ABSTRACT
Rhipicephalus microplus causes huge losses in cattle. Host genetic background greatly affects the immune efficiency in resistance or susceptibility to tick infestation, which is one of the many factors that play a role on that trait. We performed a systematic review of genome-wide association studies (GWAS) for tick resistance in cattle resulting in 1353 candidate genes for post-GWAS analyses. From those, genes showing possible structural variants from the bovine genome were classified by the Variant Effect Predictor from Ensembl. Ninety-two candidate genes showed potential structural variants in 5′ UTR and coding region and were used for functional annotation. Enriched biological processes (e.g. regulation of eosinophil chemotaxis, RIG-I signalling pathway and monocyte differentiation) and candidate genes (e.g. DAPK2, PUM1, ACIN1, INPP5D) linked with immune system function were identified and thus associated with tick resistance. Besides, gene-transcription factors (TFs) networks were obtained from TFs associated with immune system (FOXO3, PPARG, STAT3, NFKB1, GATA3 and ARNT) and the candidate genes associated with tick resistance in cattle highlighted (e.g. OR4L1, PNP, LRRIQ1, GIMAP8, MYO6, MEP1A and LRFN2). Thus, promising candidate genes with a possible functional role for tick resistance in cattle are presented for further in vitro and/or in vivo analyses.
1. Introduction
Cattle production is greatly impacted by the ectoparasite Rhipicephalus microplus infestation, which deeply impacts the animal health resulting in losses of meat, milk and leather production (Gueretz et al. Citation2020). Infestation control is largely based on the use of chemicals, which may include compounds with high toxicity and environmental persistence (Bai and Ogbourne Citation2016), and consequently harmful to human, animal and environment health. In addition, the recurrent and massive use of these compounds cause ticks to acquire resistance against the active principles used (Garcia et al. Citation2019), hindering the viability of their control. In this way, alternative strategies aiming to mitigate the losses caused by ectoparasites infestation must be offered for both, dairy and beef production.
It is known that Bos indicus breeds are genetically more resistant to ticks than Bos taurus breeds, probably due to naturally selected genes throughout their evolutionary process (Ayres et al. Citation2015). This indicates that resistance to tick infestation may be a potential factor to be studied in animal breeding programmes (Cardoso et al. Citation2014). Hence, genome-wide association studies (GWAS) identifying genomic regions associated with tick resistance (Neto et al. Citation2010; Porto Neto et al. Citation2010; Mapholi et al. Citation2016; Sollero et al. Citation2017; Otto et al. Citation2018) have been carried out. However, analyses of post-GWAS covering biological knowledge of the genes underlying this trait, as well as the possible potentially associated variants, are still scarce.
Post-GWAS analyses using gene networks have been carried out in the main livestock species including bovine (Verardo et al. Citation2016; Otto et al. Citation2018; Littiere et al. Citation2020). However, the use of these tools combined with sequencing data may be a promising way to identify the most candidate genes. After the advent of the next-generation sequencing technology, genome resequencing has increased the chances of finding variants in genes associated with important phenotypes (Schneeberger and Weigel Citation2011), and the structural variants are a promising class. Structural variants are usually determined as a DNA region of approximately 50 base pairs (bp) (Conrad et al. Citation2010) to 1 kb or more (Freeman et al. Citation2006). They can be classified into inversions and translocations, segmental duplications (SDs) and copy number variants (CNVs) which are unbalanced genomic events (Feuk et al. Citation2006).
A previous study described that CNVs might strongly influence gene expression through deletions/duplications changes in the position and function of cis-regulatory elements (Spielmann et al. Citation2018). For example, deletions variants located in genes involved with the human innate immune system affected the clinical infection caused by the Andean orthohantavirus (Ribeiro et al. Citation2019). Since the immune system is one of the important mechanisms that confers bovine resistance or susceptibility to tick infestation (Turner et al. Citation2022), it is possible that candidate genes underlying the immune response may show structural variants. Thus, in this study, we identified candidate genes for tick resistance in cattle that may present structural variants to perform functional annotations via gene networks (biological processes and transcription factors networks) and highlight the most candidate genes for further studies.
2. Materials and methods
2.1. Systematic review and identification of candidate genes
A systematic review was carried out in May 2020 to identify candidate genes related to tick resistance in cattle. We searched for articles using the Google Scholar search engine (https://scholar.google.com.br) in terms to maximize the search by not discriminating specifics databases, and thus assuming to consider all available articles database. The search consisted of combinations of keywords (in English) with the following criteria terms: (A) term related to the evaluated trait (‘tick resistance’, ‘tick’, ‘inflammatory response’); (B) type of association test (‘GWAS’, ‘genome wide association’) and (C) species/breed (‘Holstein’, ‘Gir’, ‘Girolando’, ‘cattle’). It is important to note that the use of breed term was first intended to search for a Brazilian national breed without compromising the final results, in terms that we used the ‘cattle’ keyword, and then focused on studies of all breeds. Two independent reviewers (1 and 2) searched for peer-reviewed papers using the queries described. Discrepancies in the judgment were resolved by consensus among the reviewers. The first step in selecting articles was to check whether the article contained (in the abstract, title or keywords) the keywords used in each combination. The articles that met these criteria were selected, and if duplicated, articles were removed. In addition, we confirmed that: (1) the article was published from a peer-reviewed journal; (2) GWAS was performed for traits related to tick resistance; (3) cattle were used as a model; (4) the consistent position of the gene and the methodology used were provided and (5) full text available. In addition, articles that did not provide enough information about the associated markers or windows (e.g. marker names and / or ‘rs’ and / or complete genomic coordinates) were also excluded from this step.
Each article was defined as a group for the identification of candidate genes. The positions of significant markers in each group were updated according to the ARS-UCD1.2 genome assembly, and the search for candidate genes carried out following the flanking region defined in each study. The search for the genes was carried out through the NCBI database (https://www.ncbi.nlm.nih.gov/gene/?term = bovine). A distance of 22 kb upstream and 22 kb downstream from each significant marker was considered for studies that did not show the flanking region for candidate genes identification, since it is the half of the average distance between markers in the SNPChip used in the studies. By checking this distance, we identified the presence of any gene that could be related with a QTL or a significant marker, as proposed by Verardo et al. (Citation2016).
2.2. Identification of structural variants associated with genes identified in systematic review
The public Ensembl database was used (‘ftp://ftp.ensembl.org/pub/release-101/variation/vcf/bos_taurus/’, accessed on November 18, 2020) to search bovine structural variants. Aiming to select target variants, we first classified the variants according to their potential function using the web interface of Ensembl Variant Effect Predictor (VEP) tool (McLaren et al. Citation2016), prioritizing structural variants located in the coding and non-coding regions of the bovine genome. Only variants associated with known genes identified in systematic review were selected in this step.
2.3. Functional analysis of candidate genes for tick resistance
From the sets of candidate genes that may present structural variants, we selected only the genes showing variants in the 5′ untranslated region (5′ UTR) and coding regions according to VEP classification. Thus, only genes containing such target variants were used for the construction of gene networks.
We first obtained a biological process network. From each group, we used the ClueGO application of Cytoscape (Bindea et al. Citation2009) to highlight biological processes associated with tick resistance. The analyses of ClueGO were based on a hypergeometric test with Bonferroni correction.
In addition, we also built a gene-transcription factor (gene-TF) network. Thus, the promoter sequences of the candidate genes of each group were analysed using the TFM-Explorer programme (http://bioinfo.lifl.fr/TFM/TFME) to search for transcription factors binding sites (TFBS). This programme uses weighting matrices from the JASPAR vertebrate database (Sandelin et al. Citation2004) to detect potential TFBS, calculating a scoring function as described in Touzet and Varré (Citation2007). From each group, the promoter region was defined by selecting the sequences at 3000 bp upstream and 300 bp downstream (FASTA format) from the transcription start site, based on the bovine genome assembly ARS-UCD1.2. This data was used as an input to TFM-Explorer. The list of transcription factors (TFs) obtained was analysed in Cytoscape 3.7.2 (Shannon et al. Citation2003) via BinGO plug-in (Maere et al. Citation2005) to determine which terms of gene ontology were significantly over-represented, assuming the standard analysis and multiple correction test.
Based on the biological processes associated with tick resistance, besides the literature review, the main TFs (key TFs) in each group related with the studied trait were selected. By this way, key TFs with known roles in biological processes related with tick resistance trait such as immune system were used to build gene-TFs networks. This approach has already been used in other species, including bovine (Verardo et al. Citation2016; Otto et al. Citation2018; Littiere et al. Citation2020), being that this judicious review of each TF is taken to avoid false associations between TF and the studied trait.
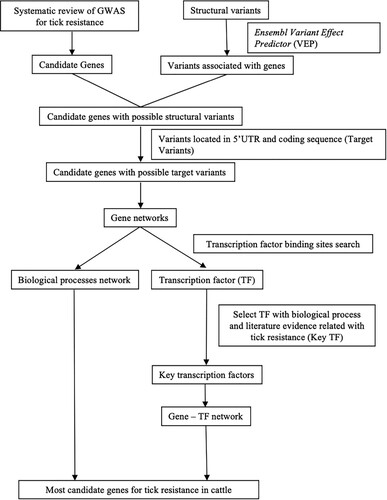
Through the Network Analyzer tool in Cytoscape v.3.7.2, promising candidate genes were highlighted according to the number of TFBS and, consequently, the number of connections/lines (edges) in each node (gene and TF), and the most connected genes in the gene-TF network were determined. Genes with more TFBS for the most representative key TFs were highlighted in the gene-TF network. Thus, the networks of biological processes and gene-TF highlighted candidate genes associated with tick resistance in cattle. A summary of whole analysis is shown in .
3. Results
3.1. Systematic review
The systematic review process is summarized in . The searches performed with 24 keyword combinations returned 43 articles, 23 found by reviewer 1 and 20 found by reviewer 2 (Supplementary Material – Table S1). Of those, 36 were duplicated between combinations of keywords and removed. The remaining seven articles proceeded to the full reading stage and three of them (Kim et al. Citation2015 ; Neto et al. Citation2011 ; Turner et al., Citation2010) were removed by not meeting the selection criteria (e.g. not be a tick resistance GWAS study or did not provide proper information about marker names and / or ‘rs’ and / or complete genomic coordinates). Thus, four articles were selected and defined as groups (from 1 to 4) for functional analysis, as shown in . Information about genotyping and quality control of each selected paper are presented in Supplementary Material – Table S2. Based on the significant SNPs/windows and their respective positions (bp) on the chromosomes described in each selected article, a total of 1353 candidate genes were identified (Supplementary Material – Table S3).
Figure 2. Flowchart showing the pipeline used to select articles based on specific criteria for systematic review.
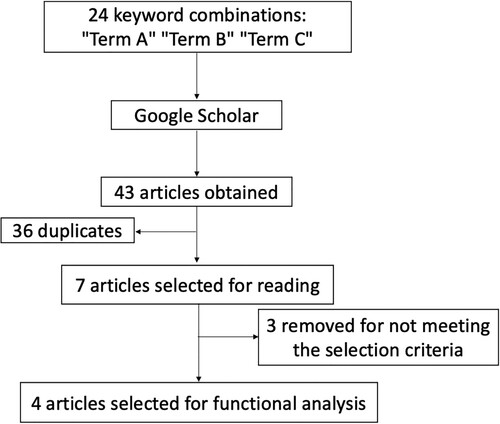
Table 1. Information of country, breed, biological samples, and traits selected from articles related to tick resistance in cattle.
3.2. Structural variants associated with genes
The Ensembl public database showed 462,845 structural variants in cattle. From them, 448,045 were classified according to their effect, using the VEP, which were associated with 14,593 genes. These variants were in gene regions such as 5′UTR, coding regions, introns and intergenic. The genes identified with these variants were used in the next step of the functional analyses.
3.3. Functional analysis of candidate genes for tick resistance
From the candidate genes identified on systematic review (1353) and those with possible structural variants identified by the VEP (14,593), a merged list was obtained with 626 remaining genes (Supplementary Material – Tables S4–S7). From them, considering the four groups, a total of 92 candidate genes showed possible target structural variants in the coding region and 5′UTR (Supplementary Material – Tables S8–S11). The target variants were classified as ‘modifier’ and ‘high’, showing impacts on the protein according to the VEP classification. In addition, we observed that the target variants located in candidate genes for tick resistance were defined as duplication, tandem duplication, gain of copy number, loss of copy number and deletion.
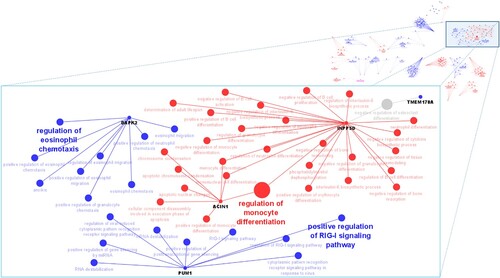
Each group of candidate genes for tick resistance displaying possible target structural variants were used to obtain genes-biological processes networks. These networks highlighted different processes associated with tick resistance in cattle (). Some of highlighted processes were the regulation of eosinophil chemotaxis associated with the DAPK2 gene (identified in group 2), positive regulation of RIG-I signalling pathway associated with PUM1 gene (identified in group 2) and regulation of monocyte differentiation that was in common between ACIN1 and INPP5D genes (identified in group 1).
Figure 3. Functional networks between genes and biological processes associated with tick resistance in cattle. Main interaction networks between biological processes and genes (nodes) are shown in zoom. Blue nodes are associated with group 2 and red nodes are associated with group 1. Grey node represents process shared between the genes of the two groups. The size of the node characterizes the enrichment of the process according to ClueGO Cytoscape plug-in (Bindea et al. Citation2009). The terms that are most enriched by subnet are in bold.
According to TFM-Explorer programme, 65 TF were related to the candidate genes (Supplementary Material – Tables S12–S15). Based on biological processes (Supplementary Material – Tables S16–S19) and literature evidence, six key TF associated with tick resistance in cattle were selected (). These key TF were used to generate gene-TF networks for each group (Supplementary Material – Figures S1–S4). Based on that, a merged network was built () enabling to highlight promising candidate genes. Thus, from the 92 candidate genes with possible target variants, promising candidate genes for tick resistance in cattle were observed (e.g. CACNA1A, LRRIQ1, NKD2, GIMAP8, WIPF3, LRRC4C, LOC112449525, LOC536229, MYO6, SPHKAP, OR4K2, OR4F1, NOR4F1, NOR4F1, NOR469, SORCS3, PNP, MEP1A, LRFNE). A summary of the main highlighted candidate genes number in each group after gene network analysis is presented as a workflow (Supplementary Material – Figure S5)
Figure 4. Gene-transcription factors network for tick resistance in cattle. Transcription factors (yellow diamond nodes) associated with candidate genes (circle nodes in red: group 1; blue: group 2; light pink: group 3; green: group 4) for tick resistance in cattle. The yellow diamond node with the purple border is associated with three groups (1, 2 and 3) while the others are only associated with the group of the border colour. Dark pink square nodes represent the biological processes associated with transcription factors. The size of the nodes represents the enrichment as a function of transcription factors binding sites number.
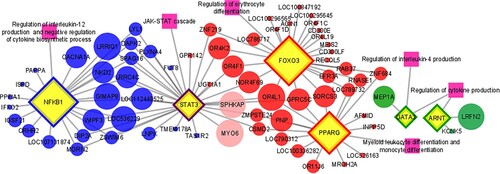
Table 2. Key transcription factors (TF) for tick resistance in cattle. Most representatives TF associated with genes identified for each group, based on their biological processes and evidence in the literature.
4. Discussion
4.1. Systematic review
According to the systematic literature review results, we observed that GWAS for tick resistance in cattle is little explored still, in terms that only four scientific articles were identified meeting the established criteria in our survey. Among them, we observed three studies in Brazil (Hereford and Braford breeds, and Gir × Holstein F2 animals) and one in South Africa with Nguni breed. The locally adapted Nguni breed is classified as Bos taurus africanus (Nyamushamba et al. Citation2017) and known by a good fertility and resistance to diseases (Mapiye et al. Citation2020). When compared to exotic temperate breeds, it has lower levels of nematode infestation (Ndlovu et al. Citation2009), tick count (Spickett et al. Citation1989; Muchenje et al. Citation2008; Marufu et al. Citation2011) and seroprevalence for Anaplasma marginale and Babesia bigemina (Marufu et al. Citation2010). In Brazil, the Gir × Holstein F2 cattle are mostly used in milk production, as they adapt to different production systems and are quite efficient (Ribeiro et al. Citation2017). According to Otto et al. (Citation2018), candidate genes for tick resistance in these crossbred cattle were identified and, according to the origin of allele, resistant cattle showed two alleles of Gir breed, while the susceptible ones possess alleles from Holstein breed.
In addition, Hereford and Braford cattle are predominant beef breeds in the southern region of Brazil (Piccoli et al. Citation2020). Studies using these breeds have shown that tick resistance is a trait with a moderate heritability estimate (0.13–0.19) and shows a positive genetic correlation with production traits (Cardoso et al. Citation2015; Biegelmeyer et al. Citation2017). Knowing that ticks cause enormous damage to cattle, especially in tropical and subtropical regions, where the climate is favourable for their development, such as Brazil and South Africa, studies of the genetic architecture of tick resistance trait is extremely important.
Thus, from the systematic review of this study, it was possible to shed light on the GWAS for tick resistance in cattle, from different regions and breeds of beef and dairy cattle. Several genomic regions associated with tick counts in cattle have been identified (Mapholi et al. Citation2016; Sollero et al. Citation2017; Mota et al. Citation2018; Otto et al. Citation2018). Thus to better understand the genetic architecture of this complex trait, a post-GWAS analysis of candidate genes combined with identification of possible target structural variants was performed for the first time in the literature.
4.2. Functional analysis of candidate genes for tick resistance with possible structural variants
From 1353 candidate genes obtained via literature review, 626 showed possible variants in several gene regions. Among them, 92 genes showed target structural variants in the 5′ UTR and coding regions. French and Edwards (Citation2020) demonstrated that genetic variants in the 5′ UTR region have a potential effect on affecting the expression or function of genes, as they can alter regulatory elements that influence their interaction with 5′ UTR, and consequently impact on post-transcriptional and translation processes. In addition, variants located in coding regions hold the potential to alter the protein sequence, which may lead to loss and alteration of the protein’s function (Zhao et al. Citation2019). Since genes identified from GWAS can contribute to the identification of inter-individual variant differences in phenotypes (Wang et al. Citation2019), it is possible that target structural variants may be located in the 92 candidate genes and can have the potential to influence the expression of tick resistance in cattle. Thus, we performed functional analyses of these genes aiming to highlight the most candidate genes associate with the studied trait.
From the biological processes networks, it was possible to observe that the ACIN1 and INPP5D genes from group 1 and DAPK2 and PUM1 genes from group 2, were enriched and associated with biological processes with probable involvement in cattle tick resistance. The ACIN1 gene encodes the Apoptotic Chromatin Condensation Inducer 1 protein, which has been shown to be important in the nuclear matrix (Mendonça et al. Citation2018) and apoptosis processes, contributing to several functions in the immune system, mainly in the adaptive response (Nagata and Tanaka Citation2017). For example, in humans, cells that are potentially cancerous and contaminated by viruses are eliminated from the body through apoptosis (Bedoui et al. Citation2020). The INPP5D gene belonging to the inositol Polyphosphate-5-Phosphatase family that is expressed in haematopoietic cells (Ware et al. Citation1996) and plays a role in several pathways, such as microglial activation, neuroinflammation, and immune response (Malik et al. Citation2015; López González et al. Citation2016; Yoshino et al. Citation2017). Moreover, Kong et al. (Citation2019) integrated proteomics and microRNA from the plasma of Jersey cattle, in response to high altitude hypoxia (HAH), inferred that miR-155-5p regulates the immune response of B cells by targeting INPP5D to HAH stress adaptation. In general, both ACIN1 and INPP5D shared important biological processes of the immune system in our analysis, such as regulation of monocyte differentiation. Monocytes perform different functions in the course of infection, inflammation, injury and healing (Okabe and Medzhitov Citation2016). When these undergo differentiation, their effector functions are potentiated, being able to induce long-term adaptive immune responses (Witte et al. Citation2018). Thus, ACIN1 located in BTA10 and INPP5D in BTA 3 are potential candidate genes for tick resistance in cattle by playing important roles in the adaptive immune system.
Considering the selected genes from group 2, we also observed biological processes that influence the immune system, such as regulation of eosinophil chemotaxis associated with the DAPK2 gene and positive regulation of RIG-I signalling pathway associated with the PUM1 gene. DAPK2 belongs to the death-associated protein kinase family of serine–threonine kinase, encoding the protein kinase 2 associated with cell death (Humbert et al. Citation2017). It shows functions in the regulation of apoptosis, autophagy and inflammation (Geering Citation2015), and is mainly expressed in neutrophil and eosinophil cells in humans (Humbert et al. Citation2017). It also plays an important role in granulocytes differentiation (Rizzi et al. Citation2007; Humbert et al. Citation2014). Granulocyte chemotaxis is one of the essential elements of innate immune response to injury or infection, making the response more efficient, in terms that this process recruits and attracts the necessary cells to the area of inflammation (Geering et al. Citation2014). Studies involving the immune responses in the skin of cattle infested with ticks have indicated a strong hypersensitivity reaction with increased resistance of cattle to ticks, including the infiltration of eosinophils and the concentration of histamine in the tick fixation site (Schleger et al. Citation1976; Kemp and Bourne Citation1980; Schleger et al. Citation1981). In a histological study of the tick bite location, a high infiltration of basophils, mast cells and eosinophils were observed in the skin of resistant Bos indicus cattle, while in susceptible Bos taurus animals, a greater recruitment of neutrophils was observed (Tabor et al. Citation2017).
Moreover, the PUM1 gene encodes the pumilio RNA binding family member 1 protein, and acts as an important regulator of key genes of the innate immune system (Liu et al. Citation2017). In addition, the biological process positive regulation of the RIG-I signalling pathway found in our study is essential to the innate immune system to identify cells that have been contaminated by viruses (Kell et al. Citation2015 ). Interestingly, the flavivirus is one pathogen that can be transmitted by R. microplus tick (Souza et al. Citation2018), and RIG-I signalling pathway seems to play an important role in the immune system regarding the recognition and elimination of these pathogens by preventing their invasion (Guo et al. Citation2018).
In this study, we also identified TF related to candidate genes for tick resistance. Among the 65 identified TF, six were highlighted as key TF associated with the immune system (FOXO3, PPARG, STAT3, NFKB1, GATA3 and ARNT) which were used to build a gene-TF network. Among them, STAT3 was identified in common to groups 1, 2 and 3 and, consequently, in three breeds: Nguni (group 1), Hereford and Braford (groups 2 and 3). The STAT3 is a downstream effector of several cytokines, an important regulator of immune responses via JAK/STAT signalling (Yan et al. Citation2018). From group 1, the FOXO3 was found associated with the regulation of erythrocyte differentiation and PPARG was associated with the differentiation of myeloid leukocytes and monocytes. In a study with mice, modulation of FOXO3 activity showed that it could increase or prevent antibody-mediated immune response, playing a role in regulating the differentiation and function of T helper cells (Qi et al. Citation2020). In addition, PPARG plays a role in differentiating several immune system cells such as macrophages, monocytes, dendritic cells, T and B lymphocytes and platelets (Croasdell et al. Citation2015; Christofides et al. Citation2020). Piper et al. (Citation2009) suggested that resistant cattle (Bos indicus) show a greater tendency to potentiate a consistent T cell-mediated response against R. microplus.
Another well-enriched TF in the gene-TF network was the NFKB1, which was associated with genes of the group 2. This TF belongs to nuclear factor-κB (NF-κB) family that regulates a diversity of genes with functions in the development of immune system, inflammation, innate and adaptive immune responses (Mulero et al. Citation2019). Moré et al. (Citation2019), analyzing differentially expressed genes in Braford cattle, demonstrated that in susceptible cattle, members of the NF-kB family were the mainly expressed. This gene family has also been observed to be expressed in tick fixation sites in Holstein cattle (Piper et al. Citation2008). In our study, NFKB1 was associated with biological processes such as regulation of the production of interleukin-12 and negative regulation of cytokine biosynthetic process, what could be a potential TF associated with candidate genes for tick resistance in cattle.
In the group 4, GATA3 was associated with the regulation of interleukin-4 (IL4) production process. This TF is known to act as a regulator of the innate and adaptive immune system through the differentiation of memory cells such as T CD4 + and T CD8 +, Th2 and innate lymphocytes (Wan Citation2014). Rodriguez-Valle et al. (Citation2013) demonstrated that susceptible Holstein cattle exposed to R. microplus showed lower expression of cytokine Th2 related to IL4 if compared with the resistant Brahman cattle. In addition, IL4 is a key regulator of humoral and adaptive immune responses (Tabor et al. Citation2017). Also observed in group 4, ARNT is expressed by several cells of the immune system and it is known to integrate actions of the environment and metabolism in the immune response (Gutiérrez-Vázquez and Quintana Citation2018). In mice, increased expression of ARNT mainly in myeloid cells, in addition to contributing to the immune system, also indicates the possibility of an excellent therapeutic strategy to improve wound healing (Scott et al. Citation2014). The wound healing process is regulated by several growth factors and cytokines released at the injury site (Kurahashi and Fujii Citation2015). In resistant x susceptible Braford cattle, after the tick infestation, the proteolysis processes in the degradation of the connective tissue and remodelling of the extracellular matrix, inflammation through cytokine signalling and the platelet-endothelium-leukocyte interactions were enriched only in resistant cattle (Moré et al. Citation2019). Thus, it is suggested that GATA3 and ARNT through the regulation of cytokines production may be influencing the healing process, in response to injury caused by ticks in cattle, thus contributing to their resistance to ticks.
Additionally, we also observed genes enriched by binding sites for the highlighted TFs (e.g. OR4L1, PNP, LRRIQ1, GIMAP8, MYO6, MEP1A and LRFN2). From them, OR4L1 and PNP were identified on group 1. The OR4L1 gene belongs to the family of olfactory receptors (Malnic et al. Citation2004). Lee et al. (Citation2018) identified this gene among the 30 main genes encoding proteins with differential methylation due to Rh2 Ginsenoside, which is known to increase the activity of cells in the immune system. In addition, the purine nucleoside phosphorylase (PNP) gene, in BTA10, is well recognized in several studies about its function in the immune system (Ghodke-Puranik et al. Citation2017; Albar et al. Citation2020; Tecle et al. Citation2021). In humans, variants found in this gene that cause the reduction or absence of its activity, were found in autoimmune diseases, gradual immune T-cell insufficiency, B-cell dysfunction, and linked to high susceptibility to infections (Albar et al. Citation2020). Tecle et al. (Citation2021) indicated that purine homeostasis in the host regulates resistance to contamination from intracellular and extracellular pathogens. Thus, OR4L1 and PNP are highlighted here as promising candidate gene for tick resistance in cattle.
In the group 2, the GIMAP8 and LRRIQ1 genes were highlighted. GIMAP8 is one of the members of GTPase family of immunity-associated proteins (GIMAPs), strongly expressed in the terminal stages of T and B cell development (Berg et al. Citation2021). Robbertse et al. (Citation2018) demonstrated that in bovine tick fixation areas, B lymphocytes are important mediators of immune responses, due to their considerable influx and significant increase in CD3 + T lymphocytes, observed in tick-resistant breeds. The LRRIQ1 gene encodes the leucine-rich repeat and IQ motif containing 1 protein, which indicates evidence that this gene acts on the immune system, recognizing pathogen effector proteins and triggering localized cell death, to limit infection in animal and plant cells (Horsefield et al. Citation2019). Moreover, proteins containing leucine-rich repeats are also known to function as intracellular immune receptors, protecting hosts against pathogen invasion (Mermigka et al. Citation2020). In this way, we suggest that GIMAP8, in BTA4, and LRRIQ1, in BTA5, may be promising candidate genes since they play an important role in cattle tick resistance.
The myosin VI gene (MYO6) was identified in the group 3 and is involved in a wide range of cellular processes such as endocytosis, exocytosis, autophagy and regulation of actin dynamics (Jonge et al. Citation2019). In inflammation situations, it plays a role in the epithelial barrier through endosome/lysosome fusion in epithelial cells (Liao et al. Citation2013). Mota et al. (Citation2018) also highlighted MYO6 as a potential candidate for tick resistance in cattle, due to its possible action on the epithelial barrier, which corroborates our findings in gene-TF network results. Thus, we suggest that the MYO6 gene in BTA9 is a potential candidate gene for resistance to ticks in cattle by playing a role in the epithelial barrier of these animals.
In the group 4, the meprin A subunit alpha (MEP1A) gene is expressed in several mammalian cells and organs, such as kidney, intestine, lung, bladder, skin, cancer and immune cells, proteolytically processing various inflammatory mediators such as cytokines and chemokines (Herzog et al. Citation2019). In Brangus cattle, MEP1A was identified in the bovine leukocyte antigen (BoLA) region on BTA23, which is responsible for encoding genes associated with the adaptive immune system and may be related to the adaptation of these animals to subtropical environments (Goszczynski et al. Citation2018). In addition, in the same group 4, the LRFN2 gene was initially described to be expressed in the brain (Brouwer et al. Citation2019), but studies have identified its expression in non-neural tissues and associated with the risk of lung cancer (Jin et al. Citation2012; Bhat et al. Citation2020). In addition, a member of his family, the LRFN4 gene, has been found to function in immune cells in monocyte/macrophage migration (Konakahara et al. Citation2011). Thus, we suggest that the MEP1A and LRFN2 genes in BTA23 are promising candidate genes for tick resistance in cattle.
It is important to note that even though we have focused on immune system-related processes in this study, there are other factors such as skin thickness and coat colours that may also play a role in tick resistance. Foster et al. (Citation2007) suggested that the skin thickness appears to play an important role on host resistance to ticks. Moreover, Gasparin et al. (Citation2007) analyzing contemporary group, revealed that lighter coloured animals are more resistant than dark coloured ones. However, in terms to address the physiological role in host tick resistance, we focused on immune system processes.
In cattle, the immune system is one of the important mechanisms that confer resistance or susceptibility to tick infestation. Ribeiro et al. (Citation2019) observed that deletions variants found in genes involved with the human innate immune system affected the clinical infection caused by the Andean orthohantavirus. The variants used in this present study were deletion, duplication, tandem duplication, gain in copy number and loss in copy number, besides the possibility to be located in genes associated with the immune system, we may infer that these genes are strong candidates to be associated with tick resistance. Based on that, we present a robust post-GWAS analyses approach via the systematic review followed by functional analyses of candidate genes with possible variants for important economical traits in livestock.
5. Conclusions
Candidate genes associated with tick resistance in cattle presenting possible target variants were identified. The gene networks allowed the identification of enriched biological processes related to the immune system and promising candidate genes associated with tick resistance in cattle. Furthermore, the gene-transcription factors networks allowed the identification of putative main TF and candidate genes for tick resistance in cattle. Thus, promising candidate genes with a possible functional role for tick resistance in cattle are presented for further in vitro and/or in vivo analyses.
Supplemental Material
Download Zip (5.8 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Albar R, Alesa A, Saqa D, Abdulaziz J, Batouk M, Alesa A. 2020. Purine nucleoside phosphorylase (PNP) deficiency: a case report. Egypt J Hosp Med. 81:1967–1969. doi:10.21608/ejhm.2020.123480.
- Ayres DR, Pereira RJ, Boligon AA, Baldi F, Roso VM, Albuquerque LG. 2015. Genetic parameters and investigation of genotype environment interactions in Nellore Hereford crossbred for resistance to cattle ticks in different regions of Brazil. J Appl Genet. 56:107–113. doi:10.1007/s13353-014-0238-5.
- Bai SH, Ogbourne S. 2016. Eco-toxicological effects of the Avermectin family with a focus on abamectin and ivermectin. Chemosphere. 154:204–214. doi:10.1016/j.chemosphere.2016.03.113.
- Bedoui S, Herold MJ, Strasser A. 2020. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat Rev Mol Cell Biol. 21:678–695. doi:10.1038/s41580-020-0270-8.
- Berg HE, Blackburn PR, Baughn LB, Ketterling RP, Xu X, Greipp PT, Hoppman NL, Smadbeck JB, Vasmatzis G, Shi M, et al. 2021. Identification of a novel KMT2A/GIMAP8 gene fusion in a pediatric patient with acute undifferentiated leukemia. Genes Chromosom Cancer. 60:108–111. doi:10.1002/gcc.22902.
- Bhat G, Verma S, Bhat A, Shah R, Sethi I, Dar KA, Abrol D, Bhat A, Raina R, Kumar R. 2020. Genetic variant rs2494938 of LRFN2 gene is associated with non-small cell lung cancer risk in north-Indian population. 3 Biotech 10, 1–6. doi:10.1007/s13205-020-02403-1.
- Biegelmeyer P, Gulias-Gomes CC, Roso VM, Dionello NJL, Cardoso FF. 2017. Tick resistance genetic parameters and its correlations with production traits in Hereford and Braford cattle. Livest Sci. 202:96–100. doi:10.1016/j.livsci.2017.05.019.
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W, Pagès F, Trajanoski Z, Galon J. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinform. 25:1091–1093. doi:10.1093/bioinformatics/btp101.
- Brouwer M, Farzana F, Koopmans F, Chen N, Brunner JW, Oldani S, Li WK, Weering JRT, Smit AB, Toonen RF, Verhage M. 2019. SALM 1 controls synapse development by promoting F-actin/PIP2-dependent neurexin clustering. EMBO J. 38:e101289. doi:10.15252/embj.2018101289.
- Cardoso CP, Silva BF, Gonçalves DS, Tagliari NJ, Saito ME, Amarante AF. 2014. Resistência contra ectoparasitas em bovinos da raça Crioula Lageana e meio-sangue Angus avaliada em condições naturais. Pesq Vet Bras. 34:141–146. doi:10.1590/S0100-736X2014000200008.
- Cardoso FF, Gomes CCG, Sollero BP, Oliveira MM, Roso VM, Piccoli ML, Higa RH, Yokoo MJ, Caetano AR, Aguilar I. 2015. Genomic prediction for tick resistance in Braford and Hereford cattle. J Anim Sci. 93:2693–2705. doi:10.2527/jas.2014-8832.
- Christofides A, Konstantinidou E, Jani C, Boussiotis VA. 2020. The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism. 114:154338. doi:10.1016/j.metabol.2020.154338.
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews D, Barnes C, Campbell P, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature. 464:704–712. doi:10.1038/nature08516.
- Croasdell A, Duffney PF, Kim N, Lacy SH, Sime PJ, Phipps RP. 2015. PPARγ and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015, 1–20. doi:10.1155/2015/549691.
- Dev A, Iyer S, Razani B, Cheng G. 2010. NF-κB and innate immunity. NF-kB Health Dis 349, 115–143. doi:10.1007/82_2010_102.
- Feuk L, Carson AR, Scherer SW. 2006. Structural variation in the human genome. Nat Rev Genet. 7:85–97. doi:10.1038/nrg1767.
- Foster A, Jackson A, D′Alteiro GL. 2007. Skin diseases of South American camelids. In Pract. 29:216–223. doi:10.1136/inpract.29.4.216.
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, et al. 2006. Copy number variation: new insights in genome diversity. Genome Res. 16:949–961. http://www.genome.org/cgi/doi/10.1101/gr.3677206.
- French JD, Edwards SL. 2020. The role of noncoding variants in heritable disease. Trends Genet. 36:880–891. doi:10.1016/j.tig.2020.07.004.
- Garcia MV, Rodrigues VDS, Koller WW, Andreotti R. 2019. Biologia e importância do carrapato Rhipicephalus (Boophilus) microplus. Embrapa Gado de Corte-Capítulo em livro científico (ALICE). http://www.alice.cnptia.embrapa.br/alice/handle/doc/1107093.
- Gasparin G, Miyata M, Coutinho LL. 2007. Mapping of quantitative trait loci controlling tick [Rhipicephalus (Boophilus) microplus] resistance on bovine chromosomes 5, 7 and 14. Anim. Genet. 38:453–459.
- Geering B. 2015. Death-associated protein kinase 2: regulator of apoptosis, autophagy and inflammation. Int J Biochem Cell Biol. 65:151–154. doi:10.1016/j.biocel.2015.06.001.
- Geering B, Stoeckle C, Rožman S, Oberson K, Benarafa C, Simon HU. 2014. DAPK2 positively regulates motility of neutrophils and eosinophils in response to intermediary chemoattractants. J Leukoc Biol. 95:293–303. https://jlb.onlinelibrary.wiley.com/toc/19383673/0/0.
- Ghodke-Puranik Y, Dorschner JM, Vsetecka DM, Amin S, Makol A, Ernste F, Osborn T, Moder K, Chowdhary V, Eliopoulos E, et al. 2017. Lupus-associated functional polymorphism in PNP causes cell cycle abnormalities and interferon pathway activation in human immune cells. Arthritis Rheum. 69:2328–2337. doi:10.1002/art.40304.
- Goszczynski DE, Corbi-Botto CM, Durand HM, Rogberg-Muñoz A, Munilla S, Peral-Garcia P, Cantet RJC, Giovambattista G. 2018. Evidence of positive selection towards Zebuine haplotypes in the BoLA region of Brangus cattle. Animal. 12:215–223. doi:10.1017/S1751731117001380.
- Gueretz JS, Barth AL, Marani NBR, Moreira F, Júnior JMO, Bianchi I, Gallina T, Schwegler E. 2020. Remoção manual de Rhipicephalusmicroplus (Acari: Ixodidae) em bovinos, como uma alternativa ao uso de carrapaticidas em pequenos rebanhos. Braz J Dev. 6:19060–19066. doi:10.34117/bjdv6n4-174.
- Guo HY, Zhang XC, Jia RY. 2018. Toll-like receptors and RIG-I-like receptors play important roles in resisting flavivirus. J Immunol Res. 2018:1–7. doi:10.1155/2018/6106582.
- Gutiérrez-Vázquez C, Quintana FJ. 2018. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 48:19–33. doi:10.1016/j.immuni.2017.12.012.
- Herzog C, Haun RS, Kaushal GP. 2019. Role of meprin metalloproteinases in cytokine processing and inflammation. Cytokine. 114:18–25. doi:10.1016/j.cyto.2018.11.032.
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai JS, Rank MX, et al. 2019. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science. 365:793–799. doi:10.1126/science.aax1911.
- Humbert M, Federzoni EA, Britschgi A, Schläfli AM, Valk PJ, Kaufmann T, Haferlach T, Behre G, Simon HU, Torbett BE, et al. 2014. The tumor suppressor gene DAPK2 is induced by the myeloid transcription factors PU. 1 and C/EBPα during granulocytic differentiation but repressed by PML-RARα in APL. J Leukoc Biol. 95:83–93. doi:10.1189/jlb.1112608.
- Humbert M, Federzoni EA, Tschan MP. 2017. Distinct TP73–DAPK2–ATG5 pathway involvement in ATO-mediated cell death versus ATRA-mediated autophagy responses in APL. J Leukoc Biol. 102:1357–1370. doi:10.1189/jlb.1A0317-132R.
- Jin G, Ma H, Wu C, Dai J, Zhang R, Shi Y, Lu J, Miao X, Wang M, Zhou Y, et al. 2012. Genetic variants at 6p21. 1 and 7p15. 3 are associated with risk of multiple cancers in Han Chinese. Am J Hum Genet. 91:928–934. doi:10.1016/j.ajhg.2012.09.009.
- Jonge JJ, Batters C, O'loughlin T, Arden SD, Buss F. 2019. The MYO 6 interactome: selective motor-cargo complexes for diverse cellular processes. FEBS letters. 593(13):1494–1507. doi:10.1002/1873-3468.13486.
- Kell AM, Gale Jr M. 2015. RIG-I in RNA virus recognition. Virology. 479:110–121. doi:10.1016/j.virol.2015.02.017.
- Kemp DH, Bourne A. 1980. Boophilus microplus: the effect of histamine on the attachment of cattle-tick larvae–studies in vivo and in vitro. Parasitology. 80:487–496. doi:10.1017/S0031182000000950.
- Kim ES, Sonstegard TS, Silva MV, Gasbarre LC, Van Tassell CP. 2015. Genome-wide scan of gastrointestinal nematode resistance in closed Angus population selected for minimized influence of MHC. PLoS One. 10:e0119380. doi:10.1371/journal.pone.0119380.
- Konakahara S, Saitou M, Hori S, Nakane T, Murai K, Itoh R, Shinsaka A, Kohroki J, Kawakami T, Kajikawa M, Masuho Y. 2011. A neuronal transmembrane protein LRFN4 induces monocyte/macrophage migration via actin cytoskeleton reorganization. FEBS Lett. 585:2377–2384. doi:10.1016/j.febslet.2011.06.011.
- Kong Z, Zhou C, Li B, Jiao J, Chen L, Ren A, Jie H, Tan Z. 2019. Integrative plasma proteomic and microRNA analysis of Jersey cattle in response to high-altitude hypoxia. J Dairy Sci. 102:4606–4618. doi:10.3168/jds.2018-15515.
- Kurahashi T, Fujii J. 2015. Roles of antioxidative enzymes in wound healing. J Dev Biol. 3:57–70. doi:10.3390/jdb3020057.
- Lee H, Lee S, Jeong D, Kim SJ. 2018. Ginsenoside rh2 epigenetically regulates cell-mediated immune pathway to inhibit proliferation of MCF-7 breast cancer cells. J Ginseng Res. 42:455–462. doi:10.1016/j.jgr.2017.05.003.
- Liao YW, Wu XM, Jia J, Wu XL, Hong T, Meng LX, Wu XY. 2013. Myosin VI contributes to maintaining epithelial barrier function. J Biomed Sci. 20:1–7. doi:10.1186/1423-0127-20-68.
- Littiere TO, Castro GH, Rodriguez MDPR, Bonafé CM, Magalhães AF, Faleiros RR, Vieira JIG, Santos CG, Verardo LL. 2020. Identification and functional annotation of genes related to horses’ performance: from GWAS to post-GWAS. Animals (Basel). 10:1173. doi:10.3390/ani10071173.
- Liu Y, Qu L, Liu Y, Roizman B, Zhou GG. 2017. PUM1 is a biphasic negative regulator of innate immunity genes by suppressing LGP2. Proc Natl Acad Sci. 114:E6902–E6911. doi:10.1073/pnas.1708713114.
- López González I, Garcia-Esparcia P, Llorens F, Ferrer I. 2016. Genetic and transcriptomic profiles of inflammation in neurodegenerative diseases: Alzheimer, Parkinson, Creutzfeldt-Jakob and tauopathies. Int J Mol Sci. 17:206. doi:10.3390/ijms17020206.
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 21:3448–3449. doi:10.1093/bioinformatics/bti551.
- Malik M, Parikh I, Vasquez JB, Smith C, Tai L, Bu G, LaDu MJ, Fardo DW, Rebeck WG, Estus S. 2015. Genetics ignite focus on microglial inflammation in Alzheimer’s disease. Mol Neurodegener. 10:1–12. doi:10.1186/s13024-015-0048-1.
- Malnic B, Godfrey PA, Buck LB. 2004. The human olfactory receptor gene family. Proc Natl Acad Sci. 101:2584–2589. doi:10.1073/pnas.0307882100.
- Mapholi NO, Maiwashe A, Matika O, Riggio V, Bishop SC, MacNeil MD, Banga C, Taylor JF, Dzama K. 2016. Genome-wide association study of tick resistance in South African Nguni cattle. Ticks Tick-Borne Dis. 7:487–497. doi:10.1016/j.ttbdis.2016.02.005.
- Mapiye O, Chikwanha OC, Makombe G, Dzama K, Mapiye C. 2020. Livelihood, food and nutrition security in Southern Africa: what role do indigenous cattle genetic resources play? Diversity (Basel). 12:74. doi:10.3390/d12020074.
- Marufu MC, Chimonyo M, Dzama K, Mapiye C. 2010. Seroprevalence of tick-borne diseases in communal cattle reared on sweet and sour rangelands in a semi-arid area of South Africa. Vet J. 184:71–76. doi:10.1016/j.tvjl.2009.02.014.
- Marufu MC, Chimonyo M, Mapiye C, Dzama K. 2011. Tick loads in cattle raised on sweet and sour rangelands in the low-input farming areas of South Africa. Trop Anim Health Prod. 43:307–313. doi:10.1007/s11250-010-9690-3.
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. 2016. The ensemble variant effect predictor. Genome Biol. 17:1–14. doi:10.1186/s13059-016-0974-4.
- Mendonça GA, Morandi Filho R, Souza ET, Gaggini TS, Silva-Mendonça MCA, Antunes RC, Beletti ME. 2018. Isolation and identification of proteins from swine sperm chromatin and nuclear matrix. Anim Reprod. 14:418–428. doi:10.21451/1984-3143-AR816.
- Mermigka G, Amprazi M, Mentzelopoulou A, Amartolou A, Sarris PF. 2020. Plant and animal innate immunity complexes: fighting different enemies with similar weapons. Trends Plant Sci. 25:80–91. doi:10.1016/j.tplants.2019.09.008.
- Moré DD, Cardoso FF, Mudadu MA, Malagó-Jr W, Gulias-Gomes CC, Sollero BP, Ibelli AMG, Coutinho LL, Regitano LC. 2019. Network analysis uncovers putative genes affecting resistance to tick infestation in Braford cattle skin. BMC Genom. 20:1–20. doi:10.1186/s12864-019-6360-3.
- Mota RR, Silva FF, Lopes PS, Tempelman RJ, Sollero BP, Aguilar I, Cardoso FF. 2018. Analyses of reaction norms reveal new chromosome regions associated with tick resistance in cattle. Animal. 12:205–214. doi:10.1017/S1751731117001562.
- Muchenje V, Dzama K, Chimonyo M, Raats JG, Strydom PE. 2008. Tick susceptibility and its effects on growth performance and carcass characteristics of Nguni, Bonsmara and Angus steers raised on natural pasture. Animal. 2:298–304. doi:10.1017/S1751731107001036.
- Mulero MC, Huxford T, Ghosh G. 2019. NF-κB, IκB, and IKK: integral components of immune system signaling. Struct Immunol 1172, 207–226. doi:10.1007/978-981-13-9367-9_10.
- Nagata S, Tanaka M. 2017. Programmed cell death and the immune system. Nat Rev Immunol. 17:333–340. doi:10.1038/nri.2016.153.
- Ndlovu T, Chimonyo M, Muchenje V. 2009. Monthly changes in body condition scores and internal parasite prevalence in Nguni, Bonsmara and Angus steers raised on sweetveld. Trop Anim Health Prod. 41:1169–1177. doi:10.1007/s11250-008-9297-0.
- Neto LP, Piper EK, Jonsson NN, Barendse W, Gondro C. 2010. Meta-analysis of genome wide association and gene expression studies to identify candidate genes for tick burden in cattle. In: 9th world congress of genetics applied to livestock production. (p. 0664).
- Neto LRP, Jonsson NN, Michael JD, Barendse W. 2011. Molecular genetic approaches for identifying the basis of variation in resistance to tick infestation in cattle. Vet Parasitol. 180:165–172. doi:10.1016/j.vetpar.2011.05.048.
- Nyamushamba GB, Mapiye C, Tada O, Halimani TE, Muchenje V. 2017. Conservation of indigenous cattle genetic resources in Southern Africa’s smallholder areas: turning threats into opportunities – a review. Asian-Australas J Anim Sci. 30:603–621. doi:10.5713/ajas.16.0024.
- Okabe Y, Medzhitov R. 2016. Tissue biology perspective on macrophages. Nat Immunol. 17:9–17. doi:10.1038/ni.3320.
- Otto PI, Guimarães SE, Verardo LL, Azevedo ALS, Vandenplas J, Soares AC, Sevillano CA, Veroneze R, Pires MFA, Freitas C, et al. 2018. Genome-wide association studies for tick resistance in Bos Taurus× Bos indicus crossbred cattle: A deeper look into this intricate mechanism. J Dairy Sci. 101:11020–11032. doi:10.3168/jds.2017-14223.
- Piccoli ML, Brito LF, Braccini J, Oliveira HR, Cardoso FF, Roso VM, Sargolzaei M, Schenkel FS. 2020. Comparison of genomic prediction methods for evaluation of adaptation and productive efficiency traits in Braford and Hereford cattle. Livest Sci. 231:103864. doi:10.1016/j.livsci.2019.103864.
- Piper EK, Jackson LA, Bagnall NH, Kongsuwan KK, Lew AE, Jonsson NN. 2008. Gene expression in the skin of Bos Taurus and Bos indicus cattle infested with the cattle tick, Rhipicephalus (Boophilus) microplus. Vet Immunol Immunopathol. 126:110–119. doi:10.1016/j.vetimm.2008.06.011.
- Piper EK, Jonsson NN, Gondro C, Lew-Tabor AE, Moolhuijzen P, Vance ME, Jackson LA. 2009. Immunological profiles of Bos Taurus and Bos indicus cattle infested with the cattle tick, Rhipicephalus (Boophilus) microplus. Clin Vaccine Immunol. 16:1074–1086. doi:10.1128/CVI.00157-09.
- Porto Neto LR, Bunch RJ, Harrison BE, Prayaga KC, Barendse W. 2010. Haplotypes that include the integrin alpha 11 gene are associated with tick burden in cattle. BMC gene. 11(1):1–13.
- Qi H, Tian D, Li M, Zhang C, Jin H, Liu L, Zhao X, Ma L, Zhao W, Ge Q, et al. 2020. Foxo3 promotes the differentiation and function of follicular helper T cells. Cell Rep. 31:107621. doi:10.1016/j.celrep.2020.107621.
- Quintana FJ, Sherr DH. 2013. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Ver. 65:1148–1161. doi:10.1124/pr.113.007823.
- Ribeiro GE, Leon LE, Perez R, Cuiza A, Vial PA, Ferres M, Mertz GJ, Vial C. 2019. Deletions in genes participating in innate immune response modify the clinical course of andes orthohantavirus infection. Viruses. 11:680. doi:10.3390/v11080680.
- Ribeiro, LS, Goes, TJF, Torres, RA, Araújo, CV, Reis, RB, Saturnino, HM, 2017. Desempenhos produtivo e reprodutivo de um rebanho F1 Holandês x Gir em Minas Gerais. Arq Bras Med Vet Zootec. 69, 1624-1634. doi:10.1590/1678-4162-9076.
- Rizzi M, Tschan MP, Britschgi C, Britschgi A, Hügli B, Grob TJ, Leupin N, Mueller BU, Simon HU, Ziemiecki A, et al. 2007. The death-associated protein kinase 2 is up-regulated during normal myeloid differentiation and enhances neutrophil maturation in myeloid leukemic cells. J Leukoc Biol. 81:1599–1608. doi:10.1189/jlb.0606400.
- Robbertse L, Richards SA, Clift SJ, Barnard AC, Leisewitz A, Crafford JE, Maritz-Olivier C. 2018. Comparison of the differential regulation of T and B-lymphocyte subsets in the skin and lymph nodes amongst three cattle breeds as potential mediators of immune-resistance to Rhipicephalus microplus. Ticks Tick Borne Dis. 9:976–987. doi:10.1016/j.ttbdis.2018.03.034.
- Rodriguez-Valle M, Moolhuijzen P, Piper EK, Weiss O, Vance M, Bellgard M, Lew-Tabor A. 2013. Rhipicephalus microplus lipocalins (LRMs): genomic identification and analysis of the bovine immune response using in silico predicted B and T cell epitopes. Int J Parasitol. 43:739–752. doi:10.1016/j.ijpara.2013.04.005.
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32:91–94. doi:10.1093/nar/gkh012.
- Schleger AV, Lincoln DT, Kemp DH. 1981. A putative role for eosinophils in tick rejection. Experientia. 37:49–50. doi:10.1007/BF01965562.
- Schleger AV, Lincoln DT, McKenna RV, Kemp DH, Roberts J. 1976. Boophilus microplus: cellular responses to larval attachment and their relationship to host resistance. Aust J Biol Sci. 29:499–512. doi:10.1071/BI9760499.
- Schneeberger K, Weigel D. 2011. Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci. 16:282–288. doi:10.1016/j.tplants.2011.02.006.
- Scott C, Bonner J, Min D, Boughton P, Stokes R, Cha KM, Walters ST, Maslowski K, Sierro F, Grey ST, et al. 2014. Reduction of ARNT in myeloid cells causes immune suppression and delayed wound healing. Am J Physiol Cell Physiol. 307:C349–C357. doi:10.1152/ajpcell.00306.2013.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. http://www.genome.org/cgi/doi/10.1101/gr.1239303.
- Sollero BP, Junqueira VS, Gomes CC, Caetano AR, Cardoso FF. 2017. Tag SNP selection for prediction of tick resistance in Brazilian Braford and Hereford cattle breeds using Bayesian methods. Genet Sel Evol. 49:1–15. doi:10.1186/s12711-017-0325-2.
- Souza WMD, Fumagalli MJ, Torres Carrasco ADO, Romeiro MF, Modha S, Seki MC, Gheller JM, Daffre S, Nunes MRT, Murcia PR, et al. 2018. Viral diversity of Rhipicephalus microplus parasitizing cattle in southern Brazil. Sci. Rep. 8:1–10. doi:10.1038/s41598-018-34630-1.
- Spickett AM, De Klerk D, Enslin CB, Scholtz MM. 1989. Resistance of Nguni, Bonsmara and Hereford cattle to ticks in a bushveld region of South Africa. http://hdl.handle.net/2263/42216.
- Spielmann M, Lupiáñez DG, Mundlos S. 2018. Structural variation in the 3D genome. NaverRev Genet. 19:453–467. doi:10.1038/s41576-018-0007-0.
- Tabor AE, Ali A, Rehman G, Rocha Garcia G, Zangirolamo AF, Malardo T, Jonsson NN. 2017. Cattle tick Rhipicephalus microplus-host interface: a review of resistant and susceptible host responses. Front Cell Infect Microbiol. 7:506. doi:10.3389/fcimb.2017.00506.
- Tecle E, Chhan CB, Franklin L, Underwood RS, Hanna-Rose W, Troemel ER. 2021. The purine nucleoside phosphorylase pnp-1 regulates epithelial cell resistance to infection in C. elegans. PLoS Pathog. 17:e1009350. doi:10.1371/journal.ppat.1009350.
- Touzet H, Varré JS. 2007. Efficient and accurate P-value computation for position weight matrices. Algorithms Mol Biol. 2:1–12. doi:10.1186/1748-7188-2-15.
- Turner N, Abeysinghe P, Peiris H, Vaswani K, Sadowski P, Cameron N, McGhee N, Logan J, Mitchell MD. 2022. Proteomic profiling of plasma-derived small extracellular vesicles: a novel tool for understanding the systemic effects of tick burden in cattle. J Anim Sci. 100(2):skac015. doi:10.1093/jas/skac015.
- Verardo LL, Lopes MS, Wijga S, Madsen O, Silva FF, Groenen MAM, Knol EF, Lopes PS, Guimarães SEF. 2016. After genome-wide association studies: gene networks elucidating candidate genes divergences for number of teats across two pig populations. J Anim Sci. 94:1446–1458. doi:10.2527/jas.2015-9917.
- Wan YY. 2014. GATA3: a master of many trades in immune regulation. Trends Immunol. 35:233–242. doi:10.1016/j.it.2014.04.002.
- Wang H, Lou D, Wang Z. 2019. Crosstalk of genetic variants, allele-specific DNA methylation, and environmental factors for complex disease risk. Front Genet. 9:695. doi:10.3389/fgene.2018.00695.
- Ware MD, Rosten P, Damen JE, Liu L, Humphries RK, Krystal G. 1996. Cloning and characterization of human SHIP, the 145-kD inositol 5-phosphatase that associates with SHC after cytokine stimulation. Blood. 88:2833–2840. doi:10.1182/blood.V88.8.2833.bloodjournal8882833.
- Witte SF, Luk F, Parraga JMS, Gargesha M, Merino A, Korevaar SS, Shankar AS, O'Flynn L, Elliman SJ, Roy D, et al. 2018. Immunomodulation by therapeutic mesenchymal stromal cells (MSC) is triggered through phagocytosis of MSC by monocytic cells. Stem Cells. 36:602–615. doi:10.1002/stem.2779.
- Yan Z, Gibson SA, Buckley JA, Qin H, Benveniste EN. 2018. Role of the JAK/STAT signaling pathway in regulation of innate immunity in neuroinflammatory diseases. Clin Immunol. 189:4–13. doi:10.1016/j.clim.2016.09.014.
- Yoshino Y, Yamazaki K, Ozaki Y, Sao T, Yoshida T, Mori T, Mori Y, Ochi S, Iga JI, Ueno SI. 2017. INPP5D mRNA expression and cognitive decline in Japanese Alzheimer’s disease subjects. J Alzheimer's Dis. 58:687–694. doi:10.3233/JAD-161211.
- Zhao Y, Feng Z, Ding Q. 2019. Type 2 diabetes variants in the SLC16A11 coding region are not loss-of-function mutations. Cell Rep. 29:781–784. doi:10.1016/j.celrep.2019.09.022.