ABSTRACT
The prion protein is coded by the PRP gene which is active in the brain and several other tissues. The functions of normal PrPs are related to energy production, protein degradation and DNA replication. Prions are the essential factor for the transmissible spongiform encephalopathies (TSEs) in different mammalian species. The present study aimed to identify the nucleotide characterization of the PRP gene in six camel breeds reared in Egypt. Genomic DNA which was extracted from 80 camels belonging to six breeds reared in Egypt was used to amplify fragments at 725-bp using a specific primer for PRP gene. Alignment of Egyptian camel PRP nucleotide sequences with those of other mammalian species declared the similarity 100% between PRP sequences of Egyptian and Iranian camels and 99.7% similarity with English, German and Chinese camel populations. The nucleotide sequence of Egyptian camel was submitted to GenBank under the accession no.: MF685344. The comparison between PRP amino acids of Camelus species with other mammalian species showed two missing amino acids in Camelus and Lama glama species; Glycine at position 83 and Leucine at position 229 whereas there is a unique amino acid addition – Glycine at position 89 – in Camelus species.
1. Introduction
The prion proteins (PrPs) are small glycoproteins dominated by three alpha helices and normally found at the cell surface in most mammals (DeMarco and Daggett Citation2009). These proteins are mainly found in neuronal tissue but are also expressed in other tissues including lung, kidney, mammary glands, gastrointestinal mucosa, lymphoid cells and skeletal muscle (Smith et al. Citation2011). The functions of normal PrPs are correlated with energy production, calcium metabolism, protein degradation and DNA replication. However the abnormal prion converts normal proteins into infectious disease-producing proteins responsible for the ovine and caprine scrapie, the bovine spongiform encephalopathy and the human Creutzfeldt–Jakob diseases (Beringue et al. Citation2008; Sikorska and Liberski Citation2012 and Cassard et al. Citation2014).
Transmissible spongiform encephalopathies (TSE) is a group of neurodegenerative disorders infected different animal species. These unique disorders occur sporadically as a result of infectious or genetic factors (Tahmoorespur and Niaraki Citation2014). The pathogen – the prion – which is responsible for this disease is different from those for other diseases in the fact that it is devoid of nucleic acids (Schneider et al. Citation2008). Unique to these diseases is the prion which is a misfolded isoform of the prion protein. This pathogenic isoform can transmit disease among different hosts through the transforming of normal prion protein into the pathogenic scrapie-inducing form.
The Camelidae family divided into two genera: the Camelus and new world camels. The genus Camelus (old-world genus) consists of two species: Camelus dromedaries (one-humped camel) and the Camelus bactrianus (two-humped camel) (Tahmoorespur and Niaraki Citation2014). The habitat of one-humped camel or Arabian camel is the hot and dry climate regions such as North Africa, Ethiopia, the Near East and West Central Asia. Two-humped camels are more suited in colder weather like the cold deserts of southern areas of Soviet Union, Mongolia, East Central Asia and China (Wilson and Reeder Citation2005).
In Egypt, the number of camel reaches about 120 thousand head (SADS Citation2009; Faye Citation2015). This species is considered one of the main sources of milk and meat in different regions of Egypt including the Nile Valley and Delta a well as the desert regions where this species also used for some labours and transport. Six camel breeds are reared in Egypt; Baladi, Maghrabi, Fallahi, Sudany, Somali and Mowaled (Othman et al. Citation2017). Despite the important role of camel and its valuable contribution as a food source for many Egyptians, nothing is known about its susceptibility or resistance to the serious scrapie disease. Regarding this fact, the present work aimed to identify the nucleotide characterization of prion protein gene in Egyptian camel and comparing it with those of other camel populations and different mammalian species.
2. Material and methods
2.1. Blood samples and genomic DNA extraction
Blood samples were collected from 80 camels belonging to six breeds reared in Egypt: Baladi (19 animals), Sodany (17 animals), Somali (14 animals), Maghrabi (13 animals), Mowaled (9 animals) and Fallahi (8 animals). Genomic DNA was extracted from the whole blood according to the method described by Miller et al. (Citation1988) with minor modifications. Briefly, blood samples were mixed with cold 2x sucrose-triton and centrifuged at 5000 rpm for 15 min at 4°C. The nuclear pellet was suspended in lysis buffer, sodium dodecyl sulphate and proteinase K and incubated overnight in a shaking water bath at 37°C. Nucleic acids were extracted with a saturated NaCl solution. The DNA was picked up and washed in 70% ethanol. The DNA was dissolved in 1× TE buffer. The DNA concentration was determined using Nano Drop1000 Thermo Spectrophotometer and then diluted to a working concentration of 50 ng/μl.
2.2. Polymerase chain reaction (PCR) amplification of the PrP gene and sequencing
The PCR primers used to amplify the PrP gene were synthesized according to Hussain et al. (Citation2017). The PCR amplifications were conducted in 50 µl volume containing 5 µl of 10× buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 µM each primer, 1.5 U Taq DNA polymerase and 100 ng camel genomic DNA. The thermocycler conditions used for amplification were: initial denaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 45 s, annealing at 54°C for 45 s and extension at 72°C for 45 s, followed by final extension at 72°C for 10 min. Amplified PCR products were electrophoresed in 2% agarose gel stained with ethidium bromide and visualized under the Gel Documentation system. The amplified products were purified with a DNA purification kit (ExoSap-IT USB Corporation) according to the manufacturer’s instructions to remove residual primers and dNTPs. Sequencing was performed in Macrogen Incorporation (Seoul South Korea).
Forward primer: 5′ – ATCCTGGTTCTCTTTGTGGT – 3′
Reveres primer: 5′ – CCCACTATGAGGAAA ATGAG – 3′
2.3. Data analysis
Prion protein gene sequences of tested camels were aligned using the BioEdit software (Hall Citation1999) in order to identify the nucleotide variations among them. Neighbour joining (NJ) tree for PRP sequences of tested camel breeds and the phylogeny tree between our camels and other camel populations as well as some mammalian species were constructed using Mega version 5.0 software (Tamura et al. Citation2011).
3. Results and discussion
The prion protein is coded by the PRP gene which is active in the brain and several other tissues (Chen et al. Citation2003). The proposed functions of the prion protein are the transport of copper into cells, the protection of brain cells and neuronal survival and the formation of synapses (Collinge et al. Citation1994). Prions are considered the essential factor for the transmissible spongiform encephalopathies (TSEs) in different mammalian species like scrapie in small ruminants, spongiform encephalopathies in bovine (BSE) and Creutzfelt–Jakob disease in humans (CJUD) (Harrison et al. Citation2010). TSEs occurred as inherited sporadic or infectious diseases. The last form – infectious – is happened due to the lack of nucleic acids (Prusiner Citation1998).
TSE disease agent contains abnormal isoform of the prion protein which is named PRPSc. This unnatural isoform was supposed the principal component for causing the transmissible spongiform encephalopathies disease (Tahmoorespur and Niaraki Citation2014). Under the normal conditions, the central nervous system produces the usual PRP isoform PRPC which is encoded by the host. Under the unusual conditions and after the infection of prion, this normal isoform is transformed to disease isoform through a translation mechanism involved the formational modifications (Harrison et al. Citation2010).
Despite the essential role of prion protein in scrapie, its molecular structure is still poorly understood (Surewicz and Apostol Citation2011; Diaz-Espinoza and Soto Citation2012). The present study aimed to identify the nucleotide characterization of the PRP gene in six camel breeds reared in Egypt and comparing it with those of other camel populations as well as different mammalian species.
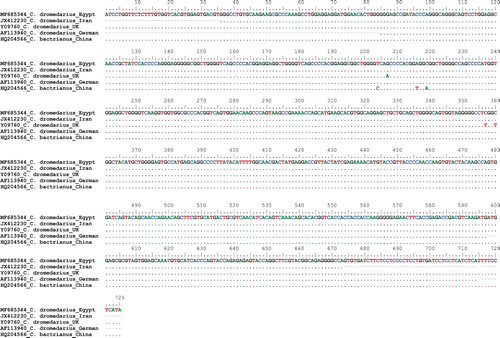
Genomic DNA which was extracted from 80 camels belonging to six breeds reared in Egypt was used to amplify fragments at 725-bp using a specific primer for PRP gene. The PCR products were purified and sequenced. The comparison of nucleotide sequences for tested 80 camels – using BioEdit software – showed that all tested camels have 100% similarity among them. The nucleotide sequence of Egyptian camel was submitted to GenBank under the accession no.: MF685344. Alignment of Egyptian camel PRP nucleotide sequence with those of other four different camel populations as well as some mammalian species () declared the similarity 100% between PRP sequences of Egyptian and Iranian camels. Also, the alignment results showed the presence of three nucleotide variations between Egyptian and each of other three (English German and Chinese) camel populations with 99.7% similarity (). The nucleotide variations were at positions 207, 357 and 360 in English camels and at 204, 216 and 219 in Chinese camel whereas there were three missing nucleotides in German camels at positions 598–600.
Table 1. Nucleotide difference counts and similarity percent of PRP sequences among different camel populations.
The comparison between PRP gene sequences of Camelus species with other mammalian species showed the high conservation of some nucleotides among Camelus species. With the exception of Lama glama species – which is closed taxonomically species to Camelus species – most of the other compared mammalian species presented conserved nucleotides different from those of Camelus species (). Searching among these nucleotides may help us to fishing out the nucleotide variations which make the camel more resistance against BSE disease comparing with other mammalian species.
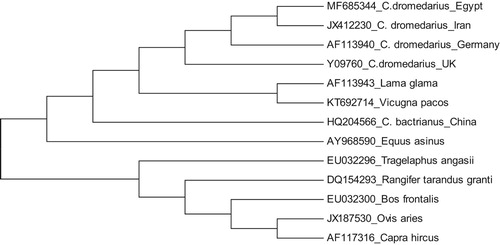
The phylogeny tree between PRP gene sequences of different camel populations including Egyptian camel and other mammalian species was done using Mega 5 software (). The tree declared that the PRP sequences are highly conserved between taxonomically related species. All dromedaries camels reared in Egypt, Iran, Germany and the UK are closely related to each other especially the Egyptian and Iranian camels which showed the complete similarity in PRP sequences. This finding supports the fact that the domestication of livestock including sheep, goat and camel began since around 8000 years in the Fertile Crescent and enter Egypt via Isthmus Suez and Sinai (Othman et al. Citation2016). The hypothesis of PRP sequence conservation between closely related species was declared from this Neighbour joining tree where for example the PRP sequences of sheep (Ovis aries) and goat (Capra hircus) are highly conserved.
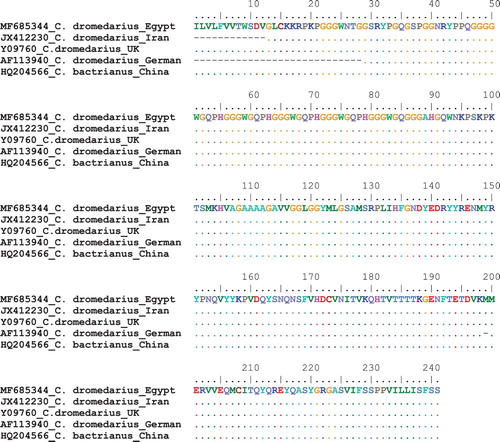
The deduced amino acid for PRP gene sequences for dromedaries camels; Egyptian, Iranian, German and England camels in addition of Chinese bactrianus camel were aligned together (). The result showed the 100% similarity between them in spite of the few DNA sequences among them as shown in . These variations in PRP sequences did not lead to the changes in the deduced amino acids. This finding declared the high conservation of PRP sequences and amino acids between different camel populations. The only variation was reported in German camels where there is a three nucleotide deletion ATG at positions 598–600 which leads to the missing of amino acid Methionine at position 199.
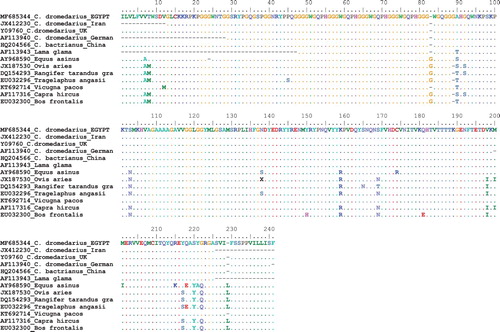
The comparison between PRP gene amino acid of Camelus species with other mammalian species showed some variations in amino acids between them (). The most valuable PRP amino acids variations between camels and other species were at positions 90, 92, 103, 159, 169, 198, 220 and 222. Two missing amino acids were found in Camelus and Lama glama species; Glycine at position 83 and Leucine at position 229 whereas there is a unique amino acid addition – Glycine at position 89 – in Camelus species which is missing in all other mammalian species compared in this study. These findings agree with the results of Tahmoorespur and Niaraki (Citation2014). These variations in nucleotide sequences and amino acid structure may be associated with resistance of camels against TSE because there is no any report about the presence of prion disease in camels (Kaluz et al. Citation1997).
In conclusion, the finding of this study showed the highly conserved nucleotide sequences of the PRP gene among Camelus species and the unique amino acid structure of camels. These genetic variations may be associated with camel resistance against TSE diseases which are infected other livestock like sheep goat and cattle.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Beringue V , Vilotte JL , Laude H. 2008. Prion agent diversity and species barrier. Vet Res. 39:47–76. doi: 10.1051/vetres:2008024
- Cassard H , Torres JM , Lacroux C , Douet JY , Benestad SL , Lantier F , Lugan S , Lantier I , Costes P , Aron N , et al. 2014. Evidence for zoonotic potential of ovine scrapie prions. Nat Commun. 5:5821–5829. doi: 10.1038/ncomms6821
- Chen SA , Mange A , Dong L , Lehmann S , Schachner M. 2003. Prion protein as trans-interacting partner for neurons is involved in neurite outgrowth and neuronal survival. Mol Cell Neurosci. 22:227–233. doi: 10.1016/S1044-7431(02)00014-3
- Collinge J , Whittington MA , Sidle KC , Smith CJ , Palmer MS , Clarke AR , Jefferys JG. 1994. Prion protein is necessary for normal synaptic function. Nature. 370:295–297. doi: 10.1038/370295a0
- DeMarco ML , Daggett V. 2009. Characterization of cell surface prion protein relative to its recombinant analogue: insights from molecular dynamics stimulations of diglycosylated, membrane-bound human prion protein. J Neurochem. 109:60–73. doi: 10.1111/j.1471-4159.2009.05892.x
- Diaz-Espinoza R , Soto C. 2012. High-resolution structure of infectious prion protein: the final frontier. Nat Struct Mol Biol. 19:370–377. doi: 10.1038/nsmb.2266
- Faye B. 2015. Role, distribution and perspective of camel breeding in the third millennium economies. Emir J Food Agric. 27(4):318–327. doi: 10.9755/ejfa.v27i4.19906
- Hall TA. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Harrison PM , Khachane A , Kumar M. 2010. Genomic assessment of the evolution of the prion protein gene family in vertebrates. Genome. 95:268–277. doi: 10.1016/j.ygeno.2010.02.008
- Hussain N , Johal H , Bhandari M. 2017. An evidence-based evaluation on the use of platelet rich plasma in orthopedics - a review of the literature. SICOT J. 3:57–63. doi: 10.1051/sicotj/2017036
- Kaluz S , Kaluzova M , Flint APF. 1997. Sequencing analysis of prion genes from red deer and camel. Gene. 199:283–286. doi: 10.1016/S0378-1119(97)00382-X
- Miller SA , Dykes DD , Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res. 16(3):1215. doi: 10.1093/nar/16.3.1215
- Othman OE , Abd El-Kader HA , Alam SS , Abd El-Aziem SH. 2017. Cytochrome b conservation between six camel breeds reared in Egypt. J Genet Engin Biotech. 15:1–6. doi: 10.1016/j.jgeb.2017.04.006
- Othman OE , Payet-Duprat N , Harkat S , Laoune A , Maftah A , Lafri M , Da Silva A. 2016. Sheep diversity of five Egyptian breeds: genetic proximity revealed between desert breeds. Local sheep breeds diversity in Egypt. Small Rumin Res. 144:346–352. doi: 10.1016/j.smallrumres.2016.10.020
- Prusiner SB. 1998. Prions. Proc Nat Acad Sci (USA). 95:13363–13383. doi: 10.1073/pnas.95.23.13363
- SADS . 2009. Sustainable Agricultural Development Strategy towards 2030. Agricultural Research and Development Council. Arab Republic of Egypt, Ministry of Agriculture and Land Reclamation. Oct. 2009.
- Schneider K , Fangerau H , Michaelsen B , Raab WH. 2008. Brain Res Bull. 77:343–355. doi: 10.1016/j.brainresbull.2008.09.012
- Sikorska B , Liberski PP. 2012. Human prion diseases: from Kuru to variant Creutzfeldt-Jakob disease. Subcell Biochem. 65:457–496. doi: 10.1007/978-94-007-5416-4_17
- Smith CB , Booth CJ , Pedersen JA. 2011. Fate of prions in soil: a review. J Environ Qual. 40:449–461. doi: 10.2134/jeq2010.0412
- Surewicz WK , Apostol MI. 2011. Prion protein and its conformational conversion: a structural perspective. Top Curr Chem. 305:135–167. doi: 10.1007/128_2011_165
- Tahmoorespur M , Niaraki SJ. 2014. Analysis of sequence variations of prion protein gene in dromedary camels in Iran. J Appl Anim Res. 42:238–243. doi: 10.1080/09712119.2013.842481
- Tamura K , Peterson D , Peterson N , Stecher G , Nei M , Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. doi: 10.1093/molbev/msr121
- Wilson DE , Reeder DM. 2005. Mammal species of the world. Washington : Smithsonian Institution Press.