Abstract
To estimate the phylotaxonomic position of Tianzhu white yak (Poephagus grunniens), multiple subunit genes of cytochrome c oxidase (cox) were sequenced: 1545 bp of cox1 (JF946751), 684 bp of cox2 (JN008944) and 781 bp of cox3 (JF946752). Sequence divergences of cox genes between yaks and cattle/zebus (5.7% in cox1, 7.7% in cox2 and 6.45% in cox3) were higher than those between yaks and American bisons (2.4% in cox1, 3.0% in cox2 and 2.1% in cox3). Molecular phylogenetic analysis with these genes also found that yaks and American bisons firstly clustered in one clade, indicating there was higher genetic comparability than that of cattle/zebus. The findings sustained the idea that in the choice of nomenclature yaks belong to the subgenus of Poephagus. According to the nucleotide substitution rate of cox genes among species of Bovinae, the speculated divergence time of yaks from cattle/zebus, American bisons, European bisons, Asian buffaloes/African buffaloes was 1.05–1.50 million years ago (MYA), 2.85–3.89 MYA, 2.90–3.70 MYA, 6.85–7.50 MYA, respectively. The sequential evolution of Bovinae members could be predicted that buffaloes were first to be domesticated during the end of Miocene and the early of Pliocene. In the end of Pliocene, the Bovinae genera were evolved to Bos, Bison and Poephagus. Poephagus was the latest evolved genera among the species of Bovinae.
1. Introduction
Yaks (Poephagus grunniens) are distributed across the Qinghai-Tibetan Plateau (QTP) and adjacent highlands (Zhang Citation1989; Wiener et al. Citation2003). QTP is the largest continuous high-elevation ecosystem in the world, occupying nearly 2.5 million km2 of the Asian continent and reaching an average elevation of more than 3000 m as sea level (Li et al. Citation1995). More than 14 million yaks are currently herded there and they provide many of life's necessities (food, hids, dung fuel and transport power) for the nomadic Tibetan pastoralists living in this extremely harsh region (Wiener et al. Citation2003).
Bos grunniens, Bos mutus and Poephagus mutus are all species names, which have been assigned to yaks. This variety of nomenclature reflects the uncertain relationship of the yak to other members of the Bovinae. It has been ascertained that yaks are originated from China. Based on the petrifactions found in the northern China, Siberia and Alaska, researchers suggest that the forefather of the today's domestic yak and wild yak (P. mutus) was a primitive wild yak that lived about 2.50 million years ago (MYA; Hou Citation1991; Cai Citation1992). Investigation of 20 microsatellite loci indicates that the divergence time between yaks and cattle (Bos taurus) was 0.57–1.53 MYA, between yaks and the American bisons (Bison bison) 0.39–1.04 MYA (Ritz et al. Citation2000), showing the genetic relationship between the yaks and American bisons is closer than the yaks and cattle. More molecular evidences, either from mitochondrial DNA (mtDNA) or from nuclear DNA, have been obtained, indicating that the yak and American bison form the same clade, separated from other members of Bovinae (Tu et al. Citation1998; Tu et al. Citation2002; Hassanin & Ropiquet Citation2004; Li et al. Citation2006a; Xie et al. Citation2010; Zhao et al. Citation2011; Qiu et al. Citation2012; Ma et al. Citation2013). However, so far some envidences derived from archaeology and historical records still have not been reached a consensus about the origns of the yak and its taxonomy. The ancient Tibetans in the northern Tibet had captured and domesticated wild yaks for agricultural use during the Palaeolithic era 5000–10,000 years ago (Olsen Citation1990; Wiener et al. Citation2003; Li et al. Citation2008). Chinese archaeologists discovered fossils in Tibet, Sichan and Qinghai province in the 1950s indicating that the domestic yak appeared 4500 years ago (Zhang Citation1989; Cai Citation1992). These findings were consistent with the report by Li (Citation2004) who used microsatellite markers to determine the period of yak origin.
The yak has similar morphological characteristics to American bison and cattle, so researchers focused on the phylogenetic relationship between yaks and other species of Bovinae (Li et al. Citation2006b; Gu et al. Citation2007; Xie et al. Citation2010). Gray (Citation1843) reported that yaks were identified as Poephagus according to their morphological differences between genus Bos and B. bison. Olsen (Citation1990, Citation1991) found that yaks have the same arrangement of premaxillaries, maxillaries and nasals as in Bison and are different from the structure in Bos. Geraads (Citation1992), using a matrix of 57 cranial characters and 32 taxa of fossil of different Bovinae species, concluded that the yak is close to the Bison. These results were consistent with the studies of Li et al. (Citation2005) and Hassanin and Ropiquet (Citation2004). In contrast, according to different morphological characteristics of Bovinae species, Bohlken (Citation1958, Citation1961) first grouped the yak with cattle, followed by gaur (Bos gaurus) and banteng (Bos javanicus) in one clade, which then clustered with the American bison and European bison (Bison bonasus). The same conclusion was made by Hartl et al. (Citation1988) and Ritz et al. (Citation2000) from biochemical genetics and microsatellite loci, respectively.
Tianzhu white yak, with 0.8 million, is one of the unique populations of Chinese yaks that centralized distribution in Tianzhu Tibetan Autonomous County, Gansu Province in China, which is located in the eastern end of the Qilian mountain and the northern edge of the QTP. The aim of the present study was to research the taxonomy of the Chinese yak and the phylogenetic relationship between the yaks and other species of Bovinae using nucleotide sequences of mitochondrial cytochrome c oxidase (cox) genes in Tianzhu white yak.
2. Materials and methods
2.1. Animals and sampling
Tianzhu white yaks were selected from Tianzhu White Yak Breeding Farm in Xidatan Village, Xidatan Town, Tianzhu Tibetan Autonomous County, Wuwei Prefecture, Gansu Province, which is located in the eastern end of the Qilian mountain and the northern edge of the QTP (102°02′–103°29′E; 36°29′–37°41′N). Any treatments on animals followed the guidelines stated in the Guide for the Care and Use of Agricultural Animals in Research and Teaching in Gansu Province, China. Ten mL of anticogulated carotid arterial blood was sampled from each adult female yak (n = 4).
2.2. Amplification of cox genes
Genomic DNA extraction was performed using animal blood genomic DNA extraction kit (Tiangen, China) according to the manufacturer's instruction. Primers were designed according to the nucleotide sequence of mitochondrial complete genome of P. grunniens (NC_006380) by Primer Premier 5.0 software and confirmed by primer-BLAST in http://www.ncbi.nlm.nih.gov/tools/primer-blast/ (). Gene amplification of cox1, cox2 and cox3 was performed with 5 min at 94°C for one cycle, followed by 35 cycles of denaturation for 45 sec at 94°C, 45 sec of annealing at 55°C and 1 min of extension at 72°C, with the final extension step for 10 min at 72°C for one cycle. Analyse 5 μL of each sample on a 0.9% agarose/EtBr gel. Each 20 μL reaction system was comprised of 1 μL genomic DNA (200 ng/μL), 1 μL forward primer (10 μM/μL) and 1 μL reverse primer (10 μM/μL), 1 μL dNTP mix (10 μM/μL), 2μL 10 × buffer, 0.5 μL Taq DNA polymerase (5 U/μL), 13.5 μL ddH2O. Expected target gene fragment was then subcloned to T-easy vector. Positive plasmid was sequenced in Sangon Biotech (Shanghai, China) Co., Ltd. At least three positive colonies of each fragment from each sample were sequenced.
Table 1. Nucleotide sequence of primers used in the present study (forward primers are listed first).
2.3. Data analyses
Using complete mtDNA of sheep, Ovis aries (AF010406) as an out-group taxon, phylogenetic analysis of Tianzhu white yak was performed based on the cox1 (JF946751), cox2 (JN008944) and cox3 (JF946752) comparison with representative Bovinae species, such as P. grunniens (Qinghai black breed), B. taurus (cattle), Bos indicus (zebu), B. bison (American bison), B. bonasus (European bison), Bubalus bubalis (Asian buffalo) and Synerus caffer (African buffalo; ). The analysis of sequence alignment, arrangement and the sequence divergence were carried out using DNAStar 5.02 software. The transition: transversion (Ts/Tv) bias was calculated as equation , where A, T, C, G was represented nucleotide substitution frequence, K1 and K2 were indicated the Ts/Tv rate ratio of purines and pyrimidines using the maximum parsimony method of MEGA5.0 software (Tamura et al. Citation2004, Citation2011) by selecting the Kimura 2-parameter model. Phylogenetic tree was constructed by Neighbour-joining method with bootstrap replication test (1000 replications). The divergence time of different species of Bovinae was estimated by the 2% molecular clock of the cox genes per million years (Birungi & Arctander Citation2001).
Table 2. Source of sequence for cox subunit genes in species of Bovinae and in O. aries.
3. Results and discussion
3.1. Cloning of cox1, cox2 and cox3 in Tianzhu white yak
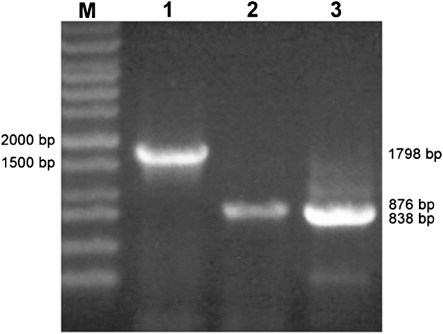
We amplified cox genes of Tianzhu white yak using genomic DNA template and primers listed in . The expected genes were subcloned and three positive colonies of each fragment from each sample were sequenced. As shown in , about 1798 bp, 876 bp and 838 bp fragments containing cox1, cox2 and cox3 were obtained from each sample (). Sequencing results indicated that the full length of cox1, cox2 and cox3 in Tianzhu white yak was 1545 bp, 684 bp and 781 bp, which has been conferred GenBank Accession Number with JF946751, JN008944 and JF946752, encoding 515, 228 and 260 amino acids, respectively. Nucleotide information showed that content of bases A and T, bases G and C is 58.06% and 41.94% in cox1, 62.57% and 37.43% in cox2, 55.31% and 44.69% in cox3, showing obviously higher contents of bases A and T than that of bases G and C.
3.2. Sequence divergence percentage among Bovinae species based on cox1, cox2 and cox3
The percentage divergences of cox1, cox2 and cox3 in Bovinae species were summarized in , and . Tianzhu white yak had the highest identity to Qinghai black breed with the minimum sequence divergence of 0.5% in cox1. The following was yak and American bison (2.4%), yak and cattle/zebu (5.7%), yak and European bison (6.2–6.3%), yak and Asian buffalo (13.8–14.1%), yak and African buffalo (14.4–14.7%). Sequence divergence between cattle and zebu, cattle/zebu and American bison/European bison, cattle/zebu and Asian buffalo/African buffalo was 1.2%, 5.3–5.7% and 14.5–15.1%, respectively (). The percentage divergence of cox2 sequence of Tianzhu white yak and Qinghai black breed was 0.6%, with the minimum value, the following order of divergence between yak and American bison, yak and European bison, yak and cattle/zebu, yak and Asian buffalo, yak and African buffalo was 3.0%, 7.4%, 7.5–7.9%, 14.4–14.8%, 15.4%, respectively (). In , the nucleotide divergence of cox3 between yak and American bison, yak and European bison, yak and cattle/zebu, yak and African buffalo, yak and Asian buffalo was 2.1%, 5.8%, 6.4–6.5%, 11.9%, 15.5%, respectively. In general, the sequence divergences between yaks and cattle/zebus (5.7% in cox1, 7.7% in cox2 and 6.5% in cox3) were higher than those between yaks and American bisons (2.4% in cox1, 3.0% in cox2 and 2.1% in cox3), indicating the genetic relationship between yaks and American bisons is closer than yaks and cattle/zebus.
Table 3. Nucleotide divergence percentage of cytochrome c oxidase subunit I (cox1) gene in species of Bovinae.
Table 4. Nucleotide divergence percentage of cytochrome c oxidase subunit II (cox2) in species of Bovinae.
Table 5. Nucleotide divergence percentage of cytochrome c oxidase subunit III (cox3) in species of Bovinae.
3.3. Phylogenetic analysis among Bovinae species based on cox1, cox2 and cox3
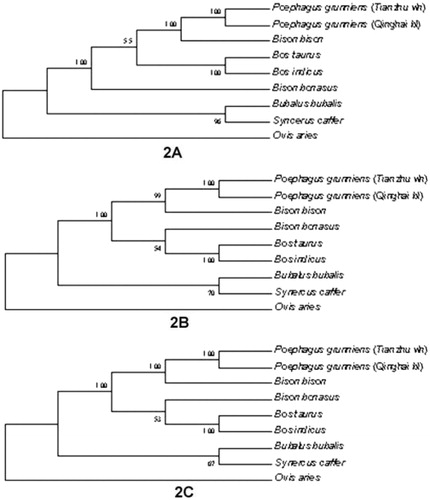
Based on the nucleotide sequence of 1545 bp of cox1, 684 bp of cox2 and 838 bp of cox3 in Tianzhou white yak, a phylogenetic tree of Bovinae was constructed using the Neighbour-joining tree method and listed as , and . Bovinae was clustered in one group, while the out-group of sheep was in another group. Among eight species of Bovinae, yaks were clustered with American bisons, but not with cattle/zebus.
3.4. Estimation of divergence time among different Bovinae species
The Ts/Tv rate of cox1, cox2 and cox3 among the different species of Bovinae was 9.042, 7.646 and 6.052, higher than the critical value 2.0 (Knight & Mindell Citation1993) and showed higher transition bias. Therefore, the molecular clock of the cox1, cox2 and cox3 gene sequence was 2% per million years in Bovinae and chosen to estimate the divergence time between different species of Bovinae. Based on cox1, the divergence time between Tianzhu white yak and Qinghai black yak, yak and American bison, yak and cattle/zebu, yak and European bison, yak and buffalo was 0.25 MYA, 1.20 MYA, 2.85 MYA, 3.15 MYA and 7.13 MYA, respectively. Based on cox2, the divergence time between Tianzhu white yak and Qinghai black yak, yak and American bison, yak and European bison, yak and cattle/zebu, yak and buffalos was 0.30 MYA, 1.50 MYA, 3.70 MYA, 3.84 MYA and 7.50 MYA, respectively. Based on cox3, the divergence time between yak and American bison, yak and European bison, yak and cattle/zebu, yak and African buffalo, yak and Asian buffalo was 1.05 MYA, 2.90 MYA, 3.25 MYA, 5.95 MYA and 7.75 MYA, respectively. These results indicated that the divergence time between yak and American bison was much later than other species of Bovinae.
The earliest report of Linnaeus (Citation1766) listed yak as Bos grunniens of Bos, which was consistent with Bohlken's results from morphological characteristics that yak and cattle first grouped, followed by gaur and banteng in one clade, then clustered with the American bison and European bison (Bohlken Citation1958, Citation1961). Gray (Citation1843) identified yak as Poephagus according to the morphological differences between genus Bos and B. bison, which was consistent with the opinions of Groves (Citation1981), Olsen (Citation1990, Citation1991), Geraads (Citation1992) from the head bone features and archaeological study, and Li et al. (2Citation006a) from the molecular research. Since the late 1980s, the sequences of mtDNA of yak have been used for the phylogenetic studies of Bovinae. Miyamoto et al. (Citation1989) sequenced the 12S rRNA and three tRNA genes, and a 247 bp partial hypervariant D-loop fragment of four taxa in the Bovinae and the results showed a similar topology of phylogeny with Groves (Citation1981), the yak grouped with the B. bison first, with an average divergence of 2.6% for the conservative rRNA/tRNA genes and 9.1% for the D-loop fragment and then followed by B. taurus (Kraus et al. Citation1992). Ward et al. (Citation1999) using a partial mtDNA control region of 667 base pairs, found that the percentage nucleotide divergence of B. indicus and B. taurus from the yak was 24.23% and 29.53%, respectively, but that of American bison and European bison from the yak was only 12.28% and 16.59%, respectively. Similar results have been obtained by Hassanin and Douzery (Citation1999), Li et al. (2Citation006a), Xie et al. (Citation2010) from complete sequence of cytochrome b (cytb), Zhao et al. (Citation2011) from cox3, Zhang et al. (Citation2012) from mtDNA D-loop, Hassanin and Douzery (Citation1999) from mtDNA 12S rRNA, aromatase cytochrome P-45. Within the nuclear genome alone, Wall et al. (Citation1992) confirmed the position of yak as more closely related to the bison and wisent than to B. taurus and B. indicus as earlier suggested by Groves (Citation1981). Hassanin and Douzery (Citation1999) sequenced the promoter segment of the lactoferrin-encoding genes of nuclear genomic DNA in the yak alongside with other Bovidae species, showing the yak clustered with the American bison first and then grouped with B. taurus in the phylogenetic tree. More similar molecular evidences have been made by Ritz et al. (Citation2000) from 20 bovine microsatellite markers, Buntjer et al. (Citation2002) from 361 fingerprinting Bovini markers of AFLP, Fan et al. (Citation2000) from exon 4 of kappa casein, Li et al. (Citation2005) from exon 2 of MHC-DRB3.
At present study, the average percent nucleotide divergence between yaks (Tianzhu white, Qinghai black) and cattle/zebus was 5.7% in cox1, 7.7% in cox2 and 6.45% in cox3, which was considerably higher than that between yaks and American bisons (2.4% in cox1, 3.0% in cox2 and 2.1% in cox3), indicating that genetic identity between yaks and American bisons is higher than that between yaks and cattle/zebus. Phylogenetic analysis based on cox1, cox2 and cox3 also showed that yaks were clustered with American bisons, but not with cattle/zebus. According to the 2% per million years mutation rate of cox genes, we speculated the divergence time between yaks and American bisons was 1.20 MYA based on cox1, 1.50 MYA based on cox2 and 1.05 MYA based on cox3, which is later than that between yaks and cattle/zebus (2.85 MYA in cox1, 3.70 MYA in cox2 and 3.25 MYA in cox3, respectively). These findings showed that the yak has the closest genetic relationship related to American bison. Therefore, it seems clear that for the domestic yak, the subgenus, Poephagus, seems more appropriate than Bos. This finding is in agreement with the results made by Wiener et al. (Citation2003), Li et al. (Citation2005), Li et al. (Citation2006a).
Using subunit genes of cytochrome c oxidase, we estimated that the divergence time of buffaloes from other species of Bovinae was 7.13–7.75 MYA, whereas the yaks from cattle/zebus, B. bisons from cattle/zebus was 2.85–3.84 MYA and 1.05–1.50 MYA, respectively, which is in agreement with the report of Xie (Citation1985), Li et al. (2Citation006a). The divergence time of cattle from zebus was 0.45–0.60 MYA, consenting with the result of Ritz et al. (Citation2000). Similar to the result of Li et al. (2Citation006a), the divergence time between yaks and American bisons was 1.7–2.05 MYA. Combination with paleontological evidence (Osborn Citation1990) and morphological characteristics (Xie Citation1985), we estimated that buffalos were first to be divided into Asian buffalos and African buffalos among the species of Bovinae during the end of Miocene (23.00–5.3 MYA) and the early of Pliocene (5.2–1.8 MYA). In the end of Pliocene, the genera of Bovinae were evolved to Bos, Bison and Poephagus. Poephagus, which branched off from the middle of Pleiocene, was the latest evolved genera among the species of Bovinae.
4. Conclusion
We cloned and sequenced multiple subunit genes of cox of Tianzhu white yak (P. grunniens): 1545 bp of cox1 (JF946751), 684 bp of cox2 (JN008944) and 781 bp of cox3 (JF946752). Sequence divergences between yaks and cattle/zebus (5.7% in cox1, 7.7% in cox2 and 6.45% in cox3) were higher than those between yaks and American bisons (2.4% in cox1, 3.0% in cox2 and 2.1% in cox3). Molecular phylogenetic analysis with these genes also found that yaks and American bisons firstly clustered in one clade, indicating there was higher genetic comparability than that of cattle/zebus. We concluded that in the choice of nomenclature yaks belong to the subgenus of Poephagus. We also speculated that buffaloes were first to be divided into Asian buffalos and African buffalos among the species of Bovinae during the end of Miocene and the early of Pliocene. In the end of Pliocene, the Bovinae genera were evolved to Bos, Bison and Poephagus. Poephagus, which branched off from the middle of Pleiocene, was the latest evolved genera among the species of Bovinae.
Funding
The work was partially financed by the project of National Natural Science Foundation of China, NSFC [grant number 31260533] and the Programme for Changjiang Scholars and Innovative Research Team in University of Ministry of Education, China [grant number IRT13091].
Additional information
Funding
References
- Birungi J, Arctander P. 2001. Molecular systematics and phylogeny of the reduncini (Artiodactyla: Bovidae) inferred from the analysis of mitochondrial cytochrome b gene sequences. J Mammal Evol. 8:125–147. 10.1023/A:1011369914909
- Bohlken H. 1958. Vergleichende Untersuchungen an Wildrindern [Comparative investigation of wild cattle] ( Tribus Bovini Simpson, 1954). Zool Jahrb. 68:113-202.
- Bohlken H. 1961. Haustiere und Zoologische Systematick [Systematic study of pets and animals]. Z Tier Zuchtungsbiol [J Anim Biochem]. 76:107–113. 10.1111/j.1439-0388.1961.tb01200.x
- Buntjer JB, Otsen M, Nijman IJ, Kuiper MT, Lenstra JA. 2002. Phylogeny of bovine species based on AFLP fingerprinting. Heredity. 88:46–51. 10.1038/sj.hdy.6800007
- Cai L. 1992. Chinese yak. Beijing: China Agriculture Press.
- Fan BL, Li N, Wu CX. 2000. Research on constructing phylogenic trees on ruminants basing on the database of milk protein genes. Acta Genet Sin. 27:485–497.
- Geraads D. 1992. Phylogenetic analysis of the tribe Bovini (Mammalia: Artiodactyla). Zool J Linn Soc. 104:193–207. 10.1111/j.1096-3642.1992.tb00922.x
- Gray JE. 1843. List of the specimens of mammalia in the collection of the British museum. London: the Trustees.
- Groves CP. 1981. Systematic relationships in the Bovinii (Artiodactyla, Bovidae). J Zool Syst Evol Res. 19:264–278. 10.1111/j.1439-0469.1981.tb00243.x
- Gu Z, Zhao X, Li N, Wu C. 2007. Complete sequence of the yak (Bos grunniens) mitochondrial genome and its evolutionary relationship with other ruminants. Mol Phylogenet Evol. 42:248–255. 10.1016/j.ympev.2006.06.021
- Hartl GB, Göltenboth R, Grilltsch M, Willing R. 1988. On the biochemistrical systematic of the Bovini. Biochem Syst Ecol. 16:575–579. 10.1016/0305-1978(88)90065-8
- Hassanin A, Ropiquet A. 2004. Molecular phylogeny of the tribe Bovini (Bovidae, Bovinae) and the taxonomic status of the Kouprey, Bos Sauveli Urbain. 1937. Mol Phylogenet Evol. 33:896–907. 10.1016/j.ympev.2004.08.009
- Hassanin A, Douzery EJP. 1999. Evolutionary affinities of the enigmatic saola (Pseudoryx nghetinhensis) in the context of the molecular phylogeny of Bovidae. Proc Biol Sci. 266:893–900. 10.1098/rspb.1999.0720
- Hou SZ. 1991. An introduction of Tibet archeology. Lhasa: Tibet People's Press.
- Knight A, Mindell DP. 1993. Substitutions bias, weighting of DNA sequence evolution, and the phylogenetic positions of fea's viper. Syst Biol. 42:18–31. 10.1093/sysbio/42.1.18
- Kraus F, Jarecki L, Miyamoto MM, Tanhauser SM, Laipis PJ. 1992. Mispairing and compensational changes during the evolution of mitochondrial ribosomal RNA. Mol Biol Evol. 9:770–774.
- Li JJ, Shi YF, Li BY. 1995. Uplift of the Qinghai-Xizang (Tibet) plateau and global change. Lanzhou: Lanzhou University Press.
- Li QF. 2004. Molecular phylogenetic analysis of yak in China [ PhD dissertation]. Nanjing: Nanjing Agricultural University.
- Li QF, Li YH, Zhao XB, Li XB, Pam ZX, Xie Z, Li N. 2005. Sequence variation at exon 2 of MHC DRB3 locus in Bovinae. J Agric Biotechnol. 13:441–446.
- Li QF, Li YX, Zhao XB, Liu ZS, Zhang QB, Song DW, Xu G, Li N, Xie Z. 2006a. Sequencing cytochrome b gene of mitochondrial DNA in yak and researching its origin and taxonomic status. Acta Vet Et Zootechnica Sin. 37:1118–1123.
- Li QF, Zhao XB, Liu HL, Li N, Xie Z. 2006b. A review of the research on taxonomic position in yak (Poephagus grunniens). Acta Zootax Sin. 31:520–524.
- Li QF, Li YX, Zhao XB, Pan ZX, Liu ZS, Zhang QB, Qu XG, Song DW, Dong LY, Li N, Xie Z. 2008. Sequencing D-loop region of mitochondrial DNA in yak and researching its taxonomic status in Bovinae. Acta Vet Zootechnica Sin. 39:1–6.
- Linnaeus C. 1766. Systema Naturae. 12th ed. Holmiae: Salvii.
- Ma ZJ, Zhong JC, Han JL, Xu JT, Liu ZN, Bai WL. 2013. Research progress on molecular genetic diversity of the yak (Bos grunniens). Hereditas. 35:151–160.
- Miyamoto MM, Tanhauser SM, Laipis PJ. 1989. Systematic relationship in the artiodactyls tribe Bovini (family Bovidae), as determined from mitochondrial DNA sequences. Syst Zool. 38:342–349.
- Olsen SJ. 1990. Fossil ancestry of the yak, its cultural significance and domestication in Tibet. Pro Acad Nat Sci Philadelphia. 142:73–100.
- Olsen SJ. 1991. Confused yak taxonomy and evidence of domestication. Illinois State Mus Sci Pap. 23:387–393.
- Osborn HF. 1990. The age of mammals in Europe, Asia and North America. New York (NY): The Macmillan Company.
- Qiu Q, Zhang GJ, Ma T, Qian WB, Wang JY, Ye ZQ, Cao CC, Hu QJ, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44:946–949.
- Ritz LR, Glowatzki-Mullis ML, MacHugh DE, Gaillard C. 2000. Phylogenetic analysis of the tribe Bovini using microsatellites. Anim Genet. 31:178–185.
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 101:11030–11035.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Tu ZC, Zhang YP, Qiu H. 1998. Mitochondrial DNA polymorphism and genetic diversity in Chinese yak. Acta Genetica Sin. 25:205–212.
- Tu ZC, Qiu H, Zhang YP. 2002. Polymorphism in mitochondrial DNA (mtDNA) of yak (Bos grunniens). Biochem Genet. 40:187–193. 10.1023/A:1015836209577
- Wall DA, Davis SK, Read BM. 1992. Phylogenetic relationships in the subfamily Bovinae (Mammalia: Artiodactyla) based on ribosomal DNA. J Mammal. 73:262–275.
- Ward TJ, Bielawski JP, Davis SK, Templeton JW, Den JN. 1999. Identification of domestic cattle hybrids in wild cattle and bison species: a general approach using mtDNA markers and the parametric bootstrap. Anim Conserv. 2:51–57. 10.1111/j.1469-1795.1999.tb00048.x
- Wiener G, Han JL, Long RJ. 2003. The yak. 2nd ed. Bangkok: Regional Office for Asia and the Pacific, Food and Agriculture Organization of the United Nations.
- Xie CX. 1985. The history of cattle and sheep raising in China. Beijing: China Agriculture Press.
- Xie YL, Li YX, Zhao XB, Zhang X, Li N, Xie Z, Liu HL, Li QF. 2010. Origins of the Chinese yak: evidence from maternal and paternal inheritance. Proceeding of the 4th International Conference Bioinformatics and Biomedical Engineering. Chengdu.
- Zhang CF, Xu LJ, Ji QM, Xin JW, Zhong JC. 2012. Genetic diversity and evolution relationship on mtDNA loop in Tibetan yaks. Acta Ecologica Sin. 32:1387–1395.
- Zhang RC. 1989. The Chinese Yak. Lanzhou: Gansu Science and Technology Press.
- Zhao SJ, Cheng ZH, Ji QM, Chai Z, Zhang CF, Xin JW, Zhong JC. 2011. Sequence analysis of mtDNA cox3 in Tibetan yaks. Sci Agri Sin. 44:4902–4910.
