ABSTRACT
Chemokine receptors play an important role in coordination of cell trafficking in many biological processes. In this study, a CC chemokine receptor 9 of cobia Rachycentron canadum (RcCCR9) was identified. Analysis of the ORF (1119 bp) of RcCCR9 revealed a predicted protein of 41.87 kDa with typical seven transmembrane domain architecture. RcCCR9 shared several conserved structural features with homologs from mammals and other fish, and had a consistent relationship with phylogenetic trees and sequence identities. Reverse transcription quantitative PCR analysis showed ubiquitous RcCCR9 transcripts in healthy cobia, mainly in immune-related organs, with the highest levels in blood and lower levels in intestines and brain. After challenge with inactivated Vibrio harveyi or viral mimic poly I:C, RcCCR9 expression was up-regulated in head kidney and down-regulated in spleen. Compared with poly I:C, V. harveyi induced a stronger up/down-regulation of CCR9 mRNA levels in the central immune organs. RcCCR9 seems to be strongly involved in host defense against bacterial infection.
Introduction
Chemokine receptors (CRs) are members of the seven-transmembrane domain superfamily of receptors and are coupled to guanine 5′-triphospate (GTP)-binding proteins (Rossi and Zlotnik Citation2003). The chemokine system is often thought as showing significant redundancy since one receptor can bind multiple ligands, and conversely, a single ligand can bind several CRs. Thus, these receptors are named using the prefixes CCR, CXCR, CX3CR, and XCR followed by an identifying number (Stone et al. Citation2017). But it is still unknown whether there is a specific group of receptors for CX chemokines. CRs, together with their ligands, play key roles in coordination of cell migration, morphogenesis, and cell activation (Chadzinska et al. Citation2014).
In mammals, 10 members of CCRs subfamily, including CCR1-10, have been identified with standard chemotactic functions (Griffith et al. Citation2014). There is increasing evidence that CCRs were associated with the pathogenesis of many diseases, and mutations in some CCRs could be beneficial for lowering disease impact and increasing the ability to antagonize some pathogens (Qidwai and Khan Citation2016). CCL25 was identified as a novel chemokine in the late 1990s, and is chemotactic for lymphocytes, dendritic cells (DCs), and activated macrophages (Vicari et al. Citation1997). Together with CCR9, the CCR9/CCL25 chemokine axis has been a focus of studies investigating its functions on lymphocytes and DCs in terms of maturation, gut-homing characteristics, and the maintenance of immunological tolerance (Wendland et al. Citation2007; Hadeiba et al. Citation2008). CCR9 has been detected in some carcinoma cells and may mediate the organ-selective metastasis (Johnson et al. Citation2010). Recent studies confirm that CCR9 serves as a novel modulator of pathological progression following myocardial infarction through NF-κB and mitogen-activated protein kinase signalling pathways (Huang et al. Citation2016).
Fish are an important group that is a bridge between those species with innate immunity only and ones with an imperfect, acquired immune network. Although CCR9 was well studied in mammals, information on the existence of this receptors in teleost fish is still limited with a few reports from rainbow trout Oncorhynchus mykiss (Dixon et al. Citation2013), miiuy croaker Miichthys miiuy (Zhu et al. Citation2013), European sea bass Dicentrarchus labrax L (Galindo-Villegas et al. Citation2013) and the orange-spotted grouper Epinephelus coioides (Yang et al. Citation2017). It is noteworthy that researchers have found that the CCL25-CCR9 interaction may play an important role in the teleost fish intestinal immunity, which was similar to that in mammals (Galindo-Villegas et al. Citation2013). In our previous study, a CC chemokine gene (RcCC3) homologous to mammalian CCL25 was identified in cobia and was shown to be associated with innate immune response (Su et al. Citation2013). Thus, obtaining its potential receptor could provide insight for better understanding the chemokine signalling in cobia as well as other teleost. In this study, a full-length cDNA of CCR9 (designated as RcCCR9) was cloned from cobia (Rachycentron canadum), a commercially important species in southern China. The mRNA expression profile of this gene was then examined in healthy and challenged specimen and its role in immune response examined.
Materials and methods
Fish collection and RNA extraction
Healthy cobia with an average body weight of 100 g were purchased from a fish farm in Xincun, Hainan province, China and kept in 1000-L tanks aerated sand filtered seawater (salinity 32 ± 1‰, pH 7.4 ± 0.4) at 30 ± 1°C. All fish were acclimatized for 2 weeks prior to experiments. Blood was collected from anaesthetized (MS-222, Sigma, USA) fish by cutting the tail and the blood cells were separated in two times the volume of anticoagulant solution (0.48% citric acid, 1.32% sodium citrate and 1.47% glucose) by centrifugation at 800 g for 5 min at 4°C and were stored in RNAlater (Ambion, USA) immediately. Heart, brain, intestine, stomach, kidney, gill, blood, spleen, liver, skin, head kidney and muscle were dissected out and stored in RNAlater at −20°C for RNA extraction. Each tissue was obtained from three independent healthy cobia. Total RNA was isolated using Trizol reagent (Invitrogen, USA) following the manufacturer’s protocol. Total RNA was incubated with RNase-free DNase I (Promega, USA) to remove any contaminating genomic DNA. First strand cDNA was synthesized from total RNA by M-MLV reverse transcriptase (Promega, USA), following the manufacturer's protocol with oligo(dT)- adaptor primer ().
Table 1. Primers used in this study.
Selection of a partial chemokine receptor sequence
The cDNA library was constructed earlier with the mixed head kidney and spleen tissues of cobia following vaccination against Vibrio harveyi, Proteus vulgaris and Streptococcus sp (Su et al. Citation2012). BLAST analysis revealed that an EST was similar to fish CCR9 gene. The EST was selected for further cloning of the novel chemokine receptor gene of cobia.
Cloning full-length cDNA for RcCCR9
On the basis of the identified EST sequence, specific primers () were designed to clone the full-length cDNA sequences by the rapid amplification of cDNA ends (RACE) approached with spleen cDNA as template. For 3′ RACE, the PCR reactions were done by semi-nested PCR with forward primers (CCR9-F1 and CCR9-F2), and reverse primer adaptor (). For 5′ RACE, the first-strand cDNA was tailed with poly(C) at the 5′ end using terminal deoxynucleotidyl transferase (Takara, Japan). PCR reactions were done by semi-nested PCR with forward primer oligo-dG and reverse primers (CCR9-R1 and CCR9-R2; ). Cycling parameters for RACE consisted of an initial denaturation period of 3 min at 94°C, followed by 35 cycles of 45 s at 94°C, 45 s at 60°C, 1 min at 72°C and, finally, a single step at 72°C for 10 min. The PCR products were purified with a QiaexII Gel Extraction Kit (Qiagen, Germany), ligated with PMD18-T vector (Takara, Japan), transformed into Escherichia coli TOP10 (Invitrogen, USA) and sequenced (BGI, China).
Sequence analysis
On the basis of the full-length cDNA sequence of RcCCR9, the nucleotide and deduced amino acid sequences were analyzed using BLAST at the NCBI (http://www.ncbi.nlm.nih.gov/blast/) and edited with DNASTAR 5.0 software. Protein analysis was performed using ExPASy online tools (http://us.expasy.org/tools). The transmembrane helices were predicted according to TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Multiple sequence alignment was done with the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Prediction of N-linked glycosylation sites was performed using NetNGlyc 1.0 (Gupta and Brunak Citation2002) while tyrosine sulfation sites were predicted using Sulfinator (Monigatti et al. Citation2002). An unrooted phylogenetic tree was constructed by the neighbor-joining method with MEGA 5 software. Confidence calculation was done by bootstrapping 1000 replications.
Immune challenge
To determine the effects of immune response on RcCCR9 expression, acclimatized fish were divided into three groups: an intraperitoneal (IP) injection of 0.2 mL of sterile phosphate-buffered saline alone (the PBS control group), IP injection of 0.2 mL of 10 mg/mL poly I:C (Sigma, USA) dissolved in PBS (the poly I:C group), or IP injection of 0.2 mL of formalin-inactivated V. harveyi suspended in PBS (1.0 × 108 colony-forming units/mL, the bacterial vaccine group). Bacterial antigen preparation, fish husbandry, IP injection and tissue sampling were as described (Su et al. Citation2012). For the PBS, poly I:C and bacterial vaccine groups, the head kidney and spleen tissues from three individuals were collected at 3, 6, 12, 24, 48 and 72 h post-injection. All samples were placed in 1.5 mL tubes containing RNAlater at −20°C for RNA extraction.
RT-qPCR analysis of RcCCR9 expression
Expression levels of RcCCR9 in different tissues of non-stimulated cobia and the head kidney and spleen tissues after immune challenge were identified by reverse transcription quantitative PCR (RT-qPCR) in a Mastercycler ep realplex Real-time PCR system (Eppendorf, Germany) with a Platinum SYBR Green qPCR SuperMix-UDG Kit (Invitrogen, USA). The house keeping gene β-actin was used as an internal control for cDNA normalization. Gene specific primers () β-actin F and β-actin F for β-actin, CCR9-qF and CCR9-qR for RcCCR9 were used to amplify specific fragment. The qPCR amplifications were done in a total volume of 20 μL containing 10 μL of 2 × Supermix, 1 μL of cDNA, 1 μL each of forward and reverse primer and 7 μL of PCR-grade water. Cycling conditions were as follows: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 61°C for 30 s. After the PCR program, data were analyzed with Mastercycler ep realplex software. Melting curve analysis of amplification products was done at the end of each PCR reaction to confirm that only one PCR product was amplified and detected. Samples were run in triplicate and the comparative C T method was used to analysis the expression level of RcCCR9. The expression levels were calculated by the 2−ΔΔCT method (Livak and Schmittgen Citation2001). Statistical analysis was done with SPSS software (version 19.0). Data are given as the mean ± standard error. Statistical significance was determined by one-way ANOVA. In all cases, significance was set at P < 0.05.
Results and discussion
Characteristics of RcCCR9
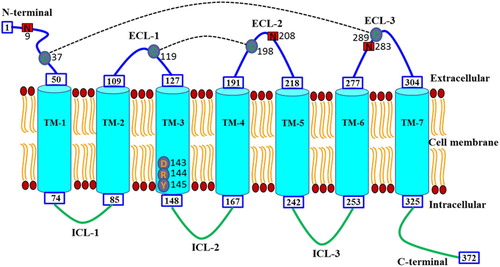
Base on the EST in the cDNA library of cobia, the full-length cDNA sequence of RcCCR9 was successfully obtained. The complete cDNA of RcCCR9 (GenBank accession number: KT748876) consisted of 1708 nucleotides, including 1119 bp ORF encoding a protein of 372 amino acids, a 5′-terminal untranslated region (UTR) of 54 bp, and a 3′ -terminal UTR of 534 bp, with a single polyadenylation site (1655AATAAA1660) (). The theoretical molecular weight of RcCCR9 was 41.87 kDa and isoelectric point was 8.57. Seven transmembrane (TM) regions including an extracellular amino-terminal domain (N-terminal), three extracellular loops (ECL), three intracellular loops (ICL) and a cytoplasmic carboxyl-terminal (C-terminal) were predicted in RcCCR9. A characteristically conserved DRY motif (143DRY145) was present in TM-3 (), which is part of one of the most conserved motifs in the seven-transmembrane domain receptors. This motif plays a pivotal role in receptor activation, being involved in the exchange of GDP with GTP and thus activating the G-protein for further downstream signalling (Mirzadegan et al. Citation2003; Oldham and Hamm Citation2008).
Figure 1. Nucleotides and the deduced amino acids sequences of RcCCR9. The start codon (ATG) and the stop codon (TAG) are marked in red. The polyadenlylation signal (AATAAA) is indicated in blue and the poly A is underlined with a horizontal line. The translation transmembrane domains are indicated in blue, and the conserved DRY motif is underlined with a red dotted line.

Figure 2. Diagrammatic representation of the domain structure and the topology of transmembrane regions in RcCCR9. The N-terminal, seven-transmembrane domains (TM1-7), three extracellular loops (ECL1-3), three intracellular loops (ICL1-3) and C-terminal are shown. The DRY motif in the TM-3 region is also indicated. Putative cysteine residues (C) involved in disulfide bonding are shown with a black dotted line. Potential N-glycosylation sites are marked in red shaded boxes.
To further characterize the newly identified cobia CCR9 molecule, a multiple alignment was constructed with selected CCR9 molecules from mammals (human and mouse) and other fish species. These CCR9 molecules are well conserved in the seven TM domains with sequences all possessing four cysteine residues (one in the N-terminal and three in the ECLs) that form two disulphide bonds, one from the N-terminal to ECL3 and one from ECL1 to ECL2 (). RcCCR9 shared a highly conserved DRY motif at the end of TM-3 as other CCR9s. Both the TM-3 region DRY motif and the seven TM domains are highly conserved in almost all organisms, which further emphasizes the evolutionary conservation of this receptor (Dixon et al. Citation2013; Zhu et al. Citation2013; Yang et al. Citation2017). The four conserved putative cysteine residues disulfide bridge played a crucial role in the conformational change of the receptor upon ligand binding in this receptor family. Previous studies had demonstrated that residues in ECLs (particularly the ECL-2) were critical for receptor activation (Ai and Liao Citation2002; Szpakowska et al. Citation2012). Similarly to grouper CCR9 (Yang et al. Citation2017), two additional cysteines were found in the intracellular C-terminal region of RcCCR9, and at least two additional cysteines were found in the same region of other piscine CCR9 genes (). It was indicated that the additional cysteine in the C-terminal region of CCR6 was crucial for receptor expression and functional activity (Ai and Liao Citation2002), but the concrete role for this receptor need to be further exploration.
Figure 3. Multiple alignment of the deduced amino acid sequences of CCR9. The multiple alignment was produced using ClustalW, and conserved amino acids shaded using BOXSHADE (version 3.21). The N-terminal, seven transmembrane domains (TM1-7), three extracellular loops (ECL1-3) and intracellular loops (ICL1-3) and the C-terminal are marked above the alignment. The four conserved cysteine residues in the extracellular domain are indicated by black arrows. The DRY motif is indicated by asterisk below the alignment. The potential N-glycosylation sites and conserved tyrosine residues in the N-terminal domain are shown with the red dotted lines and red lines, respectively. R.c (Rachycentron canadum), L.c (Larimichthys crocea), O.n (Oreochromis niloticus), P.o (Paralichthys olivaceus), S.s (Salmo salar), S.m (Scophthalmus maximus), M.m (Mus musculus) and H.s (Homo sapiens), the GenBank accession numbers are listed in .
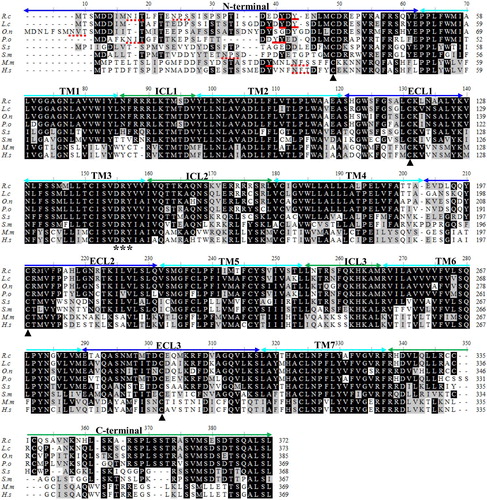
Potential N-glycosylation sites were identified in RcCCR9 at Asn9, Asn208 and Asn283 (). At least one potential N-linked glycosylation site or tyrosine residue is present at the N-terminal of these vertebrate CCR9 molecules but is missing in Salmo salar CCR9 (). There are conserved N-linked glycosylation sequences in the amino terminus of the mammalian sequences, while these are not conserved in the fish sequences, although most of the fish CCR9 sequences have putative glycosylation sequences that do not align with the mammalian ones or each other. There are approximately two tyrosine sulfation sites at the N-terminal of fish CCR9 proteins that show a degree of conservation (). Previous studies demonstrated that tyrosine residues just after the potential N-glycosylation site were important for these receptors ligands binding, internalization and scavenging (Dixon et al. Citation2013; Hewit et al. Citation2014).
Sequence identity and phylogenetic analysis
RcCCR9 deduced amino acid sequence showed a 58.20–83.02% identity with the sequences of other fish CCR9s, and shared a 40.0–41.69% identity with those of mammalian CCR9 molecules, whilst exhibiting lower amino acid sequence identities to reptile and bird CCR9 molecules (37.75–38.66%) (). A phylogenetic tree () constructed based on the amino acid sequence of the RcCCR9 with the other known vertebrate chemokine receptor sequences showed that all the CCR9 molecules form a clade separate to the other chemokine receptors. In the CCR9 clade, lineage-specific groups with over 70% bootstrap supports (i.e. mammals, birds, reptiles, amphibians and fishes) are apparent. The results showed that CCR9 genes independently evolved in aquatic and terrestrial organisms and the fish CCR9 molecules formed the corresponding group. RcCCR9 grouped with the common ancestor of Paralichthys olivaceus CCR9. The overall topology of the tree was consistent with traditional taxonomy and phylogenetic transition, showing that R. canadum had high similarity with other teleosts (Liu et al. Citation2009; Zhu et al. Citation2013; Grimholt et al. Citation2015; Yang et al. Citation2017). This result was similar with that of the amino acid identity analysis.
Figure 4. Phylogenetic analysis of partial vertebrate CCR9 and other chemokine receptors. A neighbor-joining tree was constructed using Mega 5.1 software. Numbers on the lines indicate the percentages of bootstrap values after 1000 replicates. RcCCR9 and atypical chemokine receptors are marked with black dot and triangles, respectively. The GenBank accession numbers are listed in Table S1.
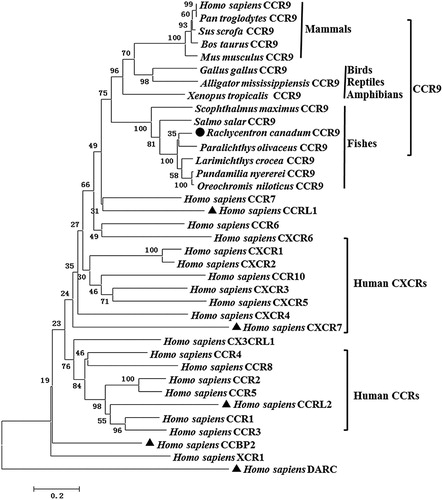
Table 2. Amino acid identity of RcCCR9 with partial vertebrate CCR9.
Tissue distribution of the RcCCR9
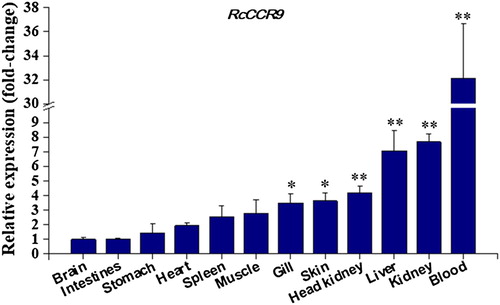
The relative expression levels of the RcCCR9 were examined in twelve tissues from three healthy fish by RT-qPCR (). The expression of the receptor was detectable in all tissues examined, although these transcripts ranged across different types of tissues, indicating that the ubiquitous expression pattern of CCR9 in cobia. The results showed that the RcCCR9 mRNA was expressed strongly in blood, highly in kidney, liver, head kidney, skin, and gill, but only weakly in muscle, spleen, heart, stomach, intestines and brain. Especially, the mRNA expression of RcCCR9 in blood (the tissue with the highest expression) was 32.2-fold greater than its expression in brain (the tissue with the lowest transcript expression). A number of previous studies demonstrated that CCR9 was more highly expressed in the head kidney and gill in grouper (Yang et al. Citation2017) and Atlantic salmon (Grimholt et al. Citation2015) than in non-immune tissues; it was also more highly expressed in the blood in rainbow trout (Dixon et al. Citation2013). These results indicate that piscine CCR9 were predominantly expressed in hematopoietic and/or immune-related tissues, such as the blood, head kidney, kidney, and gill. In mammals, CCR9 was mainly distributed in immature T lymphocytes and on the surface of intestinal cells (Tu et al. Citation2016). Recent studies have provided important data regarding the high CCR9 mRNA expression levels were detected in the thymus of grouper (Yang et al. Citation2017) and rainbow trout (Dixon et al. Citation2013), suggesting a conserved regulatory role of the receptor in T cell development. It is, however, worth noting that RcCCR9 exhibited a different tissue-specific expression profile with its potential ligand, RcCC3. Just as grouper, CCL25 and CCR9 were identified without a consistent expression profile (Yang et al. Citation2017). Therefore, it is well worth further exploring the functional reasons behind these differences in CCR9 and its specific ligand expression patterns in fish.
Figure 5. Constitutive expression of cobia RcCCR9 in twelve tissues from three healthy fish was determined by quantitative real-time RT-PCR. The mRNA expression levels were normalized to the endogenous control gene, β-actin. Data were expressed as a ratio to RcCCR9 mRNA expression in the brain. Each vertical bar represented the mean ± SE (N = 3). Significant differences in RcCCR9 expression between the brain and other tissues are indicated with an asterisk (*) for P < 0.05 or with two asterisks (**) for P < 0.01.
Expression analysis of RcCCR9 upon induction with V. harveyi and poly I:C
Mammalian CCR9 are involved in regulating the trafficking of developing T cells in the thymus and in the homing of CCR9 + memory T cells to the small intestine, most of which have been demonstrated in human and mouse (Papadakis et al. Citation2000; Wendland et al. Citation2007). However, it is important to understand the post-immune responses of CCR9 in the central immune organs (head kidney and spleen) of cobia following stimulants that are immunologically important in order to develop effective disease management strategies. After inactivated V. harveyi challenge (a), RcCCR9 expression in head kidney significantly increased from stimulation starting time to 6 h post-induction and reached the highest point, and then the expression decreased gradually to a low level in 24 h, followed by a rapid significantly increase from 48 to 72 h. In contrast, the expression in spleen remained stably from 0 to 3 h, and then the expression decreased gradually to a very low level in 24 h, followed by a rapid increase from 48 to 72 h. The expression was down-regulated and maintained significantly lower level than normal from 12 to 72 h (b). Overall, the administration of inactivated V. harveyi increased the cobia CCR9 transcripts in the head kidney, while suppressing the transcription in the spleen. Similar results have also been showed that CCR9 was significantly up-regulated in all tested tissues except in the spleen of miiuy croaker after challenging with Vibrio anguillarum (Zhu et al. Citation2013). CCR9 was a key chemokine receptor mediating the local inflammatory responses in the gastrointestinal tract (Zhang et al. Citation2015). In a previous study, the expression of CCR9 transcripts in the gut mucosal tissue of sea bass was increased after treated with the recombinant TNF-α alongside an oral vaccine against V. anguillarum (Galindo-Villegas et al. Citation2013). Unfortunately, the intestines or stomach was not used to analyze the expression of RcCCR9 following external stimuli. Further studies are required to elucidate the role of CCR9 participates in local mucosal immunity despite the low transcript levels in the gastrointestinal tract of healthy cobia.
Figure 6. Expression analysis of RcCCR9 in head kidney and spleen at different time points after stimulation by quantitative real-time RT-PCR. The mRNA expression levels were normalized to the endogenous control gene, β-actin. The RcCCR9 expression in head kidney (a) and spleen (b) after treatment with inactivated V. harveyi, respectively; the RcCCR9 expression in head kidney (c) and spleen (d) after treatment with poly I:C, respectively. Data were expressed as a ratio to RcCCR9 mRNA expression in the respective PBS control. Each vertical bar represented the mean ± SE (N = 3). Significant differences in RcCCR9 expression between stimulated and unstimulated tissues (0 h) were indicated with an asterisk (*) for P < 0.05 or with two asterisks (**) for P < 0.01.
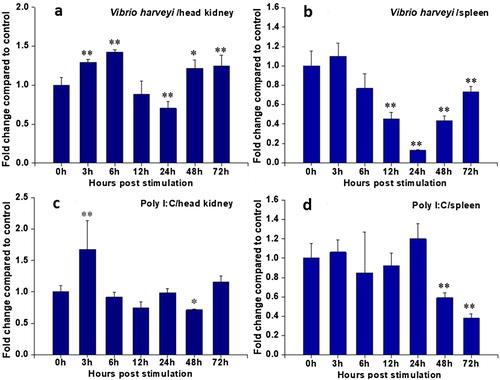
We further investigated the RcCCR9 transcriptional modulation by the viral mimic poly I:C. Following stimulation, the expression level of RcCCR9 in head kidney was significantly up-regulated at 3 h and down-regulated at 48 h but had no impact at other time points (c). No significant difference was observed in the expression of RcCCR9 in spleen before 24 h of post-treatment, but significantly decreased at 48 h and reached the hardpan at 72 h (d). Unlike RcCCR9 was down-regulated or no impact at most time points after poly I:C challenge, numerous reports showed CCR9 is up-regulated in immune-relevant tissues during pathogenic infection. For instance, a study of grouper suggested that CCR9a was significantly up-regulated in the head kidney and spleen after treatment with Cryptocaryon irritans (Yang et al. Citation2017). Similarly, in response to infectious pancreatic necrosis virus infection, trout CCR9A was up-regulated in the mid-gut and hindgut (Ballesteros et al. Citation2014). The down-regulation of RcCCR9 expression imply this receptor could be involved in the viral-induced mobilization of spleen cells to primary sites of viral encounter and also reflect the complex networks in combating different pathogens.
In mammal, the increase in CCL25 within the intestine in chronic inflammatory diseases was correlated with the increase of CCR9-expressing T cells in mesenteric lymph nodes (Papadakis et al. Citation2000). However, previous documented cobia CCL25-like gene (RcCC3) was significantly up-regulated in the head kidney and spleen after treatment with V. harveyi or poly I:C (Su et al. Citation2013). The expression pattern of RcCC9 mRNA in the central immune organs of cobia was not consistent with that of its ligand. The head kidney and spleen home a large number of myeloid lineage cells, including lymphocytes, many kinds of granulocytes, and macrophages suggesting the complicated involvement of the chemokine receptors and their ligands in fish immunity. The regulation of RcCCR9 expression in these tissues appeared to be stronger responses to the V. harveyi challenge than poly I:C. Although its roles and the mechanisms of regulation are not known at present, the findings that these distinct and inconsistent time course expression profiles of RcCCR9 are likely due to differences in defence strategies against microbial invaders.
In summary, a CCR9 gene was identified from cobia. The putative amino acid sequence contains the conserved structural characteristic of chemokine receptor. Phylogenetic analysis and sequence alignment showed that RcCCR9 is more closely related and has higher sequence identity to homologs from other fish. RcCCR9 was constitutively expressed in all examined tissues, with highest expression in blood and lower levels in intestines and brain. The stimulants representing the bacterial and viral invasions induced an up-regulation of RcCCR9 transcription in head kidney and a down-regulation of its expression in the spleen. RcCCR9 seems to be strongly involved in host defense against bacterial infection, since compared with poly I:C, V. harveyi induced a stronger up/down-regulation of CCR9 mRNA levels in the head kidney and spleen.
TAAR_1662424_Supplementary_Table
Download MS Word (18 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ai LS , Liao F. 2002. Mutating the four extracellular cysteines in the chemokine receptor CCR6 reveals their differing roles in receptor trafficking, ligand binding, and signaling. Biochem. 41:8332–8341. doi:10.1021/bi025855y.
- Ballesteros NA , Rodríguez SS , Pérezprieto SI , Aquilino C , Tafalla C. 2014. Modulation of genes related to the recruitment of immune cells in the digestive tract of trout experimentally infected with infectious pancreatic necrosis virus (IPNV) or orally vaccinated. Dev Comp Immunol. 44:195–205. doi:10.1016/j.dci.2013.12.009.
- Chadzinska M , Golbach LA , Pijanowski L , Scheer M , Kemenade BM. 2014. Characterization and expression analysis of an interferon-γ2 induced chemokine receptor CXCR3 in common carp (Cyprinus carpio L). Dev Comp Immunol. 47:68–76. doi:10.1016/j.dci.2014.07.008.
- Dixon B , Luque A , Abos B , Castro R , Gonzaleztorres L , Tafalla C. 2013. Molecular characterization of three novel chemokine receptors in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immun. 34:641–651. doi:10.1016/j.fsi.2012.12.003.
- Galindo-Villegas J , Mulero I , García-Alcazar A , Muñoz I , Peñalver-Mellado M , Streitenberger S , Scapigliati G , Meseguer J , Mulero V. 2013. Recombinant TNFα as oral vaccine adjuvant protects European sea bass against vibriosis: insights into the role of the CCL25/CCR9 axis. Fish Shellfish Immun. 35:1260–1271. doi:10.1016/j.fsi.2013.07.046.
- Griffith JW , Sokol CL , Luster AD. 2014. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 32:659–702. doi:10.1146/annurev-immunol-032713-120145.
- Grimholt U , Hauge H , Hauge AG , Leong J , Koop BF. 2015. Chemokine receptors in Atlantic salmon. Dev Comp Immunol. 49:79–95. doi:10.1016/j.dci.2014.11.009.
- Gupta R , Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 7:310–322. doi:10.1142/9789812799623_0029.
- Hadeiba H , Sato T , Habtezion A , Oderup C , Pan J , Butcher EC. 2008. CCR9 expression defines tolerogenic plasmacytoid dendritic cells able to suppress acute graft-versus-host disease. Nat Immunol. 9:1253–1260. doi:10.1038/ni.1658.
- Hewit KD , Fraser A , Nibbs RJ , Graham GJ. 2014. The N-terminal region of the atypical chemokine receptor ACKR2 is a key determinant of ligand binding. J Biol Chem. 289:12330e42. doi:10.1074/jbc.M113.534545.
- Huang Y , Wang D , Wang X , Zhang Y , Liu T , Chen Y , Tang Y , Wang T , Hu D , Huang C. 2016. Abrogation of cc chemokine receptor 9 ameliorates ventricular remodeling in mice after myocardial infarction. Sci Rep. 6:32660. doi:10.1038/srep32660.
- Johnson EL , Singh R , Singh S , Johnsonholiday C , Grizzle WE , Partridge EE , Lillard JW. 2010. CCL25-CCR9 interaction modulates ovarian cancer cell migration, metalloproteinase expression, and invasion. World J Surg Onco. 8:62. doi:10.1186/1477-7819-8-62.
- Liu Y , Chang MX , Wu SG , Nie P. 2009. Characterization of C-C chemokine receptor subfamily in teleost fish. Mol Immunol. 46:498–504. doi:10.1016/j.molimm.2008.10.003.
- Livak KJD , Schmittgen T. 2001. Analysis of relative gene expression data using real-time quantitative PCR and 2(-delta delta C(T)) method. Methods. 25:402–408. doi: 10.1006/meth.2001.1262
- Mirzadegan T , Benko G , Filipek S , Palczewski K. 2003. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry. 42:2759–2767. doi:10.1021/bi027224+ doi: 10.1021/bi027224+
- Monigatti FE , Gasteiger E , Bairoch A , Jung E. 2002. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 18:769–770. doi: 10.1093/bioinformatics/18.5.769
- Oldham WM , Hamm HE. 2008. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 9:60–71. doi:10.1038/nrm2299.
- Papadakis K , Prehn J , Nelson V , Cheng L , Binder SW , Ponath PD , Andrew DP , Targan SR. 2000. The role of thymus–expressed chemokine and its receptor CCR9 on lymphocytes in the regional specialization of the mucosal immune system. J Immunol. 165:5069–5076. doi:10.4049/jimmunol.165.9.5069.
- Qidwai T , Khan MY. 2016. Impact of genetic variations in CC chemokine receptors and ligands on infectious diseases. Hum Immunol. 77:961–971. doi:10.1016/j.humimm.2016.06.010.
- Rossi DL , Zlotnik A. 2003. The biology of chemokines and their receptors. Annu Rev Immunol. 18:217–242. doi:10.1146/annurev.immunol.18.1.217.
- Stone MJ , Hayward JA , Huang C , Huma ZE , Sanchez J. 2017. Mechanisms of regulation of the chemokine–receptor network. Int J Mol Sci. 18:342. doi:10.3390/ijms18020342.
- Su Y , Feng J , Sun X , Guo Z , Xu L , Jiang J. 2013. Characterization and transcriptional analysis of a new CC chemokine associated with innate immune response in cobia (Rachycentron canadum). Mol Biol. 47:389–398. doi: 10.1134/S0026893313030163
- Su Y , Guo Z , Xu L , Jiang J , Wang J , Feng J. 2012. Identification of a cobia (Rachycentron canadum) cc chemokine gene and its involvement in the inflammatory response. Fish Shellfish Immun. 32:204–210. doi:10.1016/j.fsi.2011.10.005.
- Szpakowska M , Fievez V , Arumugan K , Van NN , Schmit JC , Chevigné A. 2012. Function, diversity and therapeutic potential of the N-terminal domain of human chemokine receptors. Biochem Pharmacol. 84:1366–1380. doi:10.1016/j.bcp.2012.08.008.
- Tu Z , Xiao R , Xiong J , Tembo KM , Deng X , Xiong M , Liu P , Wang M , Zhang Q. 2016. CCR9 in cancer: oncogenic role and therapeutic targeting. J Hematol Oncol. 9:10. doi:10.1186/s13045-016-0236-7.
- Vicari AP , Figueroa DJ , Hedrick JA , Foster JS , Singh KP , Menon S , Copeland NG , Gilbert DJ , Jenkins NA , Bacon KB , Zlotnik A. 1997. TECK: a novel CC chemokine specifically expressed by thymic dendritic cells and potentially involved in T cell development. Immunity. 7:291–301. doi:10.1016/S1074-7613(00)80531-2.
- Wendland M , Czeloth N , Mach N , Malissen B , Kremmer E , Pabst O , Förster R. 2007. CCR9 is a homing receptor for plasmacytoid dendritic cells to the small intestine. P Natl Acad Sci USA. 104:6347–6352. doi:10.1073/pnas.0609180104.
- Yang M , Zhou L , Wang HQ , Luo XC , Dan XM , Li YW. 2017. Molecular cloning and expression analysis of CCL25 and its receptor CCR9s from epinephelus coioides, post cryptocaryon irritans, infection. Fish Shellfish Immu. 67:402–410. doi:10.1016/j.fsi.2017.06.039.
- Zhang J , Romero JA , Chan A , Goss J , Stucka S , Cross J , Chamberlain B , Varoglu M , Chandonnet H , Ryan D , Lippa B. 2015. Biarylsulfonamide CCR9 inhibitors for inflammatory bowel disease. Bioorg Med Chem Lett. 25:3661–3664. doi:10.1016/j.bmcl.2015.06.046.
- Zhu Z , Wang R , Ren L , Xu T. 2013. Characterization of the CCR3 and CCR9 genes in miiuy croaker and different selection pressures imposed on different domains between mammals and teleosts. Dev Comp Immunol. 41:631–643. doi:10.1016/j.dci.2013.06.015.
