ABSTRACT
This study’s aim was to detect NR5A2 expression and explore association between single nucleotide polymorphism (SNP) of the NR5A2 gene with reproductive traits in Jiaxing Black (JXB) sows. The expression analysis showed that the expression level of pig NR5A2 mRNA in the ovary and liver was significantly higher (p < 0.01) compared to other tissues. 10 SNPs were detected in the exons of NR5A2, and five SNPs (g.136706A > T, g.84609G > A, g.136759G > C, g.138802T > C, g.138864A > G) met Hardy Weinberg equilibrium conditions. The total number of piglets born (TBA) and number of still born piglets (NSB) of sows with GG genotype at the g.136759G > C loci were higher than the CC genotype (P < 0.05). For the g.138864A > G loci, individuals with the AA genotype had significantly higher TBA than those of sows with the AG genotype (P < 0.05). By combining genotypes, it was found that the H7H8 diplotype had the best TBA performance. Moreover, the g.138864A > G and g.136759G > C loci had significant effects on the NR5A2 mRNA expression level in the ovary. The above results indicated that SNPs within NR5A2 exons of the NR5A2 gene have a certain effect on the reproductive traits of JXB pigs.
Introduction
NR5A2 (Nuclear receptor subfamily 5, group A, member 2), also known as fetoprotein transcription factor (FTF) and liver receptor homolog-1 (LRH-1), is an important orphan receptor belonging to the NR5A subfamily of nuclear receptors. LRH-1 was first extracted from the liver of mice, and subsequent studies have reported that LRH-1 is significantly more expressed in the ovary than in the liver or other tissues (Falender et al. Citation2003; Liu et al. Citation2003). LRH-1 was later isolated from zebrafish, rats, chickens, horses, frogs and humans. Studies have elicited mRNA expression of NR5A2 in the adult ovary, more precisely in granulocytes and luteal cells rather than follicular membrane cells and interstitial cells (Falender et al. Citation2003). Further research on the characterization of the imperative and indispensable function of LRH-1 and its extensive regulatory network in the initiation of ovulation has confirmed that LRH-1 is a nuclear receptor that can be targeted by agonists or antagonists, with a considerable potential for the regulation of fertility and the treatment of infertility (Bianco et al. Citation2019). Furthermore, LRH-1 depletion has been found to prevent ovulation, cumulus expansion, and luteinization, whereas uterine depletion of LRH-1 compromises decidualization and pregnancy (Meinsohn et al. Citation2019). Aromatase, encoded by Cyp19a1, catalyzes the conversion of androgens into estrogens, which is crucial for female reproduction in mammals and other vertebrates (Shao et al. Citation2015). As a matter of fact, studies demonstrate that LRH-1 plays an essential role in upregulation of the cyp19a1a gene in the ovarian follicular cells during vitellogenesis, and the sequestration of LRH-1 in the cytoplasm may downregulate cyp19a1a expression in the mature ovary (Lu et al. Citation2014).
Relevant research indicates that despite the fact that NR5A2+/− mice have normal follicular development, ovulation, and estrogen production, they exhibit an altered luteal function (Labelle-Dumais et al. Citation2007). However, LRH-1 may also be a multifunctional steroidogenic factor in ovarian physiology (Liu et al. Citation2003). Within the follicle, NR5A2 appears to be regulated by FSH and LH, whereas its expression in the corpus luteum is regulated by prolactin. Additionally, NR5A2 has been proven to control steroid hormone biosynthesis by regulating the expression of genes related to steroidogenesis, such as the steroidogenic acute regulatory protein (STAR) (Sirianni et al. Citation2002), cholesterol side-chain cleavage (Cyp11a1) (Saxena et al. Citation2004), 17α-hydroxylase (Liu et al. Citation2003), lyase (Cyp17) (Annicotte et al. Citation2003), 3β-hydroxysteroid dehydrogenase (3β-Hsd) (Sirianni et al. Citation2002; Saxena et al. Citation2004), and 11β-hydroxylase (Cyp11b1) (Sirianni et al. Citation2002). It is worth mentioning that a 3.5 folds decrease in the expression of LRH-1 within the ovary of this kind of infertile mouse was detected (Freiman et al. Citation2001). Moreover, this conclusion is supported by in vivo experiments (mice). Further experimental results show that the reduced reproductive ability of NR5A2−/+ females arises from a reduction in circulating progesterone concentrations and can be remedied by exogenous progesterone supplementation (Labelle-Dumais et al. Citation2007). Thus, the high expression of NR5A2 in the gonads and its ability to regulate the expression of genes related to steroid synthesis point out the importance of NR5A2 on the reproductive function. The gene expression of NR5A2 in the mSSCsl (Mouse spermatogonial stem cells long-term propagated) was markedly higher in comparison to that in the mSSCsf (freshly isolated Mouse spermatogonial stem cells) (Bai et al. Citation2016). Thus, NR5A2 seems to promote the proliferation of mSSCsl. NR5A2 expression in mural granulosa cells of preovulatory follicles is essential for cumulus expansion; NR5A2 regulates cumulus expansion by controlling the expression of various genes (Bertolin et al. Citation2017). The correlation between NR5A2 and reproductive traits was confirmed by the results of the above experiments.
The current research on NR5A2 is focused on its link with human diseases such as intestinal disorders (Enteritis, intestinal cancer), breast cancer, and immune system diseases, diabetes, metabolic disease, and its regulating function on the liver. However, to the best of our knowledge, studies on NR5A2 in domestic animals are rare, let alone in pigs. Currently, related studies have discovered that different regions of the NR5A2 gene in Hu sheep exhibit polymorphisms related to the reproductive traits of Hu sheep (Li et al. Citation2015, Citation2019). Li et al. (Citation2019) reported that nucleotide polymorphisms were associated with the litter size of Hu sheep based on amplified sequence of the NR5A2 promoter. Li et al. (Citation2015) also found that single nucleotide polymorphisms (SNPs) in the coding region of the NR5A2 gene was associated with the reproductive performance of Hu sheep. Given the findings reported by the above studies, it is logic to conclude that there is a correlation between the NR5A2 gene and reproductive traits. Nonetheless, there is a limited amount of literature on the association of the NR5A2 gene with breeding traits in livestock. Moreover, in a recent study (Guo et al. Citation2020) reported that enhancing the expression of NR5A2 can stimulate progesterone synthesis in pigs. In early porcine embryo development, LRH1 inhibition upregulated the expression of the proapoptotic Bax and Casp3 genes, and decreased the expression levels of Bcl2, an anti-apoptotic gene (Guo et al. Citation2016).
To date, no research has been reported on NR5A2 gene polymorphisms related to pig reproductive traits. JXB pigs, a famous local breed in China, are known for their significant characteristics of high prolificacy, sexual precocity, high adaptation and roughage-resistance and so on (Wu et al. Citation2019). Based on these features, JXB pigs were used in this study as test animals to analyse the effect of the NR5A2 gene’s marker loci on their reproductive traits. In order to explore the association of SNPs of the NR5A2 gene with JXB pigs reproductive traits, and to find the molecular markers related to the reproductive traits of JXB pigs, the polymorphism loci within the NR5A2 gene in JXB pigs were detected by means of direct sequencing. Meanwhile, the link between different genotypes and diplotypes of a single polymorphic locus and the productive traits of JXB pigs was investigated.
Materials and methods
Animal and trait data collection
This experiment was conducted following the Chinese guidelines for animal welfare and approved by the animal welfare committee of Zhejiang University (Approval Number: 12969). All of the animal procedures were performed according to the Chinese guidelines for animal welfare and approved by Zhejiang University (Hangzhou, China). We chose JXB pigs fed by Zhejiang Qinglian Food Co., Ltd (Haining, China) as experimental subjects. Samples of ear tissues from 128 Jinxing sows were collected, soaked in 75% alcohol and stored at −20°C for DNA extraction. Eleven tissues (large intestine, kidney, lung, small intestine, heart, liver, longissimus dorsi, fat, ovary, hypophysis, hypothalamus) from five Jiaxing black sows were used for NR5A2 Tissue expression profile of the gene. Ovaries of Jiaxing black sows with g.138802T > C and g.138864A > G mutation loci were taken (five each of six genotypes). Data on the reproductive traits of 128 sows including the total number of piglets born (TBA), number of piglets born alive (NBA), Number of stillborn piglet (NSB), and litter weight at birth (LWB) were recorded and organized.
DNA and RNA extraction, cDNA synthesis
Fresh samples of tissues ear, large intestine, fat, heart, longissimus dorsi, kidney, liver, lung, ovary, small intestine, hypophysis and hypothalamus were frozen in liquid nitrogen immediately after being separated, then stored at −80°C. The total RNA content was isolated from the tissues using a TRIzol A+ Total RNA regent (TIANGEN, Beijing, China) according to the instructions of manufacture. The amount and purity of the extracted RNA were measured by the NanoDropND2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and the quality of the obtained RNA was checked by electrophoresis (Zhang et al. Citation2020). The Prime ScriptTM RT Reagent Kit with a gDNA Eraser (Takara Biotechnology Co. Ltd., Dalian, China) was used to convert RNA to cDNA following the manufacturer’s protocol.
Primer design, PCR amplification and sequencing
Genomic DNA was isolated from ear tissues using standard phenol/chloroform extraction and stored at −20°C. According to the Sus scrofa NR5A2 gene sequence (GenBank accession NC_010452), primers were designed by the Primer software version 5.0. These primers were used to amplify extron 1–10 (Table S1), with extron10 designed by 4 primers.
The PCR reaction mixture (25 μL) consisted of a 12.5 μL 2 × Taq Master Mix, 1 μL of each primer, and 2 μL template DNA. PCR was carried out with an initial denaturing step at 94°C for 5 mins and then 35 cycles at 94°C for 40s, 55°C for 30 s and 72°C for 1 min; with a final extension at 72°C for 4 mins. After that, the PCR products were distinguished using 1% agarose gel electrophoresis with a 2000bp DNA marker.
All the PCR products were purified with an agarose gel DNA recovery kit (Tiangen, Beijing, China) and the purified products were sent to Qingke Yuxi Biotechnology Co., Ltd (Hangzhou, China) for sequencing. The sequencing results were analysed using the SeqMan software to spot polymorphism sites.
Relative expression of NR5A2 mRNA
The NR5A2 gene’s primer pairs included for real-time fluorescent quantitative PCR were designed using Primer-BLAST based on the pigs NR5A2 mRNA (NM_001267893.1). Primers were synthesized by Tsingke Biotech Co., Ltd. (Hangzhou, China).
Real-time PCR was performed using a StepOnePlus Real-Time PCR System (Applied Bio systems, Foster City, CA, USA) with a 20 µL PCR mixture including 2 µL of cDNA, 10 µL of 2 × SYBR Premix Ex TaqII, 0.4 µL ROX Reference Dye (50×) 0.8 µL of forward primer, 0.8 µL of reverse primer and 6 µL of nuclease-free water. The 2 × SYBR Premix Ex TaqII, ROX Reference Dye (50×) and nuclease-free water were contained in the TB Green Premix Ex Taq TM II (Takara, Dalian, China). The reactions were incubated in a 96-well optical plate (Applied Biosystems, American) at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 30 s. Each sample was analysed in triplicate. The mRNA expression levels were normalised to the mRNA expression of pig β-actin and calculated using the 2−ΔΔCt method (Livak and Schmittgen Citation2001).
Statistical analysis
A phylogenetic tree of 8 species was constructed using the maximum likelihood method based on the nucleotide sequence of the NR5A2 gene established with the MEGA7.0 software, with chicken as an outgroup in birds. Genotype, allele frequencies and population parameters including heterozygosity (He), effective allele numbers (Ne), polymorphism information content (PIC), and Hardy-Weinberg equilibrium (HWE) were computed with Pop Gene (Version1.3.2). Linkage disequilibrium of the regions that are in accordance with Hardy-Weinberg equilibrium were explored using the HAPLOVIEW software (Version 4.2).
The relationship between SNPs and four reproduction traits (TBA, NBA, NSB, LWB) of 128 JXB sows was determined using the SPSS software (Version 20.0), The analytical results were expressed as the mean ± SD. The Phase software (version2.1) was used to analyse haplotypes and haplotype combinations. The correlation between diplotypes and the reproduction traits in 128 JXB sows were also identified. HaploView was used to analyse the linkage disequilibrium (LD) and the general linear model (GLM) was as follows: Yijk=μ+Pi+Gj+Mk+Nh+eijkh, where Yijk is the individual observation of the reproduction traits, u is overall population mean, Pi is fixed effect of parity (i = 1,2), Gj is the effect of genotypes (j = 1,2or3) or the diplotypes (i = 1,2,3,4,5), Mk is the fixed effect of season, Nh is the sire effect and eijkh is the random residual error. The additive effects (a) of the NR5A2 gene were estimated as 1/2(BB-AA) and the dominance effect (d) as AB-1/2(AA + BB).
The relative expression data were checked for normality by the Kolmogorov–Smirnov test in the SPSS v20.0 software (IBM, Chicago, IL, USA), and logarithmic transformation (log2) was used to correct the non-normal distribution (Cao et al. Citation2019). Next, one-way ANOVA was implemented to evaluate the relative expression level of the NR5A2 gene among groups. Multiple comparison results of the groups were adjusted by Bonferroni correction.
Results
Phylogenetic relationships of NR5A2 proteins among 8 species
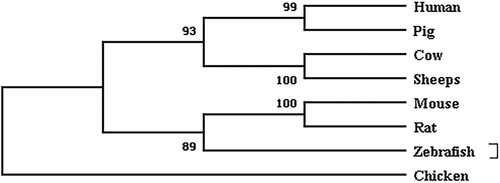
As illustrated in , the NR5A2 proteins of all 8 species fell under two subgroups. All of the mammalian and zebrafish sequences clustered into one subgroup and the chicken sequences clustered into another subgroup. It can be seen that the gene is highly homologous in several of these species.
Polymerase chain reaction amplification of NR5A2 gene exons
All 10 exons of the NR5A2 gene were successfully amplified using JXB sows’ DNA and 13 pairs of primers. Since exon 10 sequence was too long, we split it into four smaller segments for amplification, which were in turn NR5A2-Exon10-a, NR5A2-Exon10-b, NR5A2-Exon10-c, and NR5A2-Exon10-d. The sizes of the amplified fragments were approximately consistent with the target ones which were 743, 1107, 683, 820 bp in length (Figure S1), respectively. Splicing of exons and introns were consistent with the GT-AG rule (Wu et al. Citation2019).
Identification of polymorphisms in the NR5A2 gene and genetic parameters analysis
In this study, the results of sequencing and sequence comparative analysis revealed that there were ten mutations within the NR5A2 gene of JXB sows (Figure S2). One SNP named g.84609G > A was identified in exon 9 and the remaining SNPs were present in exon 10, namely: g.135751G > T, g.138806G > T, g.136703T > A, g.136706A > T, g.136759G > C, g.138771C > T, g.138933C > T, g.138802T > C, g.138864A > G. Among those that result in synonymous mutations are g.84609G > A (Arg→Arg), g.138771C > T (Pro→Pro), g.138864A > G (Cys→Cys); The missense mutations are g.135751G > T (Pro→Ser), g.136703T > A (Tyr→Cys), g.136706A > T (Phe→Tyr), g.136759G > C (Phe→Asp), g.138802T > C (Asn→Asp), g.138933C > T (Leu→Arg); The nonsense mutation is g.138806G > T (Ser→UAA). The gene heterozygosity (He), effective allele numbers (Ne) and polymorphism information content (PIC) of ten mutations were calculated, and the Chi-square test was used to determine whether or not the ten mutations in the population were in accordance with the Hardy-Weinberg equilibrium (). PIC values of g.84609G > A, g.135751G > T, g.136706A > T, g.136759G > C, g.138771C > T and g.138802T > C demonstrated that these SNPs had medium genetic diversity (0.25 < PIC < 0.5). The genotypic distribution of g.135751G > T, g.138806G > T, g.136703T > A, g.138771C > T and g.138933C > T deviated from the Hardy-Weinberg equilibrium (HWE) (p < 0.05), whereas the distribution of g.84609G > A, g.136706A > T, g.136759G > C, g.138802T > C, g.138864A > G followed the Hardy-Weinberg equilibrium (HWE) (p > 0.05), so we selected these mutations for further analysis.
Table 1. Genotypic frequency, allelic frequency and diversity parameter in the exons of the NR5A2 gene in JXB sows (MEAN ± SD).
Linkage disequilibrium analysis, haplotype and diplotype construction
D’/r2 linkage disequilibrium analysis indicated that five SNPs (g.135751G > T, g.138806G > T, g.136703T > A, g.138771C > T and g.138933C > T) exhibited a strong linkage disequilibrium (LD), thus constructing a 54 kb block1 (). Using the Phase software, we identified 14 haplotypes () and 17 haplotypes combinations (Table S2). H8 was the most abundant haplotypes (0.432) out of the 14 haplotypes, H8, H9, H11 and H12 accounted for 84% of the observations. Out of the 17 diplotypes, the frequencies of 12 diplotypes were lower than 0.03, which means that the remaining five diplotypes (H8H12, H8H11, H8H9, H6H8, and H7H8) were further analysed.
Figure 2. Analysis of blocks with 5 SNPs in the NR5A2 gene in JXB sows.
Note: The values in the squares indicate the paired linkage disequilibrium (LD) values (D’) of the SNPs. When D’ = 1, the values will not be displayed. The redder the square, the stronger the LD. The haplotype block was defined using the default setting of the Haploview software.
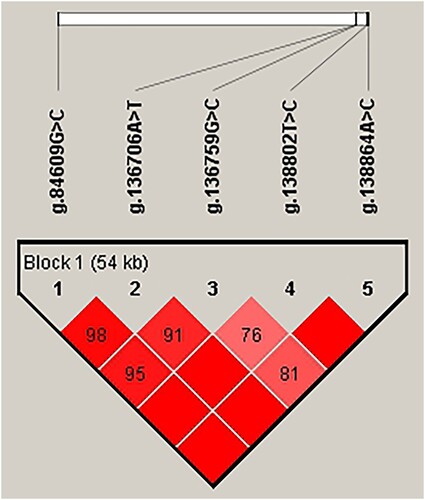
Table 2. Haplotypes based on the block 1 and frequencies in JXB sows.
Association of NR5A2 polymorphisms with reproductive traits
The association of five NR5A2 SNPs with the reproductive traits of JXB sows is displayed in . At the g.136759G > C SNP, GG subjects produced more pigs per litter in terms of TBA and NBA compared to the CC sows (p < 0.05). Animal subjects with the AA genotype in g.138864A > G had higher NBA values than those with AG genotypes (p < 0.05) and no statistically significant difference was identified for the other traits including TBA, WPB and NDB. Besides, g.84609G > A, g.136706A > T, and g.138802T > C were not correlated with any trait. Further association analysis on diplotypes was delineated in . Sows with H8H11 diplotypes had lower TBA values than those with H7H8 diplotypes (p < 0.05). There were no statistically significant difference across the variou diplotypes with respect to other traits such as TNB, NBA, NSB and LWB. Hence, as far as the TBA phenotype is concerned, the H7H8 diplotype can be regarded as the best diplotype in this population.
Table 3. Association of five SNPs of the NR5A2 gene with reproductive traits in Jiaxing Black sows (MEAN ± SD).
Table 4. Association of diplotypes of the NR5A2 gene with reproductive traits in Jiaxing Black sows (MEAN ± SD).
Relative expression levels of pig NR5A2 mRNA in 11tissues
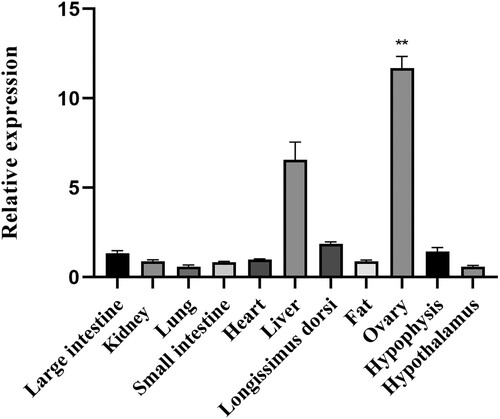
The relative expression results confirmed that NR5A2 mRNA was widely expressed in the large intestine, fat, heart, longissimus dorsi, kidney, liver, lung, ovary, small intestine, hypophysis and hypothalamus, but the level of expression was subject to tissue-specific regulation. The mRNA expression levels of pigs were significantly higher (p < 0.01) in the ovary and liver compared to other tissues ().
NR5A2 mRNA relative expression with different genotypes in the ovary
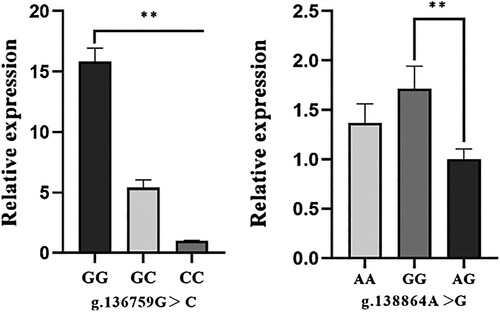
The ovary was selected to detect the expression levels of different genotypes of the (g.136759G > C, g.138864A > G) loci that were significantly associated with reproduction traits, since it exhibited the highest NR5A2 mRNA relative expression levels. The results depicted that g.136759G > C and g.138864A > G were both significantly associated with the expression level of the NR5A2 gene. As laid out in , for the g.136759G > C locus, ovaries with the GG genotype had higher (p < 0.01) NR5A2 mRNA expression levels than those with CC and GC genotypes. Similarly, ovaries with the AA genotype had higher (p < 0.01) NR5A2 mRNA expression levels than those with GG and AG genotypes in the g.138864A > G locus.
Figure 4. Relative expression levels of the pig NR5A2 mRNA in ovary with different genotypes.
Note: In the loci of g.136759G > C, g.138864A > G with the average ΔCt value of CC and AG genotype as the calibrator, respectively. Data were shown as the mean ± SD for 5 pigs of each genotype. The column with ‘**’ showed a significant difference (p < 0.01).
Discussion
Research on the physiological function of the NR5A2 gene is now well established. LRH-1, a member of the lone nuclear receptor superfamily, plays an important role in pigs’ early embryonic development (Guo et al. Citation2016). Sladitschek found that deletion of the NR5A2 gene severely impaired ectoderm differentiation in mice. The results (Labelle-Dumais et al. Citation2007) showed that during lutealization and natural gestation, NR5A2-/- mice experienced a significant decrease in progesterone production compared to wild type mice, which in turn led to a decrease in the fertility of the mice, undoubtedly indicating that this gene plays an important role in reproduction. Being two closely related homologs in the NR5A subfamily, SF-1 (NR5A1) and LRH-1 (NR5A2) (Fayard et al. Citation2004) have been previously found to be expressed in granulocytes (Falender et al. Citation2003). Both NR5A1 and NR5A2 play similar roles in the production of steroids in mouse granulocytes (Saxena et al. Citation2007). Recent studies have highlighted the potential application of the NR5A2 gene in the regulation of goat estrus, which implies that NR5A2 plays a key role in the goat ovary and is essential for luteinogenesis (An et al. Citation2018). Furthemore, C > G mutation was detected at the core promoter region-388 of the NR5A1 gene in Hu sheep, and the association analysis indicated that the number of second and third lambs of GC-type ewes was higher than that of CC-type ewes (P < 0.05) (Li et al. Citation2018). It was also found that NR5A2 mRNA levels were positively correlated with Hu sheep spawning rate and the number of lambs born. Two single nucleotide polymorphisms (T4OC and T1419C) were detected in the NR5A2 coding sequence. Also, at the third litter and average litter size, ewes with the CC genotype at the T40C locus produced more lambs than ewes with the TT genotype. Li et al. (Citation2019) amplified the NR5A2 gene’s promoter sequence and identified its core promoter region as −721 nt to −281 nt. An AT > G polymorphism of −700 nt was depicted in the core promoter region and a subsequent correlation analysis found that the second and average lambing numbers were significantly higher in GG ewes than in TG and TT ewes, with the third lambing number being significantly higher in GG ewes than in TT ewes.
In this study, 10 SNP loci were detected on the NR5A2 gene of JXB pigs, five of which were closely associated with their reproductive traits. According to the results obtained from the χ2 test, g.135751G > T, g.138806G > T, g.136703T > A, g.138771C > T, and g.138933C > T were not in agreement with the HWE, which might be due to the artificial selection in the breed. The results showed that five SNPs (g.84609G > A, g.136706A > T, g.136759G > C, g.138802T > C, g.138864A > G) were associated with the reproductive traits of JXB sows, but three of them (g.84609G > A, g.136706A > T and g.138802T > C) elicited no significant differences in breeding traits with respect to different genotypes. Indeed, even though there was no significant difference between the combined haplotype of these SNPs and relevant traits, the genotypic effect of one SNP may be influenced by other SNPs through interactions (Ren et al. Citation2014). The loci of g.136759 G > C and g.138864A > G had significant effects on the NR5A2 mRNA expression levels in the ovary, which further supports the NR5A2 gene’s influence on the reproductive traits of JXB sows. Given that the g.84609G > A, g.138933C > T, g.138802T > C, and g.138864A > G of NR5A2 gene exons 9 and 10 were of synonymous mutations, they cannot result in the change of amino acid. It is worth noting that previous studies have demonstrated that LRH-1 plays a vital role in sperm motility, survival, and cholesterol efflux. Additionally, researchers have speculated that the role of LHR-1 in sperm cells is tightly coupled with estrogen signalling, supporting the idea that LRH-1 acts as a downstream effector of the estradiol pathway influencing certain functions of sperm cells (Montanaro et al. Citation2015). It has been shown that SNPs can affect gene expression through splicing regulation, which seems to explain this phenomenon (Kelemen et al. Citation2013). Moreover, studies have reported the role of synonymous mutations on gene function and phenotype, such as synonymous mutations in the goat gene POU1F1, which are associated with milk production and new born weight (Ren et al. Citation2014). Consequently, identifying the mechanism of association between these synonymous mutations and the reproductive traits in JXB sows is an interesting work. Furthermore, the litter size of H7H8 diplotype JXB sows was significantly higher than that of H8H11 diplotype JXB sows, albeit no statistically significant discrepancy was detected relative to other diplotypes, which could be due to the low number of sows in some haplotype classes. In conclusion, based on this experiment’s results, the NR5A2 gene can be used as a target gene in livestock breeding.
The latest research articles report that NR5A2 is a downstream transcription factor of Notch signalling that regulates progesterone synthesis, and thus the NR5A2 gene is involved in suppressing Notch signalling to promote progesterone secretion by enhancing the expression of the NPC1 (NPC intracellular cholesterol transporter 1) and STAR (steroidogenic acute regulatory protein) (Guo et al. Citation2020). The NR5A2 gene core promoter-700T/G polymorphism produced a new transcription factor MTF-1 binding site that upregulated the transcriptional activity and mRNA expression of the NR5A2 gene in granulosa cells, thereby increasing the litter size of Hu sheep; presumably by regulating the steroid synthesis of NR5A2, which in turn affected ovulation and litter size (Li et al. Citation2019). This study is the first to report a correlation between the NR5A2 gene and reproductive performance in pigs, but further research is necessary to determine the exact molecular mechanism through which the polymorphism of the NR5A2 gene affects reproductive traits in pigs. All of the aforementioned evidence is enough to prove that NR5A2 polymorphisms may be used as a potential genetic marker to optimise the breeding of JXB pigs.
Conclusion
In a nutshell, this study demonstrated that the NR5A2 mRNA expression level in the ovary was the highest, and polymorphisms of the NR5A2 gene were significantly associated with the reproductive traits of Jiaxing Black pigs, which indicates that the NR5A2 gene can be used for molecular marker-assisted selection of reproductive traits in JXB pigs and to provide a theoretical basis to optimise the breeding of JXB pigs.
TAAR_2020124_Supplement
Download MS Word (614.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- An X, Ma H, Han P, Zhu C, Cao B, Bai Y. 2018. Genome-wide differences in DNA methylation changes in caprine ovaries between oestrous and dioestrous phases. J Anim Sci Biotechnol. 9:85.
- Annicotte JS, Fayard E, Swift GH, Selander L, Edlund H, Tanaka T, Kodama T, Schoonjans K, Auwerx J. 2003. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol Cell Biol. 23:6713–6724.
- Bai Y, Feng M, Liu S, Wei H, Li L, Zhang X, Shen C, Zhang S, Ma N. 2016. Differential gene expression in mouse spermatogonial stem cells and embryonic stem cells. Int J Mol Med. 38:423–432.
- Bertolin K, Meinsohn MC, Suzuki J, Gossen J, Schoonjans K, Duggavathi R, Murphy BD. 2017. Ovary-specific depletion of the nuclear receptor Nr5a2 compromises expansion of the cumulus oophorus but not fertilization by intracytoplasmic sperm injection. Biol Reprod. 96:1231–1243.
- Bianco S, Bellefleur AM, Beaulieu E, Beauparlant CJ, Bertolin K, Droit A, Schoonjans K, Murphy BD, Gevry N. 2019. The ovulatory signal precipitates LRH-1 transcriptional switching mediated by differential chromatin accessibility. Cell Rep. 28:2443–2454.e4.
- Cao H, Dong X, Mao H, Xu N, Yin Z. 2019. Expression analysis of the PITX2 gene and associations between its polymorphisms and body size and carcass traits in chickens. Animals (Basel). 9.
- Falender AE, Lanz R, Malenfant D, Belanger L, Richards JS. 2003. Differential expression of steroidogenic factor-1 and FTF/LRH-1 in the rodent ovary. Endocrinology. 144:3598–3610.
- Fayard E, Auwerx J, Schoonjans K. 2004. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 14:250–260.
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. 2001. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 293:2084–2087.
- Guo J, Zhao MH, Liang S, Choi JW, Kim NH, Cui XS. 2016. Liver receptor homolog 1 influences blastocyst hatching in pigs. J Reprod Dev. 62:297–303.
- Guo R, Chen F, Shi Z. 2020. Suppression of notch signaling stimulates progesterone synthesis by enhancing the expression of NR5A2 and NR2F2 in porcine granulosa cells. Genes (Basel). 11.
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing. Gene. 514:1–30.
- Labelle-Dumais C, Pare JF, Belanger L, Farookhi R, Dufort D. 2007. Impaired progesterone production in Nr5a2+/- mice leads to a reduction in female reproductive function. Biol Reprod. 77:217–225.
- Li Y, Zhang J, Qian Y, Meng C, Wang H, Cao S. 2018. Mutation -388 C>G of NR5A1 gene affects litter size and promoter activity in sheep. Anim Reprod Sci. 196:19–27.
- Li Y, Zhang J, Qian Y, Meng C, Wang H, Zhong J, Cao S. 2019. A T>G mutation in the NR5A2 gene is associated with litter size in Hu sheep through upregulation of promoter activity by transcription factor MTF-1. Front Genet. 10:1011.
- Li YX, Zhang J, Qian Y, Meng CH, Wang HL, Tao XJ, Zhong S, Cao SX, Li QF. 2015. Molecular characterization, expression, polymorphism of NR5A2 and its relationship with litter size in Hu sheep. Genet Mol Res. 14:12765–12775.
- Liu DL, Liu WZ, Li QL, Wang HM, Qian D, Treuter E, Zhu C. 2003. Expression and functional analysis of liver receptor homologue 1 as a potential steroidogenic factor in rat ovary. Biol Reprod. 69:508–517.
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
- Lu H, Zhang S, Liu Q, Zhang L, Zhang W. 2014. Cytoplasmic localization of Lrh-1 down-regulates ovarian follicular cyp19a1a expression in a teleost, the orange-spotted grouper Epinephelus coioides. Biol Reprod. 91:29.
- Meinsohn MC, Smith OE, Bertolin K, Murphy BD. 2019. The orphan nuclear receptors steroidogenic factor-1 and liver receptor homolog-1: structure, regulation, and essential roles in mammalian reproduction. Physiol Rev. 99:1249–1279.
- Montanaro D, Santoro M, Carpino A, Perrotta I, De Amicis F, Sirianni R, Rago V, Gervasi S, Aquila S. 2015. Human sperm liver receptor homolog-1 (LRH-1) acts as a downstream target of the estrogen signaling pathway. J Anat. 227:541–549.
- Ren G, Huang YZ, Wei TB, Liu JX, Lan XY, Lei CZ, Zhang C-L, Zhang Z-Y, Qi X-L, Chen H. 2014. Linkage disequilibrium and haplotype distribution of the bovine LHX4 gene in relation to growth. Gene. 538:354–360.
- Saxena D, Escamilla-Hernandez R, Little-Ihrig L, Zeleznik AJ. 2007. Liver receptor homolog-1 and steroidogenic factor-1 have similar actions on rat granulosa cell steroidogenesis. Endocrinology. 148:726–734.
- Saxena D, Safi R, Little-Ihrig L, Zeleznik AJ. 2004. Liver receptor homolog-1 stimulates the progesterone biosynthetic pathway during follicle-stimulating hormone-induced granulosa cell differentiation. Endocrinology. 145:3821–3829.
- Shao X, Guo Y, Xu X, Zheng Y, Wang J, Chen Z, Huang J, Huang P, Cai J, Wang X. 2015. The CYP19 RS4646 polymorphism IS related to the prognosis of stage I-II and operable stage III breast cancer. PLoS One. 10:e0121535.
- Sirianni R, Seely JB, Attia G, Stocco DM, Carr BR, Pezzi V, Rainey WE. 2002. Liver receptor homologue-1 is expressed in human steroidogenic tissues and activates transcription of genes encoding steroidogenic enzymes. J Endocrinol. 174:R13–R17.
- Wu F, Zhang W, Song QQ, Li HH, Xu MS, Liu GL, Zhang JZ. 2019. Association analysis of polymorphisms of G protein-coupled receptor 54 gene exons with reproductive traits in Jiaxing Black sows. Asian-Australas J Anim Sci. 32:1104–1111.
- Zhang W, Chen Q, Xu L, Cai J, Zhang J. 2020. The potential role of PSMA6 in modulating fat deposition in pigs by promoting preadipocyte proliferation and differentiation. Gene. 145228.