ABSTRACT
There is an increasing demand for horse milk, but the current domestic performance of the horse milk has not yet conducted a systematic in-depth study. We carried out a pilot study on 13 single nucleotide polymorphisms (SNPs) located within five genes in Kazakh horse using Kompetitive Allele Specific PCR (KASPTM). An association study with average daily milk production of 60 Kazakh mares was performed for 6/13 polymorphic SNPs. The results showed that only two SNPs rs114262445 (NUMB), rs1136347938 (CA8) out of the 6 polymorphic markers were significantly associated with milk production in Kazakh horses. These results indicate the potential of using SNPs for the NUMB and CA8 genes as candidate genes for selection and farm production outputs. This study is relevant for future horse genomic studies.
KEYWORDS:
Introduction
Kazakh horse is an autochthonous breed in central Asia, adapted to harsh environment. Milk performance and sports trait were developed by artificial selection (Bahetiguli Citation2011). Mare’s milk has a sweet taste that ranges from bluish-white to slightly brown colouration. Important components of the milk are lactose, protein, fat, vitamins, enzymes, and minerals (Mazhitova et al. Citation2015). Mare’s milk is not a new fad, but it is coming back into fashion. In Europe, Belgium, France, the Norway and Netherlands, mare’s milk is a staple food. Koumiss is the main product of mare milk, in Mongolia, Kazakhstan and Kyrgyzstan the tradition is to ferment dairy beverage called kumis, or airag in Mongolia (Akuzawa et al. Citation2011). A small proportion of people from ancient times have been producing koumiss, and appreciated it for its nutritional and health-promoting properties (Mazhitova et al. Citation2015).
Koumiss healthcare value can be summarized as the following aspects. In Russia and Mongolia set up special ‘koumiss medical center’ (Li et al. Citation2006), koumiss could combine with traditional medicine to assist treatment, such as cardiovascular diseases, alimentary system diseases, nervous system disease, tuberculosis, anaemia, gynaecological and glycuresis (Wu Citation1986). Favourable influences on the alimentary canal’s activity, the nervous system, cardiovascular diseases, Phthisis, endocrine glands and the immune system have been reported (Jaqielski Citation1872; Mccaskie Citation1878; Stoianova et al. Citation1988; Fedechko et al. Citation1995). An increasing interest has been shown for the manufacture of koumiss at an industrial level in human nutrition and health.
Recent advances in next-generation sequencing (NGS) technology have provided researchers with unprecedented tools to increase the discovery of genetic variation associated with traits of economic importance in animals (Petersen et al. Citation2013; Crisà et al. Citation2016; Stefaniuk et al. Citation2016; Purfield et al. Citation2017). But this method was rarely reported to be used to study the milk traits of horse. To date, in horses, many genes have been investigated and significantly associated with important phenotypic traits, such as coat colour variations (MC1R, PMEL) (Andersson et al. Citation2013; Corso et al. Citation2017), Morphometric Traits (WWOX, ANKRD1, ECA3, ZFAT) (Meira et al. Citation2014; Abri et al. Citation2017; Grilz-Seger et al. Citation2019; Gurgul et al. Citation2019), reproduction traits (STK31,TSPY, AEG1, CRISP1) (Giese et al. Citation2002; Sabeur et al. Citation2008). However, few genes were related to milk traits of horse. Several of the genes that we identified here confirm previously documented selection signatures in Kazakh horses, such as NUMB, ADCY8, CA8, LGALS2, SLC25A30 (Liu et al. Citation2018).
The aim of this study was to investigate the polymorphism of 13 SNPs of 5 genes on milk production using genotyping methods in Kazakh horse reared in Fuyun of Xinjiang. Furthermore, association study was performed between milk traits of horse and polymorphic SNPs. The ultimate purpose was to find and describe polymorphisms that could prove useful for the breeding of future specialized breed in Kazakh horses.
Materials and methods
Animals
Sixty Kazakh horse (5-7rd lactations) reared under grazing conditions (Fuyun: 45°–48°03′N88°10′-90°31′E).
Collecting data
We sampled milk from two months to five months of lactation, we collected every 10 days and took four time points at 12 am, 14, 16, and 18 pm, weighed and recorded the milk yield of each Kazakh horse at each time point. The mare’s milk sampling step was the same during the whole experiment. The mares and foals were separated before milked, they could to see, smell each other, but the foal was not able to suckle. The 2 teats of the mare were hand milked as completely as possible. We used four time points to calculated daily milk production, and average milk of day was calculated by 12 daily milk yield records for four months.
Selection of SNP
A total of 5 genes have been chosen based on the results of prior signatures of selection (Liu et al. Citation2018), and 13 single nucleotide polymorphisms (SNPs) of these genes were collected from the database of NCBI.
Genomic DNA extraction and genotyping
Genomic DNA was isolated using a DNA kit from GeneOnBioTech. Kompetitive Allele Specific PCR (KASPTM, LGC Genomics, Teddington, Middlesex, UK) genotyping was used for the bi-allelic discrimination of the selected 13 SNPs (). Genotype data for each animal were exported for the statistical analysis.
Table 1. Selected SNPs used in the study for genotyping the Kazakh horse.
Primer design
Software of prime 5 was used to design primer of 5 genes. Primer sequences are given in .
Table 2. Primer information of five genes.
PCR amplification
PCR amplification was conducted in a 5 μLvolume containing 2.5 μL of 2×Master Mix, primer 0.07 μl, genomic DNA 30–50 μg, ddH2O supplement of 5 μL. The PCR conditions were as follows: an initial step at 95°C (one cycle for 10 min), 10 cycles for 20 s at 95°C, 60 s at 61–55°C temperature for each primer pair, and 26 cycles for 20 s at 95°C, 60 s at 55°C.
Data analysis
The genotyping results received from LGC Genomics were analysed with Douglas software from LGC Genomics (www.lgcgroup.com). Further, a genetic association study was conducted on polymorphic SNPs. We performed factorial ANOVA (Bonferroni correction) and T-test using the software SPSS19.0. The chi-squared test (χ2) was used to determine whether the populations were in Hardy-Weinberg equilibrium (HWE). HWE analysis was performed for setting the genetic association studies.
Ethics of experimentation
All experimental procedures involving animals were approved (animal protocol numbers: 2018001)by the Animal Care and Use Committee of Xinjiang Agricultural University, Urumqi, Xinjiang, China.
Results
Result of KASP
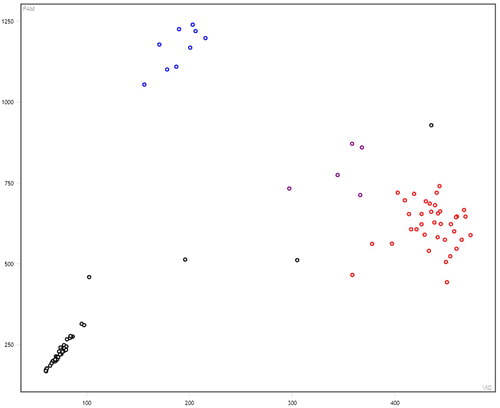
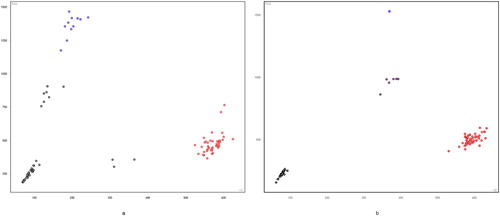
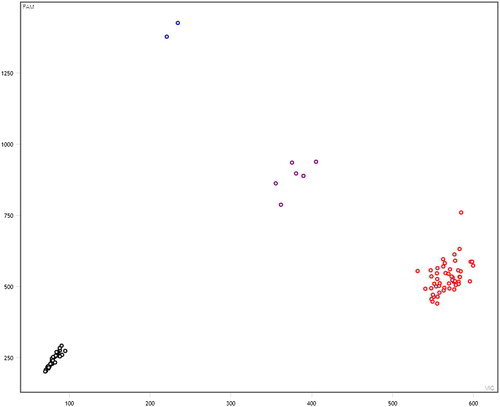
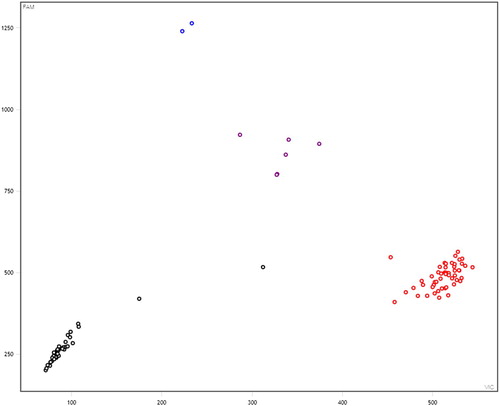
A total of 6 SNPs among the selected 13 SNPs were polymorphic across the Kazakh horse and further used for the association study (Figure S1–S5). In these figures: black represent none sample, red represents homozygous, purple represents heterozygous, and blue represents mutant homozygous (–).
Figure 1. Genotyping results of NUMB gene (g.18986402 A>G). Note: Black represent none sample, red represents homozygous, purple represents heterozygous, and blue represents mutant homozygous. The same below.
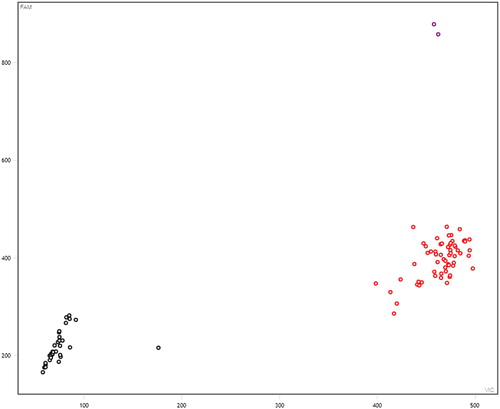
Polymorphism analysis
The different genotype frequencies are shown in . Fisher exact test of SPSS 19.0 software was used to calculation χ2 and P-value. The g.71925075 C > T locus (rs1142665928) was not in HWE, the other five SNPs were in HWE.
Table 3. List of polymorphic SNPs and genotypes frequencies in Kazakh horse breeds.
Correlation analysis of genotype and milk yield in Kazakh horse
The two SNPs had significant effects on milk yield in Kazakh horse breed ( and ).
Figure 6. A:association analysis of g.18986402G>A loci of NUMB and daily milk yield in Kazakh horse; B:association analysis of g.24074470A>G loci of CA8 and daily milk yield in Kazakh horse.
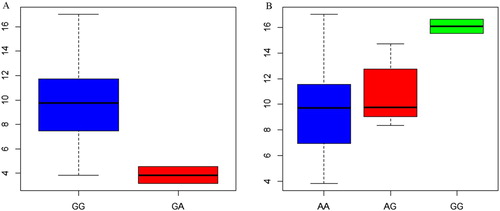
Table 4. List of SNPs found to be associated with milk yield in Kazakh horse breeds.
Discussion
The KASP genotyping system has low-labour and economic advantages over other genotyping assays (Landoulsi et al. Citation2017). The results of the current study showed that the horse NUMB g.18986402 A > G and CA8 g.24074470G > A might play an important role in affecting milk yield traits and could be linked to major genes that affect milk yield traits in horse. Current findings revealed the significant association and the effects of those loci and the investigated milk traits.
The NUMB gene was first discovered in Drosophila (Uemura et al. Citation1989). NUMB is a member of the closely conserved protein family, it plays key role in brain development, nervous system development. The cell fate determinant Numb influences developmental decisions by antagonizing the Notch signalling pathway (Mcgill et al. Citation2003; Rasin et al. Citation2007). Mammalian Numb expression promotes the ubiquitination of membrane tethered Notch1 and the degradation of the intracellular domain following receptor activation, and Notch signaling plays an important role in cellular differentiation, proliferation, and apoptotic events at all stages of development, functioning as an essential communication mechanism to direct cell fate selection of neighbouring cells (Artavanis-Tsakonas et al. Citation1999). If the Notch signaling pathway is out of regulation, the mammary epithelium will always be in a hyperplastic state, which will affect the lactation process. In China, NUMB gene was found to have an important effect on mammary gland of mice (Zhang Citation2016). The expression of this gene reached the highest level in late pregnancy; it regulates the lactation process by regulating the Notch signaling pathway, indicating that NUMB gene may play an important regulatory role in lactation (Zhang Citation2016).
In this study, mutation in the SNP g.24074470A > G of NUMB gene is associated with a decrease in milk yield of Kazakh horse. The reason may be that the expression of NUMB gene inhibits the Notch signaling pathway, thereby hindering the lactation process.
CA8 gene was found to have wide range effect on a variety of disease such as Novel Syndrome Characterized (Türkmen et al. Citation2009), femoral bone mineral density (Mori et al. Citation2009), and thermal antinociception (Fu et al. Citation2017), however, no studies on animals were reported. This gene product, carbonic anhydrase-related protein VIII (CARP VIII), is predominantly present in cerebellar Purkinje cells, where it interacts with the inositol 1,4,5-trisphosphate receptor type 1, a calcium channel (Aspatwar et al. Citation2013). Hou studied the differential gene of different lactation stage in cow, and CA8 gene was found in the lactation stage of dairy cow (Hou Citation2009). The CA8 gene may be involved in the regulation of aerobic glycolysis and increase cell viability. Therefore, so it is speculated that the product of CA8 gene promotes the absorption of milk components by increasing the activity of mammary epithelial cells. In this study, mutation in the SNP g.24074470A > G of CA8 gene has three genotypes of GG, AA and AG in the Kazakh population. Furthermore, milk yield of GG genotype was significantly higher than for the other two genotypes. So, this mutation may affect the ability of lactation in Kazakh horses.
Conclusions
In this study, 13 SNPs of five genes were genotyped using KASP genotyping assays; however, only 6 SNPs were polymorphic. We found evidence of significant associations between two intragenic SNPs and milk yield traits in Kazakh horses. These two SNPs are located within two different genes not yet reported before as candidates for horse milk yield. These two SNPs within the genes NUMB and CA8 turned out as promising molecular markers for daily milk yield in Kazakh horses.
Acknowledgements
The authors thank Yuehui Ma and Lin Jiang.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abri MAA, Posbergh C, Palermo K, Sutter NB, Eberth J, Hoffman GE, Brooks SA. 2017. Genome-wide scans reveal QTLs for withers height in horses near the ANKRD1 gene. J Equine Vet Sci. 60:67–73. doi: 10.1016/j.jevs.2017.05.008
- Akuzawa R, Miura T, Surono IS. 2011. Asian fermented milks. Encyclopedia Dairy Sci. 2:507–511. doi: 10.1016/B978-0-12-374407-4.00186-2
- Andersson LS, Wilbe M, Viluma A, Cothran G, Ekesten B, Ewart S, Lindgren G. 2013. Equine multiple congenital ocular anomalies and silver coat colour result from the pleiotropic effects of mutant PMEL. Plos One. 8:e75639. doi: 10.1371/journal.pone.0075639
- Artavanis-Tsakonas S, Rand MD, Lake RJ. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. doi: 10.1126/science.284.5415.770
- Aspatwar A, Tolvanen ME, Jokitalo E, Parikka M, Ortutay C, Harjula SK, Rämet M, Vihinen M, Parkkila S. 2013. Abnormal cerebellar development and ataxia in CARP VIII morphant zebrafish. Hum Mol Genet. 22:417–432. doi: 10.1093/hmg/dds438
- Bahetiguli MLTH. 2011. Introduction of Kazakh horse. Xinjiang Anim Husbandry. s. 1:15.
- Corso J, Hepp D, Ledur MC, Peixoto JO, Fagundes NJR, Freitas TRO. 2017. Genetic variation of the bronze locus (MC1R) in turkeys from Southern Brazil. Genet Mol Biol. 40:104–108. doi: 10.1590/1678-4685-gmb-2016-0136
- Crisà A, Ferrè F, Chillemi G, Moioli B. 2016. RNA-Sequencing for profiling goat milk transcriptome in colostrum and mature milk. BMC Vet Res. 12:264. doi: 10.1186/s12917-016-0881-7
- Fedechko IM, Hrytsko RI, Herasun BA. 1995. The anti-immunodepressive action of koumiss made from cow’s milk. Lik Sprava. 9–12:104–106.
- Fu ES, Erasso DM, Zhuang GZ, Upadhyay U, Ozdemir M, Wiltshire T, Sarantopoulos KD, Smith SB, Maixner W, Martin ER, Levitt RC. 2017. Impact of human CA8 on thermal antinociception in relation to morphine equivalence in mice. Neuroreport. 28:1215–1220. doi: 10.1097/WNR.0000000000000872
- Giese A, Jude R, Kuiper H, Raudsepp T, Piumi F, Schambony A, Guérin G, Chowdhary BP, Distl O, Töpfer-Petersen E, Leeb T. 2002. Molecular characterization of the equine testis-specific protein 1 (TPX1) and acidic epididymal glycoprotein 2 (AEG2) genes encoding members of the cysteine-rich secretory protein (CRISP) family. Gene. 299:101–109. doi: 10.1016/S0378-1119(02)01018-1
- Grilz-Seger G, Druml T, Neuditschko M, Mesarič M, Cotman M, Brem G. 2019. Analysis of ROH patterns in the Noriker horse breed reveals signatures of selection for coat color and body size. Anim Genet. 50(4):334–346. doi: 10.1111/age.12797
- Gurgul A, Jasielczuk I, Semik-Gurgul E, Pawlina-Tyszko K, Stefaniuk-Szmukier M, Szmatoła T, Polak G, Tomczyk-Wrona I, Bugno-Poniewierska M. 2019. A genome-wide scan for diversifying selection signatures in selected horse breeds. Plos One. 14(1):e0210751. doi: 10.1371/journal.pone.0210751
- Hou XM. 2009. Screening functional genes of development and lactation mammary gland in dairy Cow. Haerbing: Northeast Agricultural University.
- Jaqielski V. 1872. Koumiss in the treatment of Phthisis. Br Med J. 1(579):124–125. doi: 10.1136/bmj.1.579.124
- Landoulsi Z, Benromdhan S, Ben Djebara M, Damak M, Dallali H, Kefi R, Abdelhak S, Gargouri-Berrechid A, Mhiri C, Gouider R. 2017. Using KASP technique to screen LRRK2 G2019S mutation in a large Tunisian cohort. BMC Med Genet. 18(1):70. doi: 10.1186/s12881-017-0432-5
- Li XK, Li KX, Zou SD. 2006. Koumiss milk. China Dairy. 07:58–60.
- Liu LL, Fang C, Liu WJ. 2018. Identification on novel locus of dairy traits of Kazakh horse in Xinjiang. Gene. 677:105–110. doi: 10.1016/j.gene.2018.07.009
- Mazhitova AT, Kulmyrzaev AA, Ozbekova ZE, Bodoshev A. 2015. Amino acid and fatty acid profile of the mare’s milk produced on suusamyr pastures of the Kyrgyz republic during lactation period. Procedia-Socialand Behav Sci. 195:2683–2688. doi: 10.1016/j.sbspro.2015.06.479
- Mccaskie N. 1878. Koumiss in obstinate vomiting. Br Med J. 1:367.
- Mcgill MA, Mcglade CJ. 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 278:23196–23203. doi: 10.1074/jbc.M302827200
- Meira CT, Farah MM, Fortes MRS, Moore SS, Pereira GL, Silva JAV, Mota MDS, Curi RA. 2014. A genome-wide association study for morphometric traits in Quarter horse. J Equine Vet Sci. 34:1028–1031. doi: 10.1016/j.jevs.2014.05.011
- Mori S, Kou I, Sato H, Emi M, Ito H, Hosoi T, Ikegawa S. 2009. Nucleotide variations in genes encoding carbonic anhydrase 8 and 10 associated with femoral bone mineral density in Japanese female with osteoporosis. J Bone Miner Metab. 27:213–216. doi: 10.1007/s00774-008-0031-9
- Petersen JL, Mickelson JR, Rendahl AK, Valberg SJ, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS, et al. 2013. Genome-wide analysis reveals selection for important traits in domestic horse breeds. Plos Genet. 9:e1003211. doi: 10.1371/journal.pgen.1003211
- Purfield DC, Mcparland S, Wall E, Berry DP. 2017. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. Plos One. 12:e0176780. doi: 10.1371/journal.pone.0176780
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. 2007. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 10:819. doi: 10.1038/nn1924
- Sabeur K, Ball BA, Corbin CJ, Conley A. 2008. Characterization of a novel, testis-specific equine serine/threonine kinase. Mol Reprod Dev. 75:867–873. doi: 10.1002/mrd.20792
- Stefaniuk M, Ropka-Molik K. 2016. RNA sequencing as a powerful tool in searching for genes influencing health and performance traits of horses. J Appl Genet. 57:199–206. doi: 10.1007/s13353-015-0320-7
- Stoianova LG, Abramova LA, Ladodo KS. 1988. Sublimation-dried mare’s milk and the possibility of its use in creating infant and dietary food products. Vopr Pitan. 3:64–67.
- Türkmen S, Guo G, Garshasbi M, Hoffmann K, Alshalah AJ, Mischung C, Kuss A, Humphrey N, Mundlos S, Robinson PN. 2009. CA8 Mutations Cause a Novel Syndrome Characterized by Ataxia and Mild Mental Retardation with Predisposition to Quadrupedal Gait. PLoS Genet. 5:e1000487. doi: 10.1371/journal.pgen.1000487
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. 1989. Numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 58:349–360. doi: 10.1016/0092-8674(89)90849-0
- Wu Z. 1986. Koumiss Therapy (In Chinese). Huhhot: Inner Mongolia People’s Publication.
- Zhang Y. 2016. The function and mechanism of numb and Numbl on mammary gland development and lactogenesis. Beijing: China Agricultural University.