris-borate buffer, pH 8.2. Approximately, 35 ml of the melted gel was poured into a gel casting tray and comb was placed. After solidification, the comb was removed, the lid was put into an electrophoretic vessel and approximately 300 ml of Tris-borate buffer was poured into it. The extracted dsRNA from five positive faecal samples was subjected to AGE where the concentration of gel was varying at 0.8%, 1.2%, 1.6% and 2.0% containing ethidium bromide for visualisation in gel documentation system (Kodak 100, Model UXT 20 m 8e). The time required for extraction of dsRNA and AGE was noted.
2.4. Detection of different concentration of dsRNA of rotavirus
The extracted concentrations of dsRNA from positive faecal samples were determined by nano-drop method. Thus, the different concentrations of rotavirus, i.e. 100 ng, 200 ng, 300 ng, 400 ng, 500 ng, 600 ng, 700 ng, 800 ng, 900 ng and 1000 ng, were loaded on standardised RNA-PAGE (7% separating gel) and AGE (1.2%) separately to find out the lowest detection limit by both the methods.
2.5. Screening of the samples by RNA-PAGE and AGE method
A total of 100 diarrhoeal stool samples of children were collected from one private hospital and Nazareth Hospital, Shillong, Meghalaya, India during 2011–2012. Additionally, 42 faecal samples from piglet diarrhoeal cases were also collected from pig farms located in and around, Shillong. All the samples were collected randomly in sterile containers and kept at −70°C till further processing. All these samples were processed for the extraction of dsRNA of rotavirus and subjected to RNA-PAGE and AGE separately for detection of rotavirus. The samples have been collected from same geographical area of Meghalaya (North Eastern Region), India. The climate of the region is tropical wet and humid conditions prevailing for most of the year. The monthly maximum temperature varies between 15 and 30°C (mean, 22°C).
3. Results and discussions
3.1. RNA-PAGE
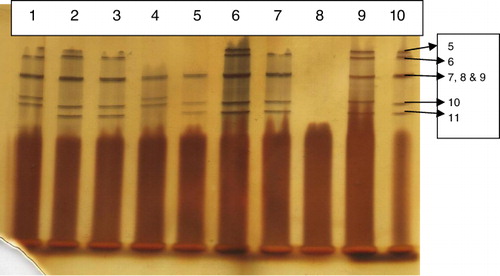
The extracted dsRNA of rotavirus from five positive faecal samples was subjected to RNA-PAGE with varying concentrations of separating gel as 7.0%, 8.5% and 12.5%; the bands of the RNA segments were clearly separated on 7.0% and 8.5% separating gel while that for 12.5% gel did not show the proper separation of the bands. However, the first four to six segments were not properly separated and stained which resulted into disappearance of these bands from all the concentrations of PAGE gels. This might be due to faulty electrophoresis and/or unstained bands and/or non-separation of the segments. It was also observed that the intensity of the bands showed increasing trend with the increase in concentration of the dsRNA in PAGE.
3.2. AGE
The extracted dsRNA of the five samples was loaded on 0.8%, 1.2%, 1.6% and 2.0% agarose gel, separately. All the 11 bands of 11 segments of dsRNA were clearly visualised in all the concentrations of gel (). Similar results were also obtained by Chudzio et al. (Citation1989). In contrast, Pšikal et al. (Citation1991) obtained only eight and nine bands in 1.5% agarose gel. However, ladder and some of the bands were not clearly visualised in 1.6% and 2.0% agarose gel, while 1.2% gel showed clear and separated bands of ladder and samples. As ethidium bromide was added during the preparation of the agarose gel, there was no separate staining procedure required. Thus, total procedure from dsRNA of rotavirus extraction till the band visualisation took 2–3 h for five samples. Further, the method was easy to perform, economical, reproducible and quick. The major advantages of AGE are the straightforward procedure, involving no complicated sample processing and preparation of specific immune sera, and its speed, both making this method usable in routine investigations (Pšikal et al. Citation1991). It was also observed that 100-ng concentration of dsRNA was sufficient to clearly visualise the samples in agarose gel (1.2%). This indicated that the samples of low virus content could also be detected by AGE method. The clarity and intensity of bands were increasing with the increase in concentration of rotaviral RNA from 100 ng to 1000 ng ().
3.3. Screening of the samples
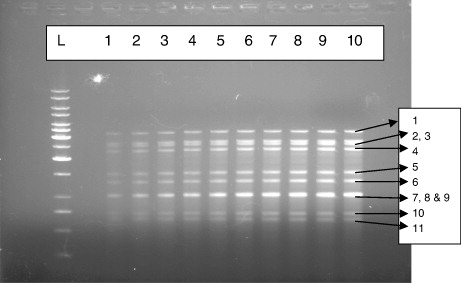
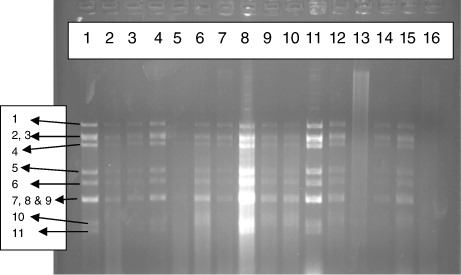
A total of 100 diarrhoeal stool samples of children and 42 of piglet diarrhoeal faecal samples were processed for extraction of genomic RNA. All the extracted dsRNA samples were loaded on 1.2% agarose gel and 7.0% RNA-PAGE for detection of rotavirus. Interestingly, 38 (38%) samples from children () and 3 (7.14%) from piglet were found to be positive for rotavirus by AGE method. However, RNA-PAGE could detect only 34 (34%) samples from children () and 3 (7.14%) from piglets. This indicated that the agarose gel method was more sensitive (91.17%) than the RNA-PAGE (81.57%). The results are in accordance with the results of Pšikal et al. (Citation1991) where it was observed that the reliable results were obtained by electrophoresis of the extracted RNA in agarose allowing a direct identification of rotaviruses, based on the characteristic arrangement of the RNA segments in the gel and on evaluation of respective electrophoretogrammes. Out of 94 samples, 43 were positive for rotavirus after examination by this method and suggested that the method was equally sensitive to ELISA (Matsui et al. Citation1990). Group A human and animal rotaviruses also displayed two electropherotypes: long and short (Matsui et al. Citation1990). Short electrophoretic patterns exhibited a larger segment 11 (encoding NSP5) that migrated more slowly and was located between gene segments 9 and 10 (Matsui et al. Citation1990). Although most group A rotaviruses have either a short or a long pattern, super-short electropherotypes have been documented. These correlations between RNA patterns and serotypes have been maintained and have become a useful epidemiologic tool (Matsui et al. Citation1990). Detailed descriptions of the correlations between electropherotype and viral antigenic and genetic properties have been reported (Gentsch et al. Citation1996; Desselberger et al. Citation2001). Differentiation of ‘long’ and ‘short’ electrophoretic pattern is quite difficult in case of AGE than that for RNA-PAGE. It was observed that out of 38 positive samples from children, 32 showed long electrophoretic patterns and remaining 6 showed short electrophoretic pattern while that for 3 piglet samples, all showed long electrophoretic pattern by both the methods, i.e. RNA-PAGE and AGE. In majority of the earlier studies, long pattern seems to be dominant (Brown et al. Citation1988; Broor et al. Citation1993; Dubal et al. Citation2013). The RNA patterns of the 38 animal strains were analysed (Pongsuwanne et al. Citation1989) using PAGE and showed long RNA profiles, except for two porcine and two bovine specimens.
3.4. Comparison between RNA-PAGE and AGE
Although both the methods detected the rotavirus, they differ in the number of positive samples. The reliable results were obtained by electrophoresis of the extracted RNA in agarose gel allowing a direct identification of rotaviruses based on the characteristic arrangement of the RNA segments in the gel and on evaluation of respective electrophoretogrammes. In case of RNA-PAGE, first four to six segments of dsRNA of rotavirus were not properly separated and stained. However, such problem was not detected in the AGE separation method. All the 11 bands of rotavirus were clearly visualised in AGE. Though all the 11 bands of rotavirus were not visualised, the clarity of the bands was superior in PAGE as compared to agarose gel (1.2%). Further, the time required for extraction of dsRNA from five samples, their PAGE running and staining by silver stain took around 6–7 h which was comparatively more than the AGE separation, took only 2–3 h for complete procedure. According to the WHO (Citation2009) manual, the PAGE method had sometimes been used to diagnose group A rotavirus infections for surveillance studies. However, this method is very labour intensive and time-consuming. Both the methods showed detection level up to 100 ng particles of dsRNA. However, while screening the stool samples from children, only 34 could be detected by RNA-PAGE and 38 by the agarose gel method. This is indicative of superiority of the agarose gel method over the RNA-PAGE method (Pšikal et al. Citation1991). Moreover, the agarose gel method was easy to perform, less laborious, economical and time-saving than the RNA-PAGE method, as it is complicated, laborious and needs expert to perform. Similarly as PAGE, AGE can be used for the differentiation of the rotavirus RNA not only within a single animal species but also between various species (Rodger & Holmes Citation1979). Thus, from our results, it can be concluded that the AGE is superior, efficient, less laborious, economical and time-saving than the RNA-PAGE method for rapid diagnosis of rotavirus from faecal samples of humans as well as animals.
Acknowledgements
The authors are thankful to the Director, ICAR Research Complex for NEH Region, Umiam, Meghalaya, India for providing the necessary facilities for the study.
Funding
The work was supported by grants from the Indian Council of Agricultural Research, New Delhi, to ZBD.
Additional information
Funding
References
- Adlhoch C, Kaiser M, Hoehne M, Mas Marques,Stefas I, Veas F and Ellerbrok H. 2011. Highly sensitive detection of the group A rotavirus using Apolipoprotein H-coated ELISA plates compared to quantitative real-time PCR. Virol J. 8: 63. 10.1186/1743-422X-8-63
- Broor S, Ghosh D, Mathur P. 2003. Molecular epidemiology of rotaviruses in India. Indian J Med Res. 118: 59–67.
- Broor S, Husain M, Chatterjee B, Chakraborty A, Seth P. 1993. Temporal variation in the distribution of rotavirus electropherotypes in Delhi, India. J Diarrhoeal Dis Res. 11: 14–18.
- Brown DWG, Mathan MM, Mathew M, Martin R, Beards GM, Mathan VI. 1988. Rotavirus epidemiology in Vellore, South India. Group, subgroup, serotype, and electrophoretype. J Clin Microbiol. 26: 2410–2414.
- Chudzio T, Kasatma S, Irvine N, Sankar-Mistry P. 1989. Rapid screening test for the diagnosis of rotavirus infection. J Clin Microbiol. 27: 2394–2396.
- Das S, Varghese V, Chaudhury S, Barman P, Mahapatra S, Kojima K, Bhattacharya SK, Krishnan T, Rathod RK, Chhotray GP, et al. 2003. Emergence of novel human group A rotavirus G12 strains in India. J Clin Microbiol. 41: 2760–2762. 10.1128/JCM.41.6.2760-2762.2003
- de Góes AC, de Moraes MT, de Castro, SW, Araújo IT, de H'alluin JC, da Silva Souza W, da Silva Junior JG, Leite JP. 2008. Development of a rapid and sensitive latex agglutination-based method for detection of group A rotavirus. J Virol Methods. 148: 211–217. 10.1016/j.jviromet.2007.11.013
- Desselberger U, Iturriza-Gomara M, Gray JJ. 2001. Rotavirus epidemiology and surveillance. Novartis Found Symp. 238: 125–152.
- Dewar J, de Beer M, Elliott E, Monaisa P, Semenva D, Steele A. 2005. Rapid detection of rotaviruses – are laboratories underestimating infection in infants? S Afr Med J. 95: 494–495.
- Dubal Z B, Bhilegaonkar KN, Barbuddhe SB, Kolhe R P, Simranpreet Kaur, Shriya Rawat, Prejit Nambier, Karunakaran M. 2013. Prevalence and genotypic (G and P) determination of porcine group A rotaviruses from different regions of India. Trop Anim Health Prod. 45: 609–615. 10.1007/s11250-012-0267-1
- Estes MK, Kapikian AZ. 2006. Rotaviruses. In: Knipe DM, Howley PM, editors. Fields virology. 5th ed., Vol. 1. Philadelphia (PA): Lippincott Williams & Wilkins; p. 1917.
- Gentsch JR, Woods, PA, Ramachandran M, Das BK, Leite JP, Alfieri A, Kumar R, Bhan MK, Glass RI. 1996. Review of G and P typing results from a global collection of strains: implications for vaccine development. J Infect Dis. 174: S30–S36. 10.1093/infdis/174.Supplement_1.S30
- Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. 1982. Rapid diagnosis of rotavirus infection by direct infection of viral nucleic acid in silver stained polyacrylamide gels. J Clin Microbiol. 16: 473–477.
- Jain V, Parashar UD, Glass RI, Bhan MK. 2001. Epidemiology of rotavirus in India. The Indian J Pediatr. 68: 855–862. 10.1007/BF02762113
- Kapikian AZ, Hoshino Y, Chanock RM. 2001. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. 4th ed. Philadelphia (PA): Lippincott Williams & Wilkins; p. 1787–1833.
- Kumar M. 2006. Polymerase chain reaction for rapid detection of important zoonotic diarrhoeal pathogens [M.V.Sc. Thesis]. Bareilly: IVRI.
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of the bacteriophage. Nature. 227: 680–685. 10.1038/227680a0
- Matsui M, Mackow ER, Matsuno S, Paul PS, Greenberg HB. 1990. Sequence analysis of gene 11 equivalents from “short” and “super short” strains of rotavirus. J Virol. 64: 120–124.
- Mukherjee A, Chattopadhyay S, Bagchi P, Dutta D, Singh NB, Arora R, Parashar UD, Gentsch JR, Chawla-Sarkar M. 2010. Surveillance and molecular characterization of rotavirus strains circulating in Manipur, North-Eastern India: Increasing prevalence of emerging G12 strains. Infect Genet Evol. 10: 311–320. 10.1016/j.meegid.2010.01.002
- Parashar UD, Burton A, Lanata C, Boschi-Pinto C, Shibuya K, Steele D, Birmingham M, Glass RI. 2009. Global mortality associated with rotavirus disease among children in 2004. The J Infect Dis. 200: S9–S15. 10.1086/605025
- Pongsuwanne Y, Taniguchi L K, Choonthanom M, Chiwakul M, Susansook T, Saguanwongse S, Jayavasu C, Urasawa S. 1989. Subgroup and serotype distributions of human, bovine, and porcine rotavirus in Thailand. J Clin Microbiol. 27: 1956–1960.
- Pšikal I., Dvorák R., Franz J., Stepánek J. 1991. Rapid identification of bovine rotavirus by electrophoresis in agarose gel. Acta Vet Brno. 60(3): 253–261. 10.2754/avb199160030253
- Rodger SM, Holmes IH. 1979. Comparison of the genomes simian, bovine and human rotaviruses by gel electrophoresis and detection of genomic variation among bovine isolates. J Virol. 30: 839–846.
- Roman E, Martinez I. 2005. Detection of rotavirus in stool samples of gastroenteritis patients. P R Health Sci J. 24: 179–184.
- Tate JE, Chitambar S, Esposito DH, Sarkar R, Gladstone B, Ramani S, Raghava SV, Sowmyanarayanan TV, Gandhe S, Arora R, Parashar UD, Kang G. 2009. Disease and economic burden of rotavirus diarrhoea in India. Vaccine. 27: F18–F24. 10.1016/j.vaccine.2009.08.098
- Temu A, Kamugisha E, Mwizamholya DL, Hokororo A, Seni J, Mshana SE. 2012. Prevalence and factors associated with Group A rotavirus infection among children with acute diarrhoea in Mwanza, Tanzania. J Infect Dev Countries. 6: 508–515.
- World Health Organisation (WHO). 2009. Manual of rotavirus detection and characterization methods. p. 3. www.who.int/vacc.