ABSTRACT
This study aimed to develop a rapid and reliable method for identifying the species origin of the meat in commercial sausages using amplification patterns for mitochondrial DNA (mtDNA). Forty commercial sausages were purchased from retail markets and subjected to mtDNA analysis. Two mtDNA markers were used for amplifying the meat source DNA. To optimize the PCR conditions, gradient PCR reactions were carried out to determine the primer annealing temperatures, and real-time PCR was done to check the minimal amount of DNA solution and to examine the cross-reaction. PCR products were observed on the gels suggesting that DNA molecules may be useful in the identification of the meat source in processed sausages. A similarity search of the DNA sequences showed that they were from pig, chicken, and fish, as described on the product labels. The real-time PCR results showed that the PCR products were observed above 10 fg (0.01 pg)/μl concentrations in pig, chicken, and processed fish meat DNA. No significant amplification was found in cross-species. This PCR-based molecular method using mtDNA markers may provide useful information for food safety and traceability purposes by supplying molecular evidence for detecting and identifying the meat sources used in sausage production.
1. Introduction
Information on the species of source animal used in meat production and processed foods is very important to consumers. To supply credible evidence for food safety and traceability, meat product labels should contain information on all the components in the product, including the meat source. Recently, molecular methods have been developed for DNA extracted from various sources and used for many purposes (e.g. economic, social, religious reasons, scientific, or industrial) (Bataille et al. Citation1999; Kusama et al. Citation2004; Girish et al. Citation2005; El-Sayed et al. Citation2010; Ali et al. Citation2012).
To detect meat species in mixed samples and processed foods, numerous analytical methods have been developed based on protein and DNA analyses. Proteins can be denatured during production by high cooking temperatures, high pressure, and other processing technologies. They have low sequence diversity among closely related species. Although protein-based methods could identify the species of origin of raw meats, protein-based methods are often inadequate for complex food products. In addition, they are less sensitive in processed materials to discriminate closely related taxa, and/or recommended high cost (Hird et al. Citation2003; Ballin et al. Citation2009; Ballin Citation2010; Sentandreu & Sentandreu Citation2014; Hou et al. Citation2015). Recently, DNA is considered to be the most appropriate molecule for species identification in foods. DNA is relatively stable at high temperatures. Therefore, DNA can be analysed not only in fresh and frozen foods, but also in processed, degraded, or mixed conditions (Lenstra Citation2003; Rodríguez et al. Citation2004; Singh & Neelam Citation2011). DNA-based methods have been developed for identifying mammalian species, and include PCR-restriction fragment length polymorphism, species-specific PCR, probe hybridization, microarrays, and DNA sequencing (Matsunaga et al. Citation1999; Mafra et al. Citation2004; López-Andreo et al. Citation2005; Kesmen et al. Citation2007; Zhang et al. Citation2007; Jonker et al. Citation2008; Koh et al. Citation2011). Mitochondrial DNA (mtDNA) is a very abundant molecule in all cells. Generally, more than 100 molecules of mtDNA are found in an animal cell, except in specific cells, such as sperm and dormant resting cells. The sequence diversity of species is sufficient to discriminate between all species identified thus far (Zehner et al. Citation1998; Cai et al. Citation2007; Chen et al. Citation2012; Santos et al. Citation2012). DNA markers are considered powerful tools for determining the presence of source species after chemical and/or physical processing in industrial production and for supplying information on each species used in meat processing and production. The DNA technique is particularly useful for identifying the species in processed meat, which can often contain unidentifiable meat products from different species (Bottero et al. Citation2003; Rao & Hsieh Citation2007; Yarmand & Homayouni Citation2010; Şakalar et al. Citation2012).
The illegal use of foreign meat sources and mislabelling of products have led to very serious social problems and decreased the consumer's choice of processed meat products. This study aimed to develop an mtDNA marker system using sequencing analysis, origin-specific PCR (OS-PCR), and real-time PCR, and examine their potential to identify the source species used in the production of commercial sausages in Korea.
2. Materials and methods
2.1. Samples and DNA isolation
Forty-eight raw meat pieces from different animal sources (pig, chicken, and fish meat) were kindly provided by three different commercial sausage manufacturers. Forty different kinds of commercial sausages (thirty-two of cooked emulsion type and eight of smoked type) were purchased from retail markets in Gyeonggi-do and Jeju-do, South Korea. Certified genomic DNA samples of species of pig, chicken, cattle, and horse were kindly provided by researchers at the Subtropical Animal Experiment Station, National Institute of Animal Science, South Korea. DNA was extracted from the meat and sausage samples using standard techniques (Sambrook et al. Citation1989), with a slight modification. Briefly, about 1 g of sausage piece was excised and rinsed with 70% ethanol-sterile water twice prior to DNA extraction. Collagenase and trypsin-ethylenediaminetetraacetic acid were added for protein lysis before adding nuclei lysis solution containing proteinase-K. The isolated DNA was diluted to 50 ng/µl concentration and used for PCR analysis as a template. To prevent human contamination during the DNA preparation, three different experimenters participated in the DNA isolation and carried out the experiments in three different laboratories.
2.2. Polymerase chain reaction
To amplify the mtDNA fragments of the commercial sausage-derived DNA samples, universal primers and origin-specific primers were designed using mitochondrial genome sequences previously deposited in the database of the National Center for Biotechnology Information (NCBI): DQ356938 from Atlantic cod (Gadus morhua, Coulson et al. Citation2006), NC_000845 from domestic pig (Sus scrofa, Lin et al. Citation1999), and AP003317 from chicken (Gallus gallus, Nishibori et al. Citation2003). Information on the primers and standard sequences is shown in . The PCR reaction was carried out in 20 μl volumes with Maxime i-Star Taq Premix (Intron Biotechnology, South Korea) containing distilled water, 200 μM of dNTPs, 2.0 units of iStar Taq DNA polymerase, 10 mM of each primer, and about 100 ng of total DNA. PCR amplification was performed under the following conditions: an initial 2 min denaturation at 94° C, followed by 35 cycles of 30 sec at 94° C, 30 sec at 62° C, and 30 sec at 72° C. The PCR products were separated on 1% agarose gels containing ethidium bromide and purified using a QIAEX II Gel Extraction Kit (Qiagen, USA). In order to determine the optimal annealing temperature of each primer pair, gradient PCR was carried out using Mastercycler Gradient (Eppendorf, Germany). The DNA concentration for PCR amplification was also checked by serial dilution samples from the 100 ng (1 × 10–7 g)/µl to 10 ag (1 × 10–17 g )/µl of genomic DNA solutions by real-time PCR using Rotor-Gene Q (Qiagen, USA) with Rotor-Gene SYBR Green PCR Kit (Qiagen, USA) followed by manufacturer's guide. Cross-reaction test was conducted by conventional PCR, gradient PCR, and real-time PCR for other species (horse, cattle, sheep, turkey, duck, quail, and pheasant).
Table 1. Primer information used in this study.
2.3. DNA sequencing and data analysis
The purified PCR product amplified using a pair of universal primers was inserted into a plasmid vector using the TOPOTM TA Cloning Kit (Invitrogen, USA) according to the manufacturer's instructions. Fifteen colonies were chosen from each transformation reaction plate, and plasmid DNA was purified using the QIAprep Spin Miniprep Kit (Qiagen, USA). Nucleotide sequencing was carried out using the DYEnamic ET Dye Terminator Kit (Amersham Biosciences, USA). Clustal W (Larkin et al. Citation2007) was used for multiple alignments of the obtained sequences and DnaSP (Librado & Rozas Citation2009) was used to determine the nucleotide diversity. A BLAST search of the NCBI's nucleotide database was conducted, and the results were compared with the unique COI sequences.
3. Results and discussion
To identify the meat sources used for the processed meat products, we examined the PCR-mediated molecular identification using universal primers and OS primer sets. PCR products, approximately 190-bp length, were found on the agarose gels from all the sausage DNA samples when amplifying with a universal primer set. The DNA sequencing results revealed no variation in the size of the PCR products amplified from the sausage DNA. The DNA sequencing results revealed 26 different nucleotide sequences in the partial fragment of the mtDNA COI gene from 600 plasmid DNA samples. The COI sequences were classified into 17 haplotypes from the results of the sequence analysis using the Clustal W and DnaSP programmes. The results of the BLAST search showed that the 17 haplotype sequences originated from those of eight species previously reported in the database. The sequence identities of our COI sequences were very similar (98.1–100%) to those deposited in the NCBI's database. shows the multiple sequence alignments for the COI sequences of pig, chicken, cod, and pollock mostly found in the sausage DNA.
Figure 1. Multiple sequence alignments for COI sequences determined in this study. Majority of pigs have two types of DNA sequences (Pig01 and Pig02), but most chickens showed an identical sequence (chicken). Two types of fish meat sources were found from cod and pollock. The underlined sequences indicate the primer sequences for the universal primer set.

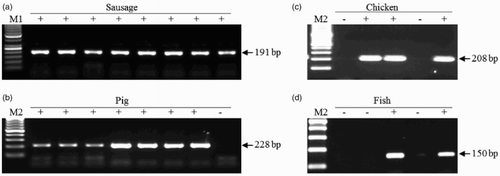
shows the PCR amplification patterns using universal primer set ((A)) and OS primer sets ((B)–(D)). PCR products were observed in all sausage DNA samples in PCR assay using a pair of universal primers ((A)). OS-PCR results showed species-specific amplification patterns corresponding to their animal species. The pig-specific PCR reaction produced 228-bp PCR bands on the gels only from the pig DNA samples but not from those of chicken DNA and fish meat DNA ((B)). In addition, the chicken- and fish meat-specific PCR reactions also produced the origin-specific amplicons (208-bp in chicken and 150-bp in fish), respectively ((C) and 2(D)).
Figure 2. PCR amplification patterns in sausage DNA samples. (A) A universal primer set 3sp_COI_F and 3sp_COI_R; (B) pig-specific Pig_CYTB_F and CYTB_R; (C) chicken-specific Gal_CYTB_F and Gal_CYTB_R; (D) fish-specific Fish_mtF and Fish_mtR. + and − indicate the presence and absence of the meat product on the labels of the sausages, respectively. M1 and M2 are DNA size markers, 50-bp and 100-bp ladder, respectively.
Specificity was evaluated by real-time PCR reactions, including DNA from pig, chicken, horse, cattle, sheep, turkey, duck and pheasant, processed fish meat and water (negative control). All cross-test reactions were amplified over a Ct value of 35 or had no detectable signals, indicating no PCR products were produced. Significant amplification signals were just observed from reactions with pig, chicken, and processed fish meat, while no amplification signals were detected with DNA from cattle, horse, sheep, goat, cattle, duck, and pheasant. Therefore, the newly developed OS primers had good specificity for pig, chicken, and fish meats. Sensitivity of OS primers was also evaluated by real-time PCR (). When the DNA amount of pig, chicken, and processed fish meat was 1 pg, the Ct values for each species were about 23.4, 24.3, and 27.6, respectively (Table S1). DNA samples from the three target meat sources (pig, chicken, and processed fish meat) were 10-fold serially diluted from 100 ng/µl to 10 ag/µl and used as templates for real-time PCR to construct standard curves. The significant amplification signals could be observed above 10 fg/µl (0.01 pg/µl) concentrations in pig and chicken and fish meat. Sausage DNA samples containing different percentages of pig, chicken, and fish were tested by the real-time PCR system. All meat mixtures yielded positive results in the real-time PCR, showing the same results from conventional OS-PCR reactions (). As shown in , the standard curves showed a linear correlation between Ct values and log-DNA concentrations. The correlation coefficients (R2) were 0.99970 (pig-specific primer set), 0.99939 (chicken-specific primer set), and 0.99827 (fish-specific primer set) ((D)–(F)), indicating significant linear relationships. Amplification efficiencies all exceeded 90%. Moreover, further electrophoresis analysis of real-time PCR products showed that there was no PCR product unexpected found from the sausage DNA samples (), and from artificial mixed DNA solutions of different species sources.
Figure 3. Real-time PCR amplification patterns of pig, chicken, and fish meat. (A–C) show amplification patterns for serial dilutions of pig (A), chicken (B), and fish meat (C). (D–F) show the standard curves for (A–C).
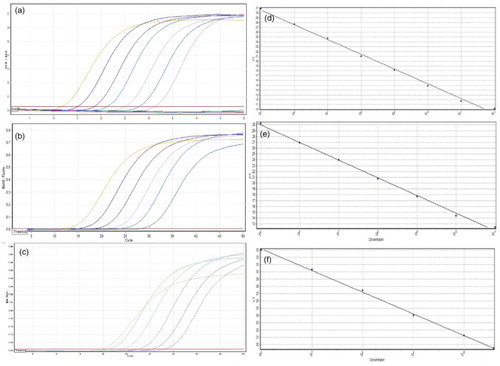
Figure 4. Electrophoresis analysis for real-time PCR products. (A) pig; (B) chicken; (C) fish meat. M is DNA size marker, 100-bp ladder. Numbers on top of the lanes indicate DNA concentrations in 1 μl solution used for PCR amplification.
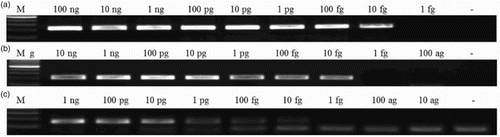
Table 2. Species identification of pig, chicken, and fish meat by DNA sequencing and PCR assays.
presents the species information listed on the sausage product labels and those identified by the similarity comparison of the COI sequences in this study. The sausage labels listed three different animal sources: pig, chicken, and fish meat. The information on the source animal on the product label of most of the sausages matched the source identified with the molecular data. The DNA sequencing results revealed no DNA from foreign mammalian species. In two DNA samples, the COI sequences corresponded to that of an unlabelled animal species (Table S2). It is likely that the unlisted species was introduced unintentionally by cross-contamination during the sausage production. The unlabelled product was identified as a fish species (Atlantic cod) by DNA sequencing. From the conventional OS-PCR and real-time PCR assays, we obtained molecular data similar to those from DNA sequencing and the records on the labels of the commercial sausages. PCR products derived from pigs and chickens were detected in 38 and 13 sausage DNA samples, respectively, which indicated the use of each animal source in the sausage production. We found PCR products derived from fish meat in eight sausage DNA samples. Six sausage samples listed the inclusion of fish on their labels. The other two did not contain any information on the inclusion of fish in the sausage production. However, the PCR bands from the fish-specific PCR assay confirmed the existence of fish-derived DNA in the sausages. Further analyses of DNA sequencing and a BLAST search showed 100% sequence identities to the sequences previously reported from Atlantic cod. As the presence of the PCR products points to the presence of DNA, we concluded that these two sausage samples contained fish-derived DNA molecules.
4. Conclusion
Species information in processed meats and processed foods is increasingly important for consumers around the world who wish to know the species of origin in processed meat products. Food manufacturers should record the meat origin on their labels of processed foods in all countries including South Korea. For religion purposes, meat species identification is especially important for Muslim and Hindu religious Halal authentication. Many molecular methods have been developed to better perform this task, including DNA extracted from various sample sources. In this study, we obtained good results with regard to the molecular identification of the species of origin in commercial sausages using a two-step approach based on universal and OS-PCR primers in conventional PCR and real-time PCR. Both the PCR results confirmed the DNA of the source animal and the species of origin in commercial sausages. We detected meat sources from animals not listed on the product labels in two samples. In addition, we could determine the minimal amounts of above 0.01 gp/ul concentrations by real-time PCR analysis for sausage DNA.
Consequently, we have developed a specific and sensitive molecular technique for detection and identification of meat components in processed food as well as raw meats. This PCR-based molecular method could be offered rapid and precise information on the origins of source animals used in the production of commercial sausages. These results suggest that this method may play an important role in quality control of meat products as providing the scientific evidence of the source species.
Supplemental data and research materials
Supplemental data for this article can be accessed at 10.1080/09712119.2015.1124334
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ali ME, Hashim U, Kashif M, Mustafa S, Che Man YB, Abd Hamid SB. 2012. Development of swine-specific DNA markers for biosensor-based halal authentication. Gen Mol Res. 11:1762–1772. doi:10.4238/2012.June.29.9
- Ballin NZ. 2010. Authentication of meat and meat products. Meat Sci. 86:577–587. doi:10.1016/j.meatsci.2010.06.001
- Ballin NZ, Vogensen FK, Karlsson AH. 2009. Species determination–can we detect and quantify meat adulteration? Meat Sci. 83:165–174. doi:10.1016/j.meatsci.2009.06.003
- Bataille M, Crainie K, Leterreux M, Durigon M, de Mazancourt P. 1999. Multiplex amplification of mitochondrial DNA for human and species identification in forensic evaluation. Forensic Sci Int. 99:165–170. doi:10.1016/S0379-0738(98)00185-6
- Bottero MT, Dalmasso IA, Nucera D, Turi RM, Rosati S, Squadrone S, Goria M, Civera T. 2003. Development of a PCR assay for the detection of animal tissues in ruminant feeds. J Food Prot. 66:2307–2312.
- Cai X, Chen H, Lei C, Wang S, Xue K, Zhang B. 2007. mtDNA diversity and genetic lineages of eighteen cattle breeds from Bos taurus and Bos indicus in China. Genetica. 131:175–183. doi:10.1007/s10709-006-9129-y
- Chen SY, Yao YG, Liu YP. 2012. Species identification of ten common farm animals based on mitochondrial 12S rRNA gene polymorphisms. Anim Biotechnol. 23:213–220. doi:10.1080/10495398.2012.696568
- Coulson MW, Marshall HD, Pepin P, Carr SM. 2006. Mitochondrial genomics of gadine fishes: implications for taxonomy and biogeographic origins from whole-genome data sets. Genome. 49:1115–1130. doi:10.1139/g06-083
- El-Sayed YS, Mohamed OI, Ashry KM, Abd El-Rahman SM. 2010. Using species-specific repeat and PCR-RFLP in typing of DNA derived from blood of human and animal species. Forensic Sci, Med, Pathol. 6:158–164. doi:10.1007/s12024-009-9117-5
- Girish PS, Anjaneyulu AS, Viswas KN, Shivakumar BM, Anand M, Patel M, Sharma B. 2005. Meat species identification by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of mitochondrial 12S rRNA gene. Meat Sci. 70:107–112. doi:10.1016/j.meatsci.2004.12.004
- Hird H, Goodier R, Hill M. 2003. Rapid detection of chicken and turkey in heated meat products using the polymerase chain reaction followed by amplicon visualisation with vistra green. Meat Sci. 65:1117–1123. doi:10.1016/S0309-1740(02)00341-8
- Hou B, Meng X, Zhang L, Guo J, Li S, Jin H. 2015. Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Sci. 101:90–94. doi:10.1016/j.meatsci.2014.11.007
- Jonker KM, Tilburg JJ, Hagele GH, de Boer E. 2008. Species identification in meat products using real-time PCR. Food Addit Contam. 25:527–533. doi:10.1080/02652030701584041
- Kesmen Z, Sahin F, Yetim H. 2007. PCR assay for the identification of animal species in cooked sausages. Meat Sci. 77:1–5. doi:10.1016/j.meatsci.2007.05.018
- Koh BRD, Kim JY, Na HM, Park SD, Kim YH. 2011. Development of species-specific multiplex PCR assays of mitochondrial 12S rRNA and 16S rRNA for the identification of animal species. Korean J Vet Service. 34:417–428. doi:10.7853/kjvs.2011.34.4.417
- Kusama T, Nomura T, Kadowaki K. 2004. Development of primers for detection of meat and bone meal in ruminant feed and identification of the animal of origin. J Food Prot. 67:1289–1292.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. doi:10.1093/bioinformatics/btm404
- Lenstra JA. 2003. DNA methods for identifying plant and animal species in food. In: Lees M, editor. Food authenticity and traceability. Cambridge: Woodhead Publishing Ltd; pp. 34–53.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. doi:10.1093/bioinformatics/btp187
- Lin CS, Sun YL, Liu CY, Yang PC, Chang LC, Cheng IC, Mao SJ, Huang MC. 1999. Complete nucleotide sequence of pig (Sus scrofa) mitochondrial genome and dating evolutionary divergence within Artiodactyla. Gene. 236:107–114. doi:10.1016/S0378-1119(99)00247-4
- López-Andreo M, Lugo L, Garrido-Pertierra A, Prieto MI, Puyet A. 2005. Identification and quantitation of species in complex DNA mixtures by real-time polymerase chain reaction. Anal Biochem. 339:73–82. doi:10.1016/j.ab.2004.11.045
- Mafra I, Ferreira IM, Faria MA, Oliveira BP. 2004. A novel approach to the quantification of bovine milk in ovine cheeses using a duplex polymerase chain reaction method. J Agric Food Chem. 52:4943–4947. doi:10.1021/jf049635y
- Matsunaga T, Chikuni K, Tanabe R, Muroya S, Shibata K, Yamada J, Shinmura Y. 1999. A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Sci. 51:143–148. doi:10.1016/S0309-1740(98)00112-0
- Nishibori M, Hanazono M, Yamamoto Y, Tsudzuki M, Yasue H. 2003. Complete nucleotide sequence of mitochondrial DNA in chickens, White Leghorn and White Plymouth Rock. Anim Sci J. 74:437–439. doi:10.1046/j.1344-3941.2003.00136.x
- Rao Q, Hsieh YH. 2007. Evaluation of a commercial lateral flow feed test for rapid detection of beef and sheep content in raw and cooked meats. Meat Sci. 76:489–494. doi:10.1016/j.meatsci.2006.12.011
- Rodríguez MA, García T, González I, Asensio L, Hernández PE, Martín R. 2004. PCR identification of beef, sheep, goat, and pork in raw and heat-treated meat mixtures. J Food Prot. 67:172–177.
- Şakalar E, Abasiyanik MF, Bektik E, Tayyrov A. 2012. Effect of heat processing on DNA quantification of meat species. J Food Sci. 77:N40–N44. doi:10.1111/j.1750-3841.2012.02853.x
- Sambrook J, Fritsch EF, Manniatis T. 1989. Isolation of high-molecular-weight DNA from mammalian cells. In: Molecular cloning: a laboratory manual, 2nd ed., New York: Cold Spring Harbor Laboratory Press. pp. 9.14–9.23.
- Santos CG, Melo VS, Amaral JS, Estevinho L, Oliveira MB, Mafra I. 2012. Identification of hare meat by a species-specific marker of mitochondrial origin. Meat Sci. 90:836–841. doi:10.1016/j.meatsci.2011.10.018
- Sentandreu MÁ, Sentandreu E. 2014. Authenticity of meat products: tools against fraud. Food Res Int. 60:19–29. doi:10.1016/j.foodres.2014.03.030
- Singh P, Neelam S. 2011. Meat species specifications to ensure the quality of meat: a review. Int J Meat Sci. 1:15–26. doi:10.3923/ijmeat.2011.15.26
- Yarmand MS, Homayouni A. 2010. Quality and microstructural changes in goat meat during heat treatment. Meat Sci. 86:451–455. doi:10.1016/j.meatsci.2010.05.033
- Zehner R, Zimmermann S, Mebs D. 1998. RFLP and sequence analysis of the cytochrome b gene of selected animals and man: methodology and forensic application. Int J Legal Med. 111:323–327. doi:10.1007/s004140050180
- Zhang C, Fowler MR, Scott NW, Lawson G, Slater A. 2007. A TaqMan real-time PCR system for the identification and quantification of bovine DNA in meals, milks and cheeses. Food Control. 18:1149–1158. doi:10.1016/j.foodcont.2006.07.018
