ABSTRACT
Polymorphisms in the prion protein gene (PRNP) have been linked with the occurrence of transmissible spongiform encephalopathies and the expression of some phenotypic traits in healthy animals including cattle. The aim of this study was to verify the 23-bp insertion/deletion (indel) within promoter region and the 12-bp indel within intron 1 of PRNP, as well as to evaluate their associations with growth traits. Two loci of PRNP were genotyped in 1558 healthy Chinese cattle sampled from 6 indigenous breeds. Moreover, phenotypic records for growth were used to find the relationship between PRNP polymorphisms and phenotypic performance. The results confirmed the occurrence of these two indel polymorphisms in the analysed breeds. Association analysis showed that the 23-bp indel was significantly related to the body length and heart girth in 18-months-old Nanyang cattle. The 12-bp indel was significantly related to the growth traits in three cattle breeds, such as the body weight in Xia'nan cattle, the daily gained weight in 12-months-old Nanyang cattle and the cannon circumference and rump length in Ji'an cattle. These findings indicated that these two indels may affect bovine growth traits, which could benefit to healthy cattle selection and breeding through marker-assisted selection.
Abbreviations AI: artificial insemination; ANOVA: analysis of variance; Bp: base pair; BSE: bovine spongiform encephalopathy; D: deletion; DD: deletion/deletion; GLM: generalized linear model; reaction; He: heterozygosity; Ho: homozygosity; HWE: Hardy–Weinberg equilibrium; I: insertion; ID: insertion/deletion; II: insertion/insertion; Indel: insertion/deletion; MAS: marker-assisted selection; Ne: effective allele numbers; PCR: polymerase chain reaction; PIC: polymorphism information content; PrP: Prion protein (PrP); PRNP: the prion protein; RP58: repressor protein with a predicted molecular mass of 58 kDa; SNP: single nucleotide polymorphism; Sp1: specific protein 1; SPSS: statistical product and service solutions; TSE: transmissible spongiform encephalopathies
Introduction
The prion protein (PrP) which is encoded by prion protein gene (PRNP) plays an important role in the transmissible spongiform encephalopathies which include the bovine spongiform encephalopathy (BSE) (Zhu et al. Citation2011; Zhao et al. Citation2015), caprine (Curcio et al. Citation2016) and ovine scrapie (Dassanayake et al. Citation2013; Li et al. Citation2017). The characteristic of TSE is the accumulation of PrPsc (pathological protease-resistant isoform), which is generated as a result of post-translational modification and changed in the stereochemical conformation of host-encoded, cellular prion proteins (PrPc) (Prusiner Citation1998). According to reports, the polymorphisms of PRNP have been shown to influence incubation and the susceptibility of these diseases such as BSE and the incubation period in humans (Shibuya et al. Citation1998; Shi et al. Citation2016), mice (Moore et al. Citation1998; Aguilar-Calvo et al. Citation2014), goats (Vitale et al. Citation2016), sheep (Stepanek and Horin Citation2017) and buffaloes (Yaman et al. Citation2017).
In cattle, various PRNP polymorphisms had been reported, including a 23-bp insertion/deletion (indel) in the promoter region (Sander et al. Citation2004), a 12-bp indel in intron 1 (Nakamitsu et al. Citation2006), a 14-bp indel in 3′-untranslated region (Hills et al. Citation2001), the number of octapeptide repeats in the open reading frame (Goldmann et al. Citation1991) and single nucleotide polymorphisms (Sander et al. Citation2005; Abe et al. Citation2006; Kues et al. Citation2006; Uchida et al. Citation2014). To date, frequencies of above-mentioned polymorphisms have been reported in cattle in USA (Seabury et al. Citation2004; Heaton et al. Citation2008), Polish Black-and-White cattle artificial insemination (AI) sires (Walawski and Czarnik Citation2003), Japanese brown cattle (Nakamura et al. Citation2007; Msalya et al. Citation2009), Asian native cattle (Shimogiri et al. Citation2010), Iranian Holstein cattle (Roshanfekr and Farid Citation2013), BSE affected cattle (Sander et al. Citation2004; Gurgul et al. Citation2012) and others. In China, polymorphisms of PRNP have never been reported in Xia'nan, Pi'nan, Ji'an and Qinchuan indigenous cattle among others.
In view of the previous reports, the polymorphisms of PRNP were mostly found to linked to BSE (Sander et al. Citation2005), in addition, some of them were also detected to be associated with some phenotypic traits in cattle (Rzewucka et al. Citation2013; Yang et al. Citation2016), sheep (Brandsma et al. Citation2004; Isler et al. Citation2006; Ioannides et al. Citation2009) and goats (Lan, Zhao, Li, et al. Citation2012; Lan, Zhao, Wu, et al. Citation2012). In detail, the prion octapeptide-repeat polymorphisms were reported to significantly affect milk traits in Latxa dairy sheep (Vitezica et al. Citation2013); In Texel sheep, the PrP genotypes were significantly associated with the litter size and weight at 135 days (Brandsma et al. Citation2004); In the ovine PRNP, the VRQ and ARQ allelic variants (locating on codons 136, 154 and 171) were associated with high scrapie susceptibility, which also led to a significant production loss (Ioannides et al. Citation2009); The 28-bp indel within the promoter region in goat PRNP was associated with production traits in Chinese native breeds (Lan, Zhao, Li, et al. Citation2012). Briefly, from the published literature, it has been shown that the 23-bp indel within the promote region and the 12-bp indel within intron 1 affected the binding sites of the transcription factor RP58 (repressor protein with a predicted molecular mass of 58 kDa) and Sp1 (specific protein 1), respectively, thus affecting the expression of PRNP or other traits (Sander et al. Citation2005; Xue et al. Citation2008). Therefore, the 23-bp indel and the 12-bp indel may be related to the phenotypic traits in cattle. However, the association between the polymorphisms of PRNP in Chinese indigenous cattle breeds and the growth traits is still limited.
Therefore, the aim of this study was to identify the 23-bp indel and the 12-bp indel of PRNP in healthy individuals of different breeds, as well as to analyse the relationship between these indels and growth traits of healthy cattle, which could benefit for healthy cattle selection and breeding through marker-assisted selection (MAS).
Material and methods
All animal experiments were conducted in accordance to the relevant laws and institutional guidelines and were approved by Northwest A&F University Institutional Animal Care and Use Committee.
Animals and growth traits
A total of 1558 healthy Chinese cattle, belonging to 6 healthy Chinese indigenous cattle breeds (Qinchuan (QC, n = 217) in Shaanxi province; Xia'nan (XN, n = 195), Pi'nan (PN, n = 250), Nanyang (NY, n = 252), Jiaxian (JX, n = 409) in Henan province; Ji'an (JA, n = 235) in Jiangxi province) were randomly sampled and included in the this study. Xia'nan and Pi'nan cattle are the crossbreeding beef cattle breeds in China, instead, Qinchuan, Nanyang, Jiaxian and Ji'an cattle are Chinese indigenous meat breeds. The growth traits in unrelated Nanyang cattle were recorded at 6, 12, 18 and 24 months, including body weight (kg), body height (cm), body length (cm), heart girth (cm), hucklebone width (cm) and average daily weight gain (kg) (Pan et al. Citation2013; Zhang et al. Citation2015). Records of growth traits in unrelated adult Qinchuan and Jiaxian cattle were also measured at similar intervals including body weight (kg), body height (cm), body length (cm), chest girth (cm), chest width (cm), chest depth (cm), cannon circumference (cm), rump length (cm), height at hip cross (cm), hucklebone width (cm), hip width (cm) and back height (cm) (Zhang et al. Citation2015). Meanwhile, growth traits in unrelated adult Ji'an, Xia'nan and Pi'nan cattle were recorded at similar intervals, which included body weight (kg), body height (cm), body length (cm), chest girth (cm), hucklebone width (cm) and the average daily weight gain (kg) (Pan et al. Citation2013; Zhang et al. Citation2015).
Genomic DNA was isolated from whole blood or ear tissues following the procedure described by Lan et al. (Citation2007) and Pan et al. (Citation2013). The concentration of genomic DNA was quantified and the working solution of each DNA samples was 50 ng/μL (Zhang et al. Citation2015).
PCR conditions and genotyping the 23-bp indel and 12-bp indel within PRNP
Based on bovine PRNP gene sequence (AC 000170.1), two indels were identified using the following known primers (23-bp-indel-foward: 5′-GTGCCAGCCATGTAAGTG-3′; 23-bp-indel-reverse: 5′-CCTATTCTGGCTATTGTTGC-3′; 12-bp-indel-forward: 5′-CTCGCAGAAGCAGGTAAATAGCC-3′; 12-bp-indel-reverse: 5′-CTAGATTCCTACACACCAC-3′) (Msalya et al. Citation2009). The 25 µL polymerase chain reaction (PCR) amplification volume contained 50 ng of genomic DNA, 0.5 µmol/L of each primer, 1 × buffer (including 1.5 mmol/L MgCl2), 200 µmol/L dNTPs (dATP, dTTP, dCTP and dGTP) and 0.625 units of Taq DNA polymerase (MBI, Vilnius, Lithuania). Touch-down PCR protocol was as follows: 5 min at 95°C; 2 cycles of 94°C for 30 s, annealing from 68°C to 52°by 2°C decrease for 30 s respectively, 72°C for 30 s; 30 cycles of 94°C for 30 s, 50°C annealing for 30 s, 72°C for 30 s; a final extension at 72°C for 10 min and subsequently cooling to 4°C. The PCR products were detected by electrophoresis in 3.5% agarose gel stained with ethidium bromide at a constant voltage (120 V) for 80 min. Then, the genotypes of the individuals were observed.
Statistical analysis
Using the SHEsis program (http://analysis.bio-x.cn), the data were analysed, including genotypic frequencies, allelic frequencies, Hardy–Weinberg equilibrium (HWE), linkage disequilibrium (LD) structure and haplotypes of two indel loci in all studied breeds. When the data were input, the platform can estimate haplotype frequency individually in controls and in samples to give the results of both single haplotype and a global data automatically. Haplotypes with low frequencies could be collected together to give one result in the case-control analysis (Shi and Liu Citation2005).
The software program SPSS software (version 18.0) (International Business Machines [IBM] Corporation, New York, USA) was used to analyse the relationship between the different genotypes and the growth traits in the cattle breeds (He et al. Citation2014). All individuals were female adults and each fed the same nutritional diet in their farm. The basic linear model was used to evaluate the relationships between genotypes and growth-related traits for each individual as follows: Yijk = μ+αi+βj+εijk, where Yijk = the observation of the growth trait (body weight, etc.) evaluated on the ith level of the fixed factor age (αi), the jth level of the fixed factor genotype (βj); where μ = the overall mean for each trait and εijk is the random error for the ijkth individual. The differences were determined using analysis of variance, which was followed by the least significant difference test. Differences between the means were considered significant at P < .05.
Results
Genotypic and allelic frequencies of 23-bp indel and 12-bp indel within bovine PRNP gene
The 23-bp indel within the promoter region of PRNP was confirmed, and three genotypes were detected (). The genotypic and allelic frequencies of 23-bp indel in all analysed breeds were listed in . The frequencies of allele deletion (D) gene ranged from 0.268 to 0.696 among the six cattle breeds. Moreover, the χ2 test showed that the 23-bp indel was in the HWE (P > .05) in the Xia'nan, Jiaxian, Ji'an and Pi'nan cattle breeds but not Qinchuan breed.
Figure 1. The structure of bovine PRNP gene, the different genotypes of two indel loci and their associations with growth traits.
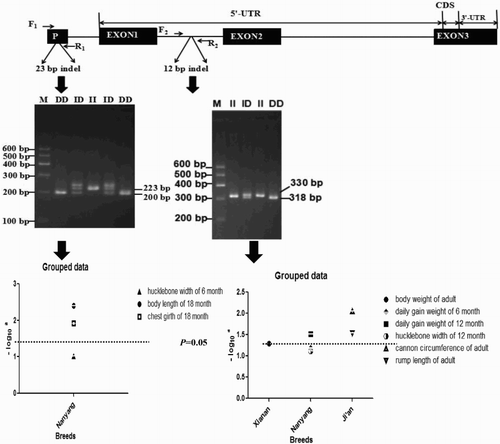
Table 1. Genotypic and allelic frequencies and population indexes of 23-bp indel for bovine PRNP.
The 12-bp indel within the intron 1 of PRNP was detected in six cattle breeds (). The allelic and genotypic frequencies of 12-bp indel were listed in . The allele insertion (I) frequencies ranged from 0.557 to 0.739 in the six cattle breeds, and the χ2 tests showed that the 12-bp indel of PRNP was in the HWE (P > .05) in Qinchuan, Nanyang, Jiaxian and Ji'an breeds except Xia'nan and Pi'nan breeds.
Table 2. Genotypic and allelic frequencies and population indexes of 12-bp indel for bovine PRNP.
The haplotypes frequencies of 23-bp indel and 12-bp indel within bovine PRNP gene
In this study, two loci in PRNP gene were analysed and assembled into haplotypes. Four common detected haplotypes and their frequencies were listed in . The frequencies of haplotype insertion/deletion (ID) were lowest in all analysed breeds except Ji'an cattle, with frequencies ranged from 0.024 (Nanyang) to 0.145 (Pi'nan). The frequencies of haplotype insertion/insertion (II) were the minimum in Ji'an cattle breed, and the frequency was 0.079. According to the analyses of LD, the D′ and r2 value between these two indel loci in the studied breeds were shown in . However, no strong LD between these two indel loci was detected in all studied breeds.
Table 3. Haplotype frequency within PRNP in cattle breeds.
Table 4. D′ and r2 values of pairwise LD of PRNP in cattle breeds.
Associations between 23-bp indel and 12-bp indel of bovine PRNP and growth traits
In the six cattle breeds, the associations between the 23-bp indel and growth traits were also analysed (). A significant relationship was observed between this indel locus and body length and heart girth in 18-month-old Nanyang cattle (P = .004 and P = .012, respectively), and the homozygous deletion genotype was predominant in this breed.
Table 5. Relationship between the two indel loci of PRNP and growth traits in three cattle breeds (LSMa ± SE) (P < .05).
In addition, the associations between the 12-bp indel and growth traits were investigated (). The Xia'nan cattles with II genotype showed heavier body weight than those with ID genotype (P = .051). In the 12-month-old Nanyang cattle group, the II homozygote at this 12-bp indel locus had heavier daily gain weight (P = .031). Moreover, in the Ji'an cattle group, deletion/deletion (DD) genotype cattle at the 12-bp indel locus had longer cannon circumference than those with ID and II genotypes (P = .009), instead, ID genotype was associated with a longer rump length (P = .029).
Discussion
Herein, we firstly confirmed the existence of 23-bp indel within the promoter region and 12-bp indel within intron 1 of PRNP in six Chinese indigenous cattle breeds: Qinchuan, Xia'nan, Pi'nan, Nanyang, Jiaxian and Ji'an.
For 23-bp indel locus, according to the reported data (), it was found that the frequencies of allele ‘D’ at a 23-bp indel locus was higher than that of allele ‘I’ in several cattle breeds worldwide (Nakamura et al. Citation2007), and a similar tendency was confirmed in Xia'nan, Pi'nan and Ji'an cattle breeds. Conversely, in three Chinese indigenous breeds (Qinchuan, Nanyang and Jiaxian), the allele ‘I’ frequency was higher than allele ‘D’ frequency. For 12-bp indel locus, the allele ‘I’ frequency was higher than allele ‘D’ frequency in all studied breeds population, which was in contradiction to the previous reported results in US sires, Mongolia native and other cattle breeds population (Seabury et al. Citation2004; Shimogiri et al. Citation2010). Notably, allele ‘I’ at these two locus was higher in Chinese indigenous breeds, and previous reports revealed that the 12-bp insertion and 23-bp insertion polymorphisms all increased the resistance to BSE (Sander et al. Citation2005; Juling et al. Citation2006; Haase et al. Citation2007; Xue et al. Citation2008; Msalya et al. Citation2010). Moreover, to date, occurrences of BSE have never been detected in China. Therefore, considering this background, this situation maybe that Chinese indigenous breeds have great resistance to disease and possess better adaptability to the unfavourable conditions, thus increasing resistance to BSE.
At present, selection for resistant PRNP genotypes and safety of animals are considered possible strategies for avoiding the prion diseases. On the other hand, the previous studies had reported that the resistant PRNP and PRND polymorphisms had different effects on growth and reproduction traits in different breeds (Lan, Zhao, Li, et al. Citation2012; Ibrahim et al. Citation2015; Yang et al. Citation2016; Li et al. Citation2017). However, the detailed associations between the PRNP polymorphisms and other economical traits remain elusive in cattle. Therefore, in this study, the potential associations of these two indel polymorphisms with growth traits were analysed. The individuals with homozygous DD genotype was superior in 23-bp indel locus, as well as the individuals with homozygous II genotype was superior in 12-bp indel locus in Nanyang cattle breed and all studied cattle breeds have lower ID haplotype frequencies. According to the previous reports, a promoter with insertion at 23-bp locus of PRNP binds strongly to RP58, while the 23-bp deletion weakens this bonds (Sander et al. Citation2005), and the 12-bp indel insertion can enhance the high expression of PRNP by binding the Sp1. Therefore, the genotype DD at 23-bp indel locus and genotype II at 12-bp indel locus may enhance PRNP expression. Up to date, de Vries et al. (Citation2004) proved significant effects for the PrP genotype on both conformation score in Texel sheep and muscle mass score in German Black Headed Mutton sheep. In addition, Stella et al. (Citation2010) found that the PrPs control the release of TNF-α factor which involved in muscle differentiation and downstream signalling pathways that affect the longevity of cells in adult muscles in many species of mammals; Smith et al. (Citation2011) cited that the low level of PrP is associated with decreased muscle mass of rat. Isler et al. (Citation2006) showed a correlation between the variation in PrP genotypes and the longissimus muscle depth of Dorset × Romanov sheep. Moreover, it had identified that overexpression of PrPC and its antiapoptosis function in gastric cancer, suggesting that PrP(C) might play a role as an effective antiapoptotic protein through Bcl-2-dependent apoptotic pathways in gastric cancer cells (Liang et al. Citation2006). Therefore, these genotypes in these two indel loci of PRNP should be considered when selecting the healthy and excellent individuals.
Briefly, genetic variability of 23-bp indel within the promoter region and 12-bp indel within intron 1 of PRNP may have the significant effects on growth traits, suggesting that these indels should be considered for eliminating or selecting excellent individuals in MAS breeding in relation to growth traits in cattle.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abe T, Hasebe H, Kobayashi E. 2006. Frequencies of bovine PrP gene polymorphisms in Holstein and Japanese black bulls in Japan. Anim Sci J. 77:395–398. doi: 10.1111/j.1740-0929.2006.00364.x
- Aguilar-Calvo P, Espinosa JC, Pintado B, Gutiérrez-Adán A, Alamillo E, Miranda A, Prieto I, Bossers A, Andreoletti O, Torres JM. 2014. Role of the goat K222-PrP(C) polymorphic variant in prion infection resistance. J Virol. 88(5):2670–2676. doi: 10.1128/JVI.02074-13
- Brandsma JH, Janss LLG, Visscher AH. 2004. Association between PrP genotypes and littersize and 135 days weight in Texel sheep. Livest Prod Sci. 85:59–64. doi: 10.1016/S0301-6226(03)00116-7
- Curcio L, Sebastiani C, Di Lorenzo P, Lasagna E, Biagetti M. 2016. Review: a review on classical and atypical scrapie in caprine: prion protein gene polymorphisms and their role in the disease. Animal. 10(10):1585–1593. doi: 10.1017/S1751731116000653
- Dassanayake RP, Truscott TC, Özyiğit MÖ, Zhuang D, Schneider DA, O'Rourke KI. 2013. Accumulation profiles of PrP(Sc) in hemal nodes of naturally and experimentally scrapie-infected sheep. BMC Vet Res. 9:82. doi: 10.1186/1746-6148-9-82
- de Vries F, Borchers N, Hamann H, Drögemüller C, Reinecke S, Lüpping W, Distl O. 2004. Associations between the prion protein genotype and performance traits of meat breeds of sheep. Vet Rec. 155:140–143. doi: 10.1136/vr.155.5.140
- Goldmann W, Hunter N, Martin T, Dawson M, Hope J. 1991. Different forms of the bovine PrP gene have five or six copies of a short, G-C-rich element within the protein-coding exon. J Gen Virol. 72(1):201–204. doi: 10.1099/0022-1317-72-1-201
- Gurgul A, Urszula C, Larska M, Polak MP, Strychalski J, Słota E. 2012. Polymorphism of the prion protein gene (PRNP) in Polish cattle affected by classical bovine spongiform encephalopathy. Mol Biol Rep. 39(5):5211–5217. doi: 10.1007/s11033-011-1318-9
- Haase B, Doherr MG, Seuberlic T, Drogemuller C, Dolf G, Nicken P, Schiebel K, Ziegler U, Groschup MH, Zurbriggen A, Leeb T. 2007. PRNP promoter polymorphisms are associated with BSE susceptibility in Swiss and German cattle. BMC Genet. 8:15. doi: 10.1186/1471-2156-8-15
- Heaton MP, Keele JW, Harhay GP, Richt JA, Koohmaraie M, Wheeler TL, Shackelford SD, Casas E, King DA, Sonstegard TS, et al. 2008. Prevalence of the prion protein gene E211 K variant in US cattle. BMC Vet Res. 4:25. doi: 10.1186/1746-6148-4-25
- He H, Zhang HL, Li ZX, Liu Y, Liu XL. 2014. Expression, SNV identification, linkage disequilibrium, and combined genotype association analysis of the muscle-specific gene CSRP3 in Chinese cattle. Gene. 535:17–23. doi: 10.1016/j.gene.2013.11.014
- Hills D, Comincini S, Schlaepfer J, Dolf G, Ferretti L, Williams JL. 2001. Complete genomic sequence of the bovine prion gene (PRNP) and its polymorphism in its promoter region. Anim Genet. 32(4):231–232. doi: 10.1046/j.1365-2052.2001.0769a.x
- Ibrahim AHM. 2015. Variability of prion protein (PrP) gene and its association with productive performance in Barki lambs. J Am Sci. 11(2):89–96.
- Ioannides IM, Mavrogenis AP, Papachristoforou C. 2009. Analysis of PrP genotypes in relation to reproductive and production traits in Chios sheep. Livest Sci. 122(2–3):296–301. doi: 10.1016/j.livsci.2008.09.022
- Isler BJ, Freking BA, Thallman RM, Heaton MP, Leymaster KA. 2006. Evaluation of associations between prion haplotypes and growth, carcass, and meat quality traits in a Dorset × Romanov sheep population. J Am Sci. 84(4):783–788.
- Juling K, Schwarzenbacher H, Williams JL, Fries R. 2006. A major genetic component of BSE susceptibility. BMC Biol. 4:33. doi: 10.1186/1741-7007-4-33
- Kues WA, Ollhoff RD, Carnwath JW, de Souza FP, Madeira HM, Niemann H. 2006. High incidence of single nucleotide polymorphisms in the prion protein gene of native Brazilian Caracu cattle. J Anim Breed Genet. 123(5):326–330. doi: 10.1111/j.1439-0388.2006.00604.x
- Lan XY, Pan CY, Chen H, Zhang CL, Li JY, Zhao M, Lei CZ, Zhang AL, Zhang L. 2007. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Rumin Res. 73(1–3):8–12. doi: 10.1016/j.smallrumres.2006.10.009
- Lan XY, Zhao HY, Li ZJ, Li AM, Lei CZ, Chen H, Pan CY. 2012. A novel 28-bp insertion–deletion polymorphism within goat PRNP gene and its association with production traits in Chinese native breeds. Genome. 55(7):547–552. doi: 10.1139/g2012-040
- Lan XY, Zhao HY, Wu CY, Hu SR, Pan CY, Lei CZ, Chen H. 2012. Analysis of genetic variability at codon 42 within caprine prion protein gene in relation to production traits in Chinese domestic breeds. Mol Biol Rep. 39(4):4981–4988. doi: 10.1007/s11033-011-1294-0
- Liang J, Pan YL, Ning XX, Sun LJ, Lan M, Hong L, Du JP, Liu N, Liu CJ, Qiao TD, Fan DM. 2006. Overexpression of PrPC and its antiapoptosis function in gastric cancer. Tumor Biol. 27(2):84–91. doi: 10.1159/000092488
- Li J, Zhu XC, Ma L, Xu HW, Cao X, Luo RY, Chen H, Sun XZ, Cai Y, Lan XY. 2017. Detection of a new 20-bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion. 11(2):143–150. doi: 10.1080/19336896.2017.1300740
- Moore RC, Hope J, McBride PA, McConnell I, Selfridge J, Melton DW, Manson JC. 1998. Mice with gene targeted prion protein alterations show that PrP Sinc and Prni are congruent. Nat Genet. 18(2):118–125. doi: 10.1038/ng0298-118
- Msalya G, Shimogiri T, Nishitani K, Okamoto S, Kawabe K, Minesawa M, Maeda Y. 2010. Indels within promoter and intron 1 of bovine prion protein gene modulate the gene expression levels in the medulla oblongata of two Japanese cattle breeds. Anim Genet. 41(2):218–221. doi: 10.1111/j.1365-2052.2009.01983.x
- Msalya G, Shimogiri T, Okamoto S, Kawabe K, Minezawa M, Namikawa T, Maeda Y. 2009. Gene and haplotype polymorphisms of the prion gene (PRNP) in Japanese Brown Japanese native and Holstein cattle. Anim Sci J. 80(5):520–527. doi: 10.1111/j.1740-0929.2009.00669.x
- Nakamitsu S, Miyazawa T, Horiuchi M, Onoe S, Ohoba Y, Kitagawa H, Ishiguro N. 2006. Sequence variation of bovine prion protein gene in Japanese cattle (Holstein and Japanese Black). J Vet Med Sci. 68(1):27–33. doi: 10.1292/jvms.68.27
- Nakamura I, Xue G, Sakudo A, Saeki K, Matsumoto Y, Ikuta K, Onodera T. 2007. Novel single nucleotide polymorphisms in the specific protein 1 binding site of the bovine PRNP promoter in Japanese Black cattle: impairment of its promoter activity. Intervirology. 50(3):190–196. doi: 10.1159/000099217
- Pan CY, Wu CY, Jia WC, Xu Y, Hu SR, Lei CZ, Lan XY, Chen H. 2013. A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle. Gene. 531(2):398–402. doi: 10.1016/j.gene.2013.09.002
- Prusiner SB. 1998. Nobel lecture: prions. Proc Natl Acad Sci USA. 95(23):13363–13383. doi: 10.1073/pnas.95.23.13363
- Roshanfekr H, Farid H. 2013. Detection of haplotype in the bovine prion protein gene (PRNP) in Najdi, Sarabi and Iranian Holstein cattle breeds. Int J Agric Crop Sci. 5(16):1725–1729.
- Rzewucka-Wójcik E, Frost A, Jędrzejczak M, Zaborski D, Pilarczyk R, Szatkowska I, Wójcik J, Grzesiak W, Dybus A. 2013. The PRNP ins/del and octapeptide repeat polymorphisms in Jersey cattle and their associations with production traits. J Appl Anim Res. 41(2):244–248. doi: 10.1080/09712119.2012.742442
- Sander P, Hamann H, Drögemüller C, Kashkevich K, Leeb T. 2005. Bovine prion protein (PRNP): promoter polymorphisms modulated PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem. 280(45):37408–37414. doi: 10.1074/jbc.M506361200
- Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T. 2004. Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics. 5(1):19–25. doi: 10.1007/s10048-003-0171-y
- Seabury CM, Womack JE, Piedrahita J, Derr JN. 2004. Comparative PRNP genotyping of US cattle sires for potential association with BSE. Mamm Genome. 15(10):828–833. doi: 10.1007/s00335-004-2400-6
- Shibuya S, Higuchi J, Shin RW, Tateishi J, Kitamoto T. 1998. Codon 219 Lys allele of PRP is not found in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 43(6):826–828. doi: 10.1002/ana.410430618
- Shi YY, Liu HE. 2005. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15(2):97–98. doi: 10.1038/sj.cr.7290272
- Shimogiri T, Msalya G, Myint SL, Okamoto S, Kawabe K, Tanaka K, Mannen H, Minezawa M, Namikawa T, Amano T, et al. 2010. Allele distributions and frequencies of the six prion protein gene (PRNP) polymorphisms in Asian native cattle, Japanese breeds, and mythun (Bos frontalis). Biochem Genet. 48(9–10):829–839. doi: 10.1007/s10528-010-9364-x
- Shi Q, Zhou W, Chen C, Zhang BY, Xiao K, Wang Y, Dong XP. 2016. Rare E196A mutation in PRNP gene of 3 Chinese patients with Creutzfeldt-Jacob disease. Prion. 10(4):331–337. doi: 10.1080/19336896.2016.1190897
- Smith JD, Moylan JS, Hardin BJ, Chambers MA, Estus S, Telling GC, Reid MB. 2011. Prion protein expression and functional importance in skeletal muscle. Antioxid Redox Signal. 15(9):2465–2475. doi: 10.1089/ars.2011.3945
- Stella R, Massimino ML, Sandri M, Sorgato MC, Bertoli A. 2010. Cellular prion protein promotes regeneration of adult muscle tissue. Mol Cell Biol. 30(20):4864–4876. doi: 10.1128/MCB.01040-09
- Stepanek O, Horin P. 2017. Genetic diversity of the prion protein gene (PRNP) coding sequence in Czech sheep and evaluation of the national breeding programme for resistance to scrapie in the Czech Republic. J Appl Genet. 58(1):111–121. doi: 10.1007/s13353-016-0354-5
- Uchida L, Heriyanto A, Thongchai C, Hanh TT, Horiuchi M, Ishihara K, Tamura Y, Muramatsu Y. 2014. Genetic diversity in the prion protein gene (PRNP) of domestic cattle and water buffaloes in Vietnam, Indonesia and Thailand. J Vet Med Sci. 76(7):1001–1008. doi: 10.1292/jvms.13-0642
- Vitale M, Migliore S, La Giglia M, Alberti P, Di Marco Lo Presti V, Langeveld JP. 2016. Scrapie incidence and PRNP polymorphisms: rare small ruminant breeds of Sicily with TSE protecting genetic reservoirs. BMC Vet Res. 12(1):921. doi: 10.1186/s12917-016-0766-9
- Vitezica ZG, Beltran de Heredia I, Ugarte E. 2013. Analysis of association between the prion protein (PRNP) locus and milk traits in Latxa dairy sheep. J Dairy Sci. 96(9):6079–6083. doi: 10.3168/jds.2013-6570
- Walawski K, Czarnik U. 2003. Prion octapeptide-repeat polymorphism in Polish Black-and-White cattle. J Appl Genet. 44(2):191–195.
- Xue G, Sakudo A, Kim CK, Onodera T. 2008. Coordinate regulation of bovine prion protein gene promoter activity by two Sp1 binding site polymorphisms. Biochem Biophys Res Commun. 372(4):530–535. doi: 10.1016/j.bbrc.2008.05.085
- Yaman Y, Karadağ O, Ün C. 2017. Investigation of the prion protein gene (PRNP) polymorphisms in Anatolian, Murrah, and crossbred water buffaloes (Bubalus bubalis). Trop Anim Health Prod. 49(2):427–430. doi: 10.1007/s11250-016-1185-4
- Yang Q, Zhang SH, Liu LL, Cao XK, Lei CZ, Qi XL, Lin FP, Qu WD, Qi XS, Liu JM, et al. 2016. Application of mathematical expectation (ME) strategy for detecting low frequency mutations: an example for evaluating 14-bp insertion/deletion (indel) within the bovine PRNP gene. Prion. 10(5):409–419. doi: 10.1080/19336896.2016.1211593
- Zhang SH, Dang YL, Zhang QF, Qin QM, Lei CZ, Chen H, Lan XY. 2015. Tetra-primer amplification refractory mutation system PCR (T-ARMS-PCR) rapidly identified a critical missense mutation (P236T) of bovine ACADVL gene significantly affecting growth traits. Gene. 559(2):184–188. doi: 10.1016/j.gene.2015.01.043
- Zhao H, Du Y, Chen S, Qing L, Wang X, Huang J, Wu D, Zhang Y. 2015. The prion protein gene polymorphisms associated with bovine spongiform encephalopathy susceptibility differ significantly between cattle and buffalo. Infect Genet Evol. 36:531–538. doi: 10.1016/j.meegid.2015.08.031
- Zhu XY, Feng FY, Xue SY, Hou T, Liu HR. 2011. Bovine spongiform encephalopathy associated insertion/deletion polymorphisms of the prion protein gene in the four beef cattle breeds from North China. Genome. 54(10):805–811. doi: 10.1139/g11-043
