ABSTRACT
Circular RNAs (circRNAs), as miRNA sponges, have been found to participate in transcriptional control and many important biological processes. Heat stress (HS) severely affects feed intake and milk production of sows. However, the expression and function of circRNAs in the hypothalamus of sows and whether they are involved in the adaptation to HS have not been explored. In this study, RNA sequencing and functional analysis were performed to explore the expression and function of circRNAs in the hypothalamus of heat-stressed sows. Toally, 38,143 circRNAs were identified from the hypothalamus of heat-stressed sows, among which 307 circRNAs (170 upregulated and 137 downregulated) were significantly differentially expressed (DE) in the HS groups compared to the control groups. Their host genes are involved in the cellular component organization or biogenesis, metabolic process, and cellular metabolic process, the MAPK signalling pathway and the TGF-β signalling pathway. And a total of 307 DE circRNAs were found to act as sponges for 457 miRNAs. This research explored the expression profiles of circRNAs in the hypothalamus of heat-stressed sows, which will provide basic data for further investigation of the biological function and the regulatory role of circRNAs in the heat-stressed sows.
1. Introduction
With the intensification of global warming, high temperatures hinder the normal growth and reproduction of animals, which would cause economic losses. High temperatures can severely affect sows feeding, reproduction, lactation and metabolism (Andreassen and Hoyheim Citation2017). Heat stress (HS) is a non-specific response of the animal body to high temperature environments, which would reduce the lactating sow's feed intake and affect mammary gland metabolism and milk production (Boulant Citation2000; Chen and Thorner Citation2007; Chauhan et al. Citation2017), which can negatively affect piglet growth during lactation and their weaning weight. In addition to the postpartum, HS can also affect sows during pregnancy, such as increasing embryo mortality and reducing farrowing rate and litter size (Fan et al. Citation2019). Analyzing the entire production cycle of sows (including animal growth), HS had a substantial economic impact on the global swine industry (Fan et al. Citation2016). The hypothalamus, as a part of the brain, plays a role in regulating body temperature, whose many areas are activated under HS (Filipowicz et al. Citation2008; Hang et al. Citation2018). The hypothalamus preoptic area (POA) is composed of neurones that are sensitive to minute modulations in hypothalamic or core temperature, which can bring thermoregulatory responses to maintain homeostasis for both internal and environmental thermal conditions (Hansen et al. Citation2013; Hao et al. Citation2016). Studying the changes in the brain proteome induced by HS several months ago, there was found that a malfunction of the hypothalamus may destroy the host physical and immune function, resulting in decreased growth performance and immunosuppression in HS pigs (Huang et al. Citation2019). There is a study found changes in the expression of heat shock proteins such as Dnajc13 and Hspcb in the hypothalamus of heat-stressed chickens (Hang et al. Citation2018). In addition, Li et al analyzed the transcriptome of the hypothalamus of heat-stressed Hu sheep and found that HS induced calcium dyshomeostasis, caused ROS accumulation, impaired the antioxidant system and innate defence, decreased growth and development, and enhanced organ damage (Huang et al. Citation2018). However, the research on the molecular mechanisms involved in the HS response in the hypothalamus of sows is limited.
Circular RNA (circRNA), as a type of non-coding RNA, belongs to the class of endogenous circular molecule generated by alternative splicing (Kisliouk et al. Citation2014). Linking the 3′- and 5′- ends of RNAs to form a covalently closed continuous loop, circRNAs cannot be degraded by ribonucleases (Kisliouk and Meiri Citation2013). Some studies have shown that circRNAs function as efficient microRNA (miRNA) sponges which can competitively terminate inhibition of their mRNA targets (Kunde et al. Citation2013; La et al. Citation2019), which suggest that circRNAs have vital roles in the fine-tuning of posttranscriptional regulation of gene expression.
In the recent years, more and more studies have been focused on various diseases (Li et al. Citation2018; Li et al. Citation2019) and some other tissues (Li et al. Citation2015; Liu et al. Citation2017; Lucy and Safranski Citation2017), but the studies on circRNA in hypothalamus is still relatively rare. Exploring the expression of circRNA in the hypothalamus of FecB ++ genotype sheep, Zhang et al found that several key hypothalamic circRNAs may participate in sheep reproduction by influencing gonadotropin-releasing hormone (GnRH) activities or affecting key gene expression indirectly or directly (Luo et al. Citation2016). Moreover, using microarrays to measure the hypothalamic circRNA expressions in heat acclimation (HA) rats, candidate biomarkers for HA (rno_circRNA_014301, rno_circRNA_010393, and rno_circRNA_011190) were identified to regulate expression levels of the genes by controlling responding miRNA activity (Filipowicz et al. Citation2008). However, little is known about the expression profile of circRNAs in the hypothalamus of sows. In this study, transcriptome sequencing was used, for the first time, to identify candidate circRNAs in the hypothalamus related to HS. This study provides a basis for further investigation of the regulatory role of circRNAs in heat-stressed sows.
2. Materials and methods
2.1. Sample preparation
All procedures involving animals were carried out with Chinese guidelines for animal welfare and approved by the Animal Care and Use Committee of Zhejiang University, Zhejiang, China (ZJU20160346). Six healthy sows (4-5 parities) during lactation were obtained from Hangzhou Daguanshan Pig Breeding Farm (Zhejiang, China), of which three sows in the control group (thermal comfort, TC, 20.25 ± 1.75℃) were raised during the summer months (from August 1st to September 3rd), the other three sows in the experimental group (heat stress, HS, 29.41 ± 1.34℃) were raised during the winter months (from November 22nd to December 24th). All the sows were raised under the same conditions during lactation except for ambient temperature. On the 28th day of lactation (piglets weaning day), these sows were slaughtered, and the hypothalamus tissues (HS: H-XQ1, H-XQ2, and H-XQ3; TC: L-XQ1, L-XQ2, and L-XQ3) were collected and frozen in liquid nitrogen and stored at −80℃.
2.2. RNA isolation
Total RNAs were extracted from the hypothalamic tissues of 6 lactating sows using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer's instructions. The concentration and purity of the total RNA were determined using Thermo Scientific Nanodrop NC2000 (Thermo Scientific, NY, USA), and the RNA integrity was accessed using RNA 6000 Nano Kit 5067–1511 of the Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA).
2.3. Library construction and sequencing
The ribosomal RNAs (rRNAs) were removed from the total RNA using Ribo-Zero rRNA Removal Kit (Illumina, Inc.), RNase R was used to remove linear RNA, and then the RNA was fragmented approximately 200–300 bp. With the circRNA fragments as templates, random hexanucleotide primers were used to synthesize first-strand cDNAs, and then second-strand cDNAs were synthesized with dUTP instead of dTTP. Subsequently, cDNAs were purified using a cDNA purification kit, and the poly chain reaction (PCR) was performed. Then, the purified library was accessed using the Agilent 2100 Bioanalyzer system. Finally, the cDNA libraries were sequenced on the Hiseq platform (Illumina Hiseq X-ten PE150) in Shanghai Personal Biotechnology Cp. Ltd (Shanghai, China).
2.4. Sequencing analysis and identification of circular RNA
The clean data (clean reads) were obtained by using Cutadapt (v1.15) to remove reads containing adapters or poly-N and other low-quality reads from the raw data. The remaining clean reads were mapped to the reference swine genome (asia.ensembl.org/index.html) using TopHat2.0.14. The 20 bp ends of the unmapped reads were aligned to the genome using Bowtie2, and the circRNA was verified by the find_circ software. Statistical analysis was conducted on the chromosome distribution and type of the identified circRNAs. Differential expression analysis of circRNAs was performed by DESeq (1.30.0). Those circRNAs with |log2 (fold change) |>1 and significant P-value<0.05 were considered as differentially expressed (DE) between HS and TC groups. The R language Pheatmap (1.0.8) software package was used for bidirectional cluster analysis of circRNAs (http://CRAN.R-project.org/package=pheatmap), calculating the distance using the Eucliden method and clustering with the hierarchical clustering method (Complete Linkage).
2.5. Functional enrichment analysis
The Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) were performed to explore the main biological functions of DE circRNAs. The GO enrichment analysis of the host genes for DE circRNAs was performed using DAVID (http://david.abcc.ncifcrf.gov/). And the KEGG pathway analysis of the host genes for DE circRNAs was determined with the KEGG database (http://www.genome.jp).
2.6. MiRNA binding site prediction
There was a study shown that circRNAs could act as miRNA sponges to regulate gene expression (La et al. Citation2019). Therefore, the miRanda software (http://www.mirbase.org/index.shtml) was used to perform miRNA prediction for circRNAs to further study the function of circRNAs. And the Cytoscape v3.4 software (Memczak et al. Citation2013) was used to construct the circRNA-miRNA interaction network.
2.7. Validation by quantitative real-time polymerase chain reaction (qRT-PCR)
The quantitative real-time polymerase chain reaction (qRT-PCR) was performed to validate the accuracy of RNA sequencing. Total RNAs were reverse transcribed into cDNA using PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Hangzhou, China) according to the manufacturer's instructions. Then the qRT-PCR reaction was performed in triplicate using an SYBR Premix Ex Taq TM (TaKaRa, Hangzhou, China) on the Thermo Scientific StepOnePlus Real-Time System (Thermo Scientific, NY, USA). All the primers were synthesized by Shanghai Personal Biotechnology Cp. Ltd (Shanghai, China), and the relative expression of DE circRNAs were quantified using 2−ΔΔCt method using GAPDH as a housekeeping gene.
3. Results
3.1. Identification of circRNA
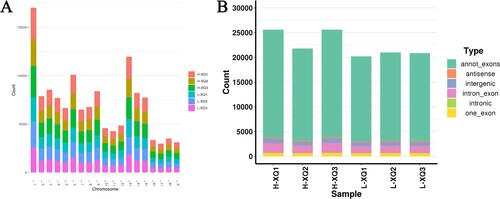
The RNA sequencing (RNA-seq) was performed to identify circRNAs in the hypothalamus involved in HS. A total of 496,979,494 clean reads were obtained after removing adapters and poly-N, as well as other low-quality reads (Table S1). More than 86% of these reads were mapped to the reference swine genome (asia.ensembl.org/index.html) using TopHat2.0.14. And 38,143 candidate circRNAs were totally identified from 6 hypothalamic samples, most of which were distributed on chromosome 1, followed by chromosomes 13 and 6 (A). In addition, these circRNAs were divided into six types based on the circRNAs results and genome annotation information identified by find_circ, with the most annot_exons types (B).
3.2. Differential expression analysis of circRNA
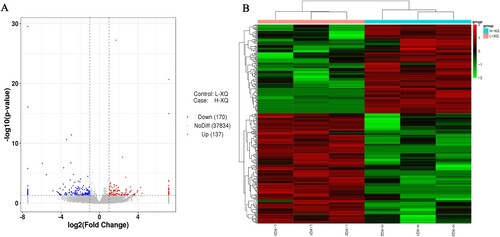
The DE circRNAs in the hypothalamus tissues of TC and HS lactating sows were identified by DEseq2, based on |log2 (fold change) |>1 and significant P-value<0.05 (Table S2). There were 307 DE circRNAs (170 upregulated circRNAs and 137 downregulated circRNAs) obtained in the HS groups compared to the control groups (A). And the heatmap showed the circRNAs significantly differentially expressed in the two groups (B).
3.3. GO and KEGG enrichment analysis of host genes
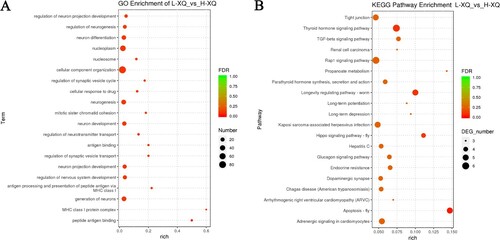
The GO enrichment and KEGG pathways analysis of the host genes for DE circRNAs were performed to further study the potential function of circRNAs (Table S3, Table S4). The GO enrichment analysis showed that most host genes were significantly enriched in cellular component organization, the regulation of nervous system development, regulation of cell development and other metabolic processes or cellular metabolic processes. (A). Meanwhile, KEGG pathways analysis indicated that most host genes of DE circRNAs were significantly assigned to regulating pluripotency of stem cells signalling pathways, cAMP signalling pathway, FoxO signalling pathway, Rap1 signalling pathway, TGF-beta signalling pathway and Tight junction (B).
3.4. CircRNA-miRNA target prediction
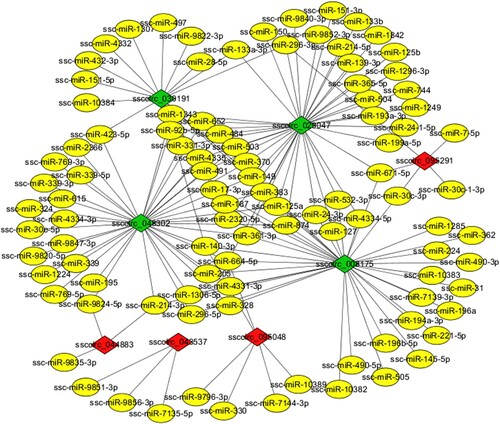
The potential miRNA binding sites were predicted to explore the functions and mechanisms of circRNAs as miRNA sponges. The greater the absolute value of free energy (the energy released when circRNA and miRNA combine to form a stable structure), the stronger the binding capacity. The binding of ssc-miR-4331-3p and ssccirc_078171 has the highest maximum matching scores and the largest free energy at the same time. In the study, a total of 307 DE circRNAs were found as sponges for 457 miRNAs (Table S5). The circRNA-miRNA target prediction results indicated that a single circRNA can target multiple miRNAs, and a single miRNA can be targeted by different circRNAs. For example, ssccirc_048082 can be targeted by 46 different miRNAs, and miR-145-3p can be targeted by 91 DE circRNAs. Eight DE circRNAs and their predictable miRNAs were selected to conduct circRNA-miRNA interaction networks (with max_energy<−25) using Cytoscape v3.4 software ().
3.5. qRT-PCR validation
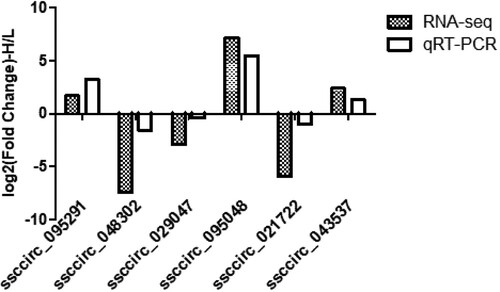
To validate the reliability of RNA sequencing results, 6 DE circRNAs (ssccirc_095291, ssccirc_048302, ssccirc_029047, ssccirc_095048, ssccirc_021722, and ssccirc_043537) were randomly selected for further verified by indicating that our sequencing results were accurate. qRT-PCR. The expression levels of these circRNAs detected by qRT-PCR were consistent with the sequencing results, indicating the reliability of our sequencing (). The primers used for qRT-PCR were available in Table S6.
4. Discussion
HS seriously affects the lactation performance of female animals. The hypothalamus, as a key brain region, centrally coordinates multiple endocrine and nervous system homeostasis functions, such as feed intake, metabolism, and the response to stress, and plays a central role in regulating systemic energy homeostasis and appetite during HS (Filipowicz et al. Citation2008; Hansen et al. Citation2013). Moreover, there has found that circRNAs had functions in animals biological and developmental processes. For example, studies have reported that circRNAs have functions during HS in the hypothalamus of mice (Filipowicz et al. Citation2008) and the pituitary of sows (Mortzfeld et al. Citation2019) and. Here, the study was the first to investigate the circRNA expression profiles of the hypothalamus in heat-stressed sows to detect potential interactions of miRNAs related to sow HS.
The hypothalamus is a region that is highly enriched with circRNAs in the mammalian brain, and circRNAs are specifically expressed in a developmental stage- and tissue-specific manner (Prunier et al. Citation1997; La et al. Citation2019). In this study, a total of 38,143 circRNAs were identified in the hypothalamic tissues. In mice, 53 circRNAs were distinctively expressed in the hypothalamus between the heat acclimation and the control group (Filipowicz et al. Citation2008). In addition, 41,863 circRNAs have been found in the hypothalamus, in which 333 (162 upregulated, while 171 downregulated) were differentially expressed in polytocous sheep in the follicular phase versus monotocous sheep in the follicular phase (Luo et al. Citation2016). Here, 307 circRNAs (170 upregulated and 137 downregulated circRNAs) were significantly differentially expressed in the HS groups compared to the control groups. Therefore, the abundance of circRNA in the hypothalamus may indicate that circRNAs have vital functions in the hypothalamus of heat-stressed sows.
CircRNAs have been reported to play a regulatory role by regulating the polymerase II transcription of their host genes in cis via specific RNA-RNA interaction (Qin et al. Citation2016). Therefore, exploring the potential functions of host genes associated with circRNAs may be useful for studying the function of circRNAs. The GO enrichment analysis showed that most host genes were enriched in the regulation of nervous system development, cellular component organization or biogenesis, regulation of cell development and other metabolic process or cellular metabolic process. And 49 host genes of DE circRNAs were associated with the cellular protein modification process. Especially, heat shock protein, also known as molecular chaperones or cell stress proteins, was a type of proteins produced by cells under stress conditions such as HS (Rybak-Wolf et al. Citation2015); in this study, the Hspb11 (heat shock protein family B 11) was linked with organelle organization and cellular component organization or biogenesis. In addition, the KEGG pathways of host genes revealed that apoptosis pathways, cAMP signalling pathway, MAPK signalling pathway, and the TGF-beta signalling pathway may participate in the hypothalamus in the process of HS. The MAPK (Mitogen-activated protein kinases) signalling pathway, was reported to be involved in cell development, stress response activation, and other molecular and cellular process (Sai et al. Citation2018). MAPK10, as a member of the MAPK gene family, was a host gene of DE sscirc_044883 and ssccirc_008385 and enriched in the MAPK signalling pathway. MAPK was found to be an integration point of a variety of biochemical signalling pathways involved in a diversity of cellular processes, such as proliferation, differentiation, growth and transcriptional regulation (Silanikove Citation2000; Shannon et al. Citation2003). And the mutation of the MAPK10 gene was reported to be involved in the process of stress-induced neuronal cell apoptosis (St-Pierre et al. Citation2003). Moreover, there was a study suggested a role of TGF-β signalling as a more general integrator of environmental signals (Sun et al. Citation2015). AKT3 (protein kinase B γ), a host gene of DE sscirc_013994, was significantly associated with the TGF-β signalling pathway. AKT3 has been found to be critical for nervous system development and to apparently regulate multiple developmental processes (Williams et al. Citation2013). The function analysis of host genes suggested the effects of HS on signalling mechanisms.
CircRNAs could act as efficient miRNA sponges, and miRNAs have been showed to participate in specific biological processes, including cellular proliferation, differentiation, apoptosis and immune response (Yang et al. Citation2005; Kunde et al. Citation2013; Xie et al. Citation2019). In the present study, 307 DE circRNAs were found as sponges for 457 miRNAs. Some of these miRNAs have been shown to participate in cell proliferation, apoptosis, and oxidative stress. For example, miR-10a is a conserved miRNA family that represses proliferation, induces apoptosis in certain cells (Yoo et al. Citation2011). It has been found that miR-10a was upregulated in porcine skeletal muscle treated by constant heat stress (Yu et al. Citation2020). According to the analysis results of miRanda, ssc-miR-10a-5p and ssc-miR-10a-3p were targeted by 30 and 14 DE circRNAs, respectively. And there were 6 pairs of circRNA-ssc-miR-10a-5p with the larger absolute value of max_energy, which meant that circRNA and miRNA combine to form a more stable structure, especially the pairs of ssccirc_048302-miR-10a-5p and ssccirc_074847-miR-10a-5p. In addition, Kisliouk et al has described miR-138 as a novel regulator of hypothalamic cell migration (Zhang et al. Citation2019). And HS during the critical period of thermal-control establishment interfered with the generation of new cells in the hypothalamus, and the miR-138 has been reported to regulate new cell generation in the hypothalamus response to HS (Zhang et al. Citation2019). Here, ssc-miR-138 was target by 52 DE circRNA, significantly interacted with sscirc_081312 and sscirc_030191. These results implicate circRNAs as a miRNA sponge in the hypothalamus and suggest that circRNA may be a vital regulatory in the HS response.
5. Conclusions
In sum, this study used transcriptome sequencing and bioinformatics analysis to explore, for the first time, the expression of circRNAs in the hypothalamus of heat-stressed sows. The GO enrichment and KEGG pathway analysis of host genes were performed to explore the function of circRNAs. Several circRNAs, such as ssccirc_008385, sscirc_013994, ssccirc_048302, and sscirc_081312, were identified through functional enrichment analysis and circRNA-miRNA target prediction. This study provides a circRNA analysis in the hypothalamus and a basis for further investigation of the regulatory role of circRNAs in heat-stressed sows.
TAAR_2005070_Supplementary_Tables
Download Zip (1.3 MB)Acknowledgments
Authors thank Qianqian Song, Fen Wu and Haiyue Cao for participating in the sample collection phase, and farm staff of Hangzhou Daguanshan Pigs Breeding Company Limited for animal care during the experimental period.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Andreassen R, Hoyheim B. 2017. miRNAs associated with immune response in teleost fish. Dev Comp Immunol. 75:77–85. doi:10.1016/j.dci.2017.02.023.
- Boulant JA. 2000. Role of the preoptic-anterior hypothalamus in thermoregulation and fever. Clin Infect Dis. 31:S157–S161.
- Chauhan NR, Kapoor M, Prabha Singh L, Gupta RK, Chand Meena R, Tulsawani R, Nanda S, Bala Singh S. 2017. Heat stress-induced neuroinflammation and aberration in monoamine levels in hypothalamus are associated with temperature dysregulation. Neuroscience. 358:79–92. doi:10.1016/j.neuroscience.2017.06.023.
- Chen RE, Thorner J. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast saccharomyces cerevisiae. Bba-Mol Cell Res. 1773:1311–1340.
- Fan L, An G, Wang S, Chen X, Liu Y, Liu Z, Ma Q, Wang J. 2019. Circular RNA expression profiling and selection of Key Circular RNAs in the hypothalamus of heat-acclimated rats. Front Physiol. 10:1112. doi:10.3389/fphys.2019.01112.
- Fan Q, Meng X, Liang H, Zhang H, Liu X, Li L, Li W, Sun W, Zhang H, Zen K, Zhang CY. 2016. miR-10a inhibits cell proliferation and promotes cell apoptosis by targeting BCL6 in diffuse large B-cell lymphoma. Protein Cell. 7:899–912. doi:10.1007/s13238-016-0316-z.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114. doi:10.1038/nrg2290.
- Hang K, Ye CY, Chen EM, Zhang W, Xue DT, Pan ZJ. 2018. Role of the heat shock protein family in bone metabolism. Cell Stress Chaperon. 23:1153–1164.
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature. 495:384–388. doi:10.1038/nature11993.
- Hao Y, Liu JR, Zhang Y, Yang PG, Feng YJ, Cui YJ, Yang CH, Gu XH. 2016. The microRNA expression profile in porcine skeletal muscle is changed by constant heat stress. Anim Genet. 47:365–369. doi:10.1111/age.12419.
- Huang J, Zhao J, Zheng Q, Wang S, Wei X, Li F, Shang J, Lei C, Ma Y. 2019. Characterization of Circular RNAs in Chinese buffalo (bubalus bubalis) adipose tissue: A focus on Circular RNAs involved in Fat deposition. Animals (Basel). 9, doi:10.3390/ani9070403.
- Huang M, Shen Y, Mao H, Chen L, Chen J, Guo X, Xu N. 2018. Circular RNA expression profiles in the porcine liver of two distinct phenotype pig breeds. Asian-Australas J Anim Sci. 31:812–819. doi:10.5713/ajas.17.0651.
- Kisliouk T, Cramer T, Meiri N. 2014. Heat stress attenuates new cell generation in the hypothalamus: a role for miR-138. Neuroscience. 277:624–636. doi:10.1016/j.neuroscience.2014.07.047.
- Kisliouk T, Meiri N. 2013. MiR-138 promotes the migration of cultured chicken embryonic hypothalamic cells by targeting reelin. Neuroscience. 238:114–124. doi:10.1016/j.neuroscience.2013.02.020.
- Kunde SA, Rademacher N, Tzschach A, Wiedersberg E, Ullmann R, Kalscheuer VM, Shoichet SA. 2013. Characterisation of de novo MAPK10/JNK3 truncation mutations associated with cognitive disorders in two unrelated patients. Hum Genet. 132:461–471. doi:10.1007/s00439-012-1260-5.
- La Y, Tang J, Di R, Wang X, Liu Q, Zhang L, Zhang X, Zhang J, Hu W, Chu M. 2019. Differential expression of Circular RNAs in polytocous and monotocous uterus during the reproductive cycle of sheep. Animals (Basel). 9, doi:10.3390/ani9100797.
- Li A, Huang WL, Zhang XX, Xie LL, Miao XY. 2018. Identification and Characterization of CircRNAs of Two Pig breeds as a New biomarker in metabolism-related diseases. Cell Physiol Biochem. 47:2458–2470.
- Li YX, Feng XP, Wang HL, Meng CH, Zhang J, Qian Y, Zhong JF, Cao SX. 2019. Transcriptome analysis reveals corresponding genes and key pathways involved in heat stress in Hu sheep. Cell Stress Chaperones. 24:1045–1054. doi:10.1007/s12192-019-01019-6.
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P. 2015. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 22:256–264. doi:10.1038/nsmb.2959.
- Liu L, Wang J, Khanabdali R, Kalionis B, Tai X, Xia S. 2017. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 14:1715–1721. doi:10.1080/15476286.2017.1367886.
- Lucy MC, Safranski TJJMR. 2017. Heat stress in pregnant sows: thermal responses and subsequent performance of sows and their offspring. Mol Reprod Dev. 84(9):946–956.
- Luo M, Li L, Xiao C, Sun Y, Wang GL. 2016. Heat stress impairs mice granulosa cell function by diminishing steroids production and inducing apoptosis. Mol Cell Biochem. 412:81–90. doi:10.1007/s11010-015-2610-0.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495:333–338.
- Mortzfeld BM, Taubenheim J, Klimovich AV, Fraune S, Rosenstiel P, Bosch TCG. 2019. Temperature and insulin signaling regulate body size in hydra by the Wnt and TGF-beta pathways. Nat Commun. 10:3257. doi:10.1038/s41467-019-11136-6.
- Prunier A, de Braganca MM, Le Dividich J. 1997. Influence of high ambient temperature on performance of reproductive sows. Livest Prod Sci. 52:123–133.
- Qin ML, Liu G, Huo XS, Tao XM, Sun XM, Ge ZH, Yang J, Fan J, Liu L, Qin WX. 2016. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 16:161–169.
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M. 2015. Circular RNAs in the mammalian brain Are highly abundant, conserved, and dynamically expressed. Mol Cell. 58:870–885.
- Sai L, Li L, Hu C, Qu B, Guo Q, Jia Q, Zhang Y, Bo C, Li X, Shao H, Ng JC. 2018. Identification of circular RNAs and their alterations involved in developing male xenopus laevis chronically exposed to atrazine. Chemosphere. 200:295–301.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504.
- Silanikove N. 2000. Effects of heat stress on the welfare of extensively managed domestic ruminants. Livest Prod Sci. 67:1–18.
- St-Pierre NR, Cobanov B, Schnitkey G. 2003. Economic losses from heat stress by US Livestock industries. J Dairy Sci. 86:E52–E77.
- Sun H, Jiang R, Xu S, Zhang Z, Xu G, Zheng J, Qu L. 2015. Transcriptome responses to heat stress in hypothalamus of a meat-type chicken. J Anim Sci Biotechnol. 6:6. doi:10.1186/s40104-015-0003-6.
- Williams AM, Safranski TJ, Spiers DE, Eichen PA, Coate EA, Lucy MC. 2013. Effects of a controlled heat stress during late gestation, lactation, and after weaning on thermoregulation, metabolism, and reproduction of primiparous sows. J Anim Sci. 91:2700–2714. doi:10.2527/jas2012-6055.
- Xie Y, Liu Y, Fan X, Zhang L, Li Q, Li S, Wang H, Xiao Y. 2019. MicroRNA-21 promotes progression of breast cancer via inhibition of mitogen-activated protein kinase10 (MAPK10). Biosci Rep. doi:10.1042/BSR20181000.
- Yang ZZ, Tschopp O, Di-Poi N, Bruder E, Baudry A, Dummler B, Wahli W, Hemmings BA. 2005. Dosage-dependent effects of Akt1/protein kinase Balpha (PKBalpha) and Akt3/PKBgamma on thymus, skin, and cardiovascular and nervous system development in mice. Mol Cell Biol. 25:10407–10418. doi:10.1128/MCB.25.23.10407-10418.2005.
- Yoo KH, Park YK, Kim HS, Jung WW, Chang SG. 2011. Identification of MAPK10 as a novel epigenetic marker for chromophobe kidney cancer. Pathol Int. 61:52–54. doi:10.1111/j.1440-1827.2010.02605.x.
- Yu TY, Yong YH, Li JY, Fang B, Hu CY, Wu LY, Liu X, Yu Z, Ma X, Patil Y, Gooneratne R. 2020. Proteomic study of hypothalamus in pigs exposed to heat stress. BMC Vet Res. 16, doi:10.1186/s12917-020-02505-1.
- Zhang H, Hu B, Xiong J, Chen T, Xi Q, Luo J, Jiang Q, Sun J, Zhang Y. 2019. Genomewide analysis of circular RNA in pituitaries of normal and heat-stressed sows. BMC Genomics. 20:1013. doi:10.1186/s12864-019-6377-7.
- Zhang Z, Tang J, He X, Zhu M, Gan S, Guo X, Zhang X, Zhang J, Hu W, Chu M. 2019. Comparative transcriptomics identify Key hypothalamic Circular RNAs that participate in sheep (ovis aries) reproduction. Animals (Basel). 9, doi:10.3390/ani9080557.