Abstract
Close-range hyperspectral images are a promising source of information in plant biology, in particular, for in vivo study of physiological changes. In this study, we investigate how data fusion can improve the detection of leaf elements by combining pixel reflectance and morphological information. The detection of image regions associated to the leaf structures is the first step toward quantitative analysis on the physical effects that genetic manipulation, disease infections, and environmental conditions have in plants. We tested our fusion approach on Musa acuminata (banana) leaf images and compared its discriminant capability to similar techniques used in remote sensing. Experimental results demonstrate the efficiency of our fusion approach, with significant improvements over some conventional methods.
1. Introduction
Close-range hyperspectral (HS) imaging is a novel research tool for biologists (Scharr et al. Citation2016). Several works have reported the design and implementation of HS imaging systems that capture reflectance information from plant leaves at close range (Mahlein et al. Citation2010; Rumpf et al. Citation2010). Most systems can be classified as pushbroom sensors in which the camera moves over the leaf and record the reflected light from a narrow section. In practice, the main obstacle to obtain high spatial resolution is the mechanical subsystem. Nevertheless, they currently provide the highest spatial and spectral resolutions. Cameras that can capture the full spectral data of a scene in one shot have become available but their resolutions are still limited (Aasen et al. Citation2015). Changes in reflectance levels occur on the leaf blade when illumination is not homogeneous or the distance from the blade to the sensor is not constant (Behmann et al. Citation2016). An example of the latter case is the midrib that appears as a linear structure of varying colors and increasing width. Another source of spectral variation is metabolic changes induced by diseases (Mahlein et al. Citation2010; Rumpf et al. Citation2010) or environmental stress (Kim et al. Citation2011). Depending on the infection type, leaves display spots or streaks of different sizes and colors. At later infection stages, these symptoms can be seen with the naked eye. At early infection stages, changes are subtle and difficult to detect. The latter is perhaps the most important research problem because limiting the spread of a disease can prevent revenue losses for farmers (Triest and Hendrickx Citation2016).
In previous works, HS leaf analysis has focused on classification of the full reflectance spectrum and so-called spectral indexes such as the normalized difference vegetation index (Jacquemoud et al. Citation2009). Adding spatial information can provide a more complete description of leaf structures. Common methods for extracting spatial information include image filtering (Benediktsson, Palmason, and Sveinsson Citation2005; Huang, Liu, and Zhang Citation2015; Liao et al. Citation2016), image segmentation (Blaschke Citation2010), and image pansharpening (e.g. principal component substitution and Bayesian methods) as in (Loncan et al. Citation2015; Mookambiga and Gomathi Citation2016). However, these methods have disadvantages such as high cost computational, significant spectral distortion, limited amount of spectral or spatial information added for object classification and blur degradation.
In this paper, we present an information fusion approach that combines spectral data from a low resolution HS image and spatial information from its corresponding high resolution RGB image. Morphological profiles are applied to extract spatial information of the leaf. Similar approaches have been exploited for pansharpening of satellite images to improve the detection of man-made structures (Liao et al. Citation2015). In contrast to the above approach, our method couples spatial exploitation and data fusion in a unified framework by enhancing the principal components of a HS image (low spatial resolution) using morphological profiles of the color image (high spatial resolution) without losing the spectral information of the original HS image.
To the best of our knowledge, this is the first attempt to apply such fusion approach in close-range HS images. Musa acuminata was chosen as experimental subject because it is an important commercial crop. In Section 2, we describe the imaging setup. Results are presented in Section 3. Future research venues are discussed in the last Section.
2. Materials and methods
2.1. Image acquisition
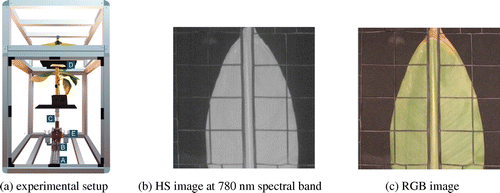
We use the pushbroom scanner described in (Ochoa et al. Citation2016). As shown in Figure (a), it consists of a high-resolution 12-bit monochrome CCD camera (B) with extended infrared sensitivity (1500 M-GE Thorlabs) attached to an spectrograph (Specim Inspector V10) with a spectral range from 364 nm to 1031 nm and nominal spectral resolution of 4.55 nm (C). These elements are mounted on a motorize slider (A). The run length of the slider is 25 cm with a step resolution of 0.5 mm. The camera is placed below the plant’s foliage as fungi and other pathogens enter by the stomata located on the leaf underside. As leaves overlap, a plastic holder (D) was used to keep them apart. Illumination is provided by two 50 W halogen lamps (E).
Spectral calibration was done by fitting known peaks of the emission spectrum of argon (Ar) and mercury (Hg) lamps. A common issue with this kind of imaging system is noise, in particular, at wavelengths near the ultraviolet and infrared regions of the spectrum. To estimate image noise levels, we measured the reflectance standard deviation for a dark reference at different exposure times. Based on these results, we set the camera’s exposure time to 200 ms, which provided an adequate trade-off between image contrast and noise.
For spatial calibration, scanning area and working distance were estimated from the optical parameters of the spectrograph lenses, the f-number of the lens was selected to f/7. The CCD sensor binning was chosen to reduce the differences in vertical and horizontal spatial resolutions. The effective scan area was 16 cm × 16 cm with a resolution of 0.5 mm per pixel. For each leaf scan, the system generates a set of 520 images of 198 × 186 pixels in the visible and infrared region (IR) of the spectrum. Finally, a high dynamic range camera is used to record 856 × 900 pixels RGB images. Examples of the system’s output can be seen in Figure (b) and (c). Since the first step in plant automatic analysis is the identification of meaningful leaf regions, we built a test data-set with the following object classes:
| (1) | Dead leaf: Necrotic areas. | ||||
| (2) | Dying leaf: Interface between healthy and necrotic areas. | ||||
| (3) | Flat blade: Region with homogeneous distance to the camera. | ||||
| (4) | Bent blade: Region with changes in distance to the camera. | ||||
| (5) | Spot: Small lesions caused either by diseases, insects or mechanical stress. | ||||
| (6) | Midrib: Central nerve of a leaf. | ||||
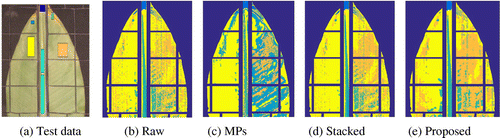
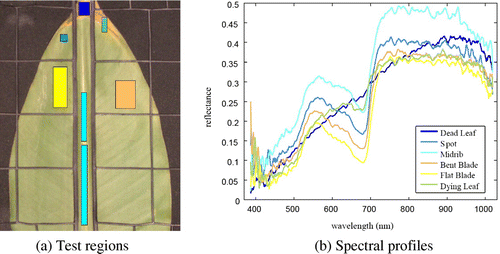
There are two classes associated to the blade because sometimes the leaf surface becomes uneven when it is held by the plastic mesh. The test datasets for each object class, highlighted with different colors, are depicted in Figure (a).
2.2. Preprocessing
Spectral data was normalized using images of white and dark standard reflectance surfaces at each scan session. The resulting image Rλ is computed as follows:
(1)
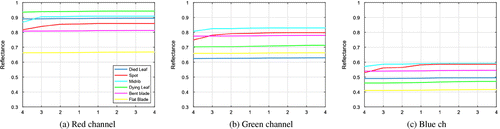
where Sλ, Dλ, and Wλ are the leaf, white, and dark pixel intensities at wavelength λ, respectively. Figure (b) shows the average normalized spectral profiles of the corresponding test regions, which are shown in Figure (a).
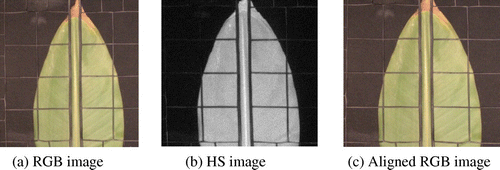
To perform multi-sensor and multi-resolution data fusion, we registered the high spatial resolution color image with respect to the low spatial resolution HS image. The junctions of the plastic holder in both images were used as control points. They are detected by averaging the response values of a line detector along rows and columns (Steger Citation1998). The response peaks were used to detect salient lines ends and a simple tracking routine was employed to find the location of junction points. From these points, the affine transformation coefficients were computed and the transformation was applied to the color image. An example of input and aligned images is depicted in Figure .
2.3. Proposed morphological information fusion
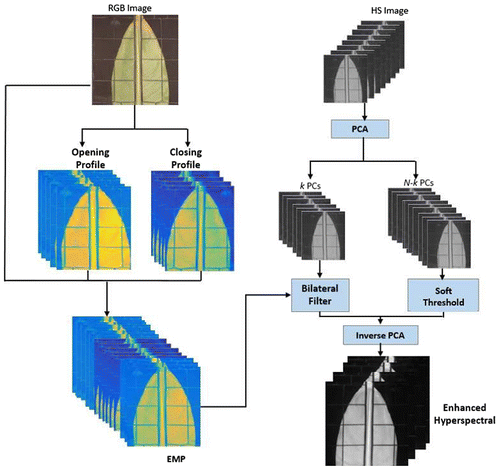
Our fusion method is aimed at obtaining an enhanced HS cube, which includes morphological information without increasing the dimensionality of the original HS cube. Figure shows an overview of the proposed method. To explore the spatial information of high spatial resolution color images, morphological profiles are built by performing opening and closing by reconstruction at several scales (Benediktsson, Palmason, and Sveinsson Citation2005). For an input image f, these operators are defined as follows:(2)
(3) where
and
are the reconstruction by dilation and erosion operators using a structural element (SE) of size n (Soille Citation2003). Opening by reconstruction removes smaller brighter objects, whereas closing by reconstruction removes smaller darker objects.
In contrast to (Liao et al. Citation2015), the extraction of morphological profiles (MP) was done on the high spatial resolution color image (instead of the principal components of the original HS image). Hence, the proposed method transfers spatial information (size, shape, texture) contained in the morphological profile of the color image to guide the spatial enhancement of the low spatial resolution HS image, while enabling spectral and spatial preservation. Also, the proposed method is very robust to image calibration as we exploit the whole spatial information instead of the channels of a panchromatic or color image.
The number of MPs images depends on the number scales and SEs types to be used. Figure (a)–(c) shows the MP images obtained with a disk-shaped SE of increasing size n = [1, 2, 4], and the arrow direction indicates larger SE sizes. Differences in relative contrast of leaf sections are clearly visible at certain scales, this suggests that geometrical and spatial information can be captured by the MPs. For a linear-shaped SE of length L and orientation θ (10°), an opening (resp. closing) deletes bright (resp. dark) objects (or object parts) which are smaller than that length in that direction. When performing such openings (or closings) with different orientations (e.g. every 10 degrees), objects which are shorter than L will be completely removed in all of these images. The maximum (resp. minimum) over all of these openings (resp. closings) will therefore remove the short objects (or object parts) and keep the long objects. Creating multiple such maximum or minimum images for different lengths L gives you the directional MP. In our experiments, the multiple color channels are used as information source.
In order to transfer the spatial information to the low spatial resolution HS image, we employ Principal Component Analysis (PCA) to decorrelate the original hyperspectral image, and separate the image content into two parts. The first several principal components (PCs) keep the most important information of a HS cube and the remaining PCs contain mainly noise. We use the spatial information (from MPs generated on high-resolution RGB image) to guide the spatial resolution enhancement of the first k PCs by using the joint bilateral filter (Tomasi and Manduchi Citation1998). We determine parameter k through our visual analysis, we found in our data-set that the first 6 PCs contain most information, thus we set k = 6. The joint bilateral filter has proven to be computationally efficient while preserving edges and smoothing flat areas (He, Sun, and Tang Citation2013). The enhanced pixel PCi is computed as follows:(5)
where Ki is a normalizing term;(6)
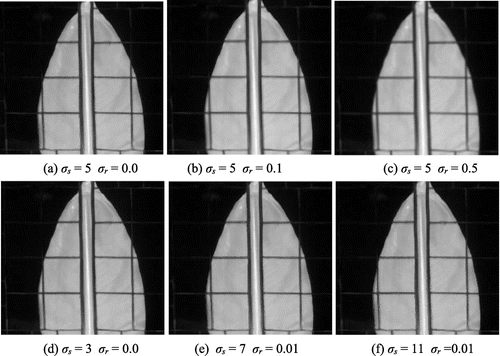
where ω is the window of size (2σs + 1) × (2σs + 1), σs is the scale of the Gaussian filter G that weights the distance between pixel locations, and σr controls the relative weight of intensity difference between guided profile pixels. Figure shows the filtering performances when using different parameter values. Larger values of σs and σr result in oversmoothing effects. The filter implementation in (Paris and Durand Citation2009) was used in our experiments. The remaining N-k PCs mainly contain noise, where N is the amount of PCs, therefore, it is not recommended to filter them because this operation will amplify the noise and considerably increases computational times. A soft-thresholding scheme is applied for denoising those PCs. To enlarge the PCs, spatial sizes same as the RGB image cubic interpolation was used. The image processing chain is summarized in Algorithm 1.
3. Results
To evaluate the gains in detection rates of the proposed fusion approach, MPs were generated for increasing values of M, from 1 to 4, and for all SEs listed in Algorithm 1. The parameters of the joint bilateral filter were set to σs = 5 and σr = 0.01. Among the different values tested, these values offered a good trade-off between denoising and detail preservation, see Figure . Filtering was applied on the first k = 6 principal components. The K-nearest neighbor classifier (K = 6) was employed in our experiments. 10% of test data-set is randomly selected as training data-set. The classifier was evaluated against the testing sets; the results were averaged over 5 runs.
We compared leaf classification rates of the proposed fusion method (Proposed), the original HS image (Raw), MPs generated on the high spatial resolution color image (MPs), and the fusion method based on stacking HS and MPs data (Stacked). Overall accuracy (OA) and average accuracy (AA) metrics were computed for each case. The first metric is the ratio of correctly classified points to the number of test data points. The AA is similar to OA but calculated for each object class. In our experiments, we found that regardless of the SE shape as M increases the classification accuracy improves. However, for M higher than 4, the improvement of the detection rates is marginal.
Table summarizes the results obtained using MPs for different SE’s shapes and M = 4. We noted that detection rates for simple SE shapes are higher than that for complex ones. Classification accuracy for the proposed method is consistently higher than that of other approaches. This indicates that our fusion method is capable of fusing the complementary information from multi-sensor and multi-resolution data without increasing the dimensionality of the original HS image. The best results correspond to our fusion method for MPs generated using the linear-shaped SE. This can be explained by the fact that leaves contain mostly low-contrast linear-like features at the midrib and veins. In general, leaves do not show a wide variety of objects in comparison to HS urban images and other types of images used in remote sensing.
Table 1. Classification results by SE shape (%).
To understand how spatial information is captured from the color image, we plotted the morphological profiles extracted using the linear-shaped SE for each object class. A point in a curve corresponds to the output of either an opening or closing by reconstruction. For M = 4, eight images are generated, 4 for the opening on the left side and 4 for the closing on the right, for each color channel, see Figure . Most curves display a small slope with the exception of the midrib and spot categories in which larger intensity variations are recorded. The range of reflectance values differ for each channel as well as the curves relative position. The characteristics can be exploited for further improvement in automatic class discrimination.
Figure 7. Average reflectance for linear-shaped SE, size 1–4, opening on the left side and closing on the right in each channel as is pointing out by the x-axis.
In the last experiment, detection accuracy was measured for each object class. The raw spectral data was included as previous work and have successfully detected leave’s objects using only spectral features. Table shows that in most classes the proposed method provides higher accuracy. For certain objects, spectral information is enough to obtain good results, for example, the dead and dying leaf categories. This is consistent with the expected differences in spectra of such leaf areas. Whereas the inclusion of spatial data improves detection rate for the other object classes. These results support our claim that the proposed fusion scheme manages to capture more morphological information than the approaches used to build the MPs and stack data-set. This effect can be observed in the classification map depicted in Figure . It is important to mention that the experiment has been repeated 5 times with different object class. The upper part of Table shows AA’s for each class in the experiment. The lower part of the table shows OA and average AA of the same experiment and the standard deviation (Std) for the 5 runs. To compare the efficiency of each method, we report the consumed time with 74.16, 273.67, 21.63, and 99.20 s for Raw, Stacked, Morph, and our proposed method, respectively. We can find that our proposed method consumes less time than Stacked and produces better results.
Table 2. Classification rates for each object class.
4. Conclusions
This work clearly demonstrates the added value of transferring spatial information (from a high spatial resolution color image) to a close range spectral data. The proposed fusion technique outperforms some conventional methods which are widely used in remote sensing literature. Although these are the initial results and our experiments focused on general leaf structures, the proposed technique can be adapted to detect objects with specific shapes and size by selecting a suitable SE.
This is of particular importance for early detection of the disease symptoms that usually appear as geometrically simple image objects. In future work, we will explore this idea on a larger data-set of infected and control plants (e.g. through UAVs). Analysis of leaf collected in the field is another promising venue to apply our technique, as mobile HS imagining is becoming reality.
Funding
This work was supported by the Flemish Interuniversity Council (VLIR), and by the FWO project [G037115N: Data fusion for image analysis in remote sensing].
Notes on contributors
Gladys Villegas is a voluntary researcher at Ghent University and Escuela Superior Politecnica del Litoral (ESPOL). Her main interests are remote sensing and precision agriculture. In particular, her interests are focused on hyperspectral image restoration, mathematical morphology, data fusion, and classification. At present, she is working on detection of vegetation diseases.
Wenzhi Liao has been working as a postdoctoral scholar at Ghent University and is a postdoctoral fellow with the Fund for Scientific Research in Flanders. His current research interests include pattern recognition, remote sensing, and image processing. In particular, he is interested in mathematical morphology, multitask feature learning, multisensor data fusion, and hyperspectral image restoration. He is a member of the IEEE Geoscience and Remote Sensing Society (GRSS) and the IEEE GRSS Data Fusion Technical Committee. He is a Senior Member of the IEEE, and serving as an Associate Editor for the IET Image Processing.
Ronald Criollo is a lecturer in the Faculty of Electrical and Computer Engineering at Escuela Superior Politecnica del Litoral University. He has been working as a researcher at the Vision and Robotics Center on many projects about image acquisition and processing.
Wilfried Philips is a full-time professor at Ghent University and head of the research group Image Processing and Interpretation, which is part of the Research Institute Interuniversity MicroElectronics Center. His research interests include image and video restoration, analysis and modeling of image reproduction systems, remote sensing, surveillance, and industrial inspection. He is a Senior Member of the IEEE.
Daniel Ochoa is a full-time professor at Escuela Superior Politecnica del Litoral University (ESPOL) He got its PhD diploma in Gent University in Computer Science. His research interests include image acquisition and processing and pattern recognition. He is the author of several SCI-indexed papers. Currently, he is the head of the Center for Artificial Vision and Robotics at ESPOL and leader of the plant disease detection project funded by VLIR.
Acknowledgements
Wenzhi Liao is a postdoctoral fellow of the Research Foundation Flanders (FWO-Vlaanderen) and acknowledges its support.
References
- Aasen, H., A. Burkart, A. Bolten, and G. Bareth. 2015. “Generating 3D Hyperspectral Information with Lightweight UAV Snapshot Cameras for Vegetation Monitoring: From Camera Calibration to Quality Assurance.” ISPRS Journal of Photogrammetry and Remote Sensing 108 (5): 245–259.10.1016/j.isprsjprs.2015.08.002
- Behmann, J., A. K. Mahlein, S. Paulus, J. Dupuis, H. Kuhlmann, E. C. Oerke, and L. Plümer. 2016. “Generation and Application of Hyperspectral 3D Plant Models: Methods and Challenges.” Machine Vision and Applications 27 (5): 611–624.10.1007/s00138-015-0716-8
- Benediktsson, J. A., J. A. Palmason, and J. R. Sveinsson. 2005. “Classification of Hyperspectral Data from Urban Areas Based on Extended Morphological Profiles.” IEEE Transactions on Geoscience and Remote Sensing 43 (3): 480–491.10.1109/TGRS.2004.842478
- Blaschke, T. 2010. “Object Based Image Analysis for Remote Sensing.” ISPRS Journal of Photogrammetry and Remote Sensing 65 (1): 2–16.10.1016/j.isprsjprs.2009.06.004
- He, K., J. Sun, and X. Tang. 2013. “Guided Image Filtering.” IEEE Transactions on Pattern Analysis and Machine Intelligence 35 (6): 1397–1409. doi:10.1109/TPAMI.2012.213.
- Huang, X., H. Liu, and L. Zhang. 2015. “Spatiotemporal Detection and Analysis of Urban Villages in Mega City Regions of China Using High-resolution Remotely Sensed Imagery.” IEEE Transactions on Geoscience and Remote Sensing 53 (7): 3639–3657.10.1109/TGRS.2014.2380779
- Jacquemoud, S., W. Verhoef, F. Baret, C. Bacour, P. J. Zarco-Tejada, G. P. Asner, C. François, and S. L. Ustin. 2009. “PROSPECT+SAIL Models: A Review of Use for Vegetation Characterization.” Remote Sensing of Environment 113 (2009): S56–S66.10.1016/j.rse.2008.01.026
- Kim, Y., D. M. Glenn, J. Park, H. K. Ngugi, and B. L. Lehman. 2011. “Hyperspectral Image Analysis for Water Stress Detection of Apple Trees.” Computers and Electronics in Agriculture 77 (2): 155–160.10.1016/j.compag.2011.04.008
- Liao, W., M. D. Mura, J. Chanussot, R. Bellens, and W. Philips. 2016. “Morphological Attribute Profiles with Partial Reconstruction.” IEEE Transactions on Geoscience and Remote Sensing 54 (3): 1738–1756.10.1109/TGRS.2015.2488280
- Liao, W., X. Huang, F. V. Coillie, S. Gautama, A. Pižurica, W. Philips, H. Liu, et al. 2015. “Processing of Multiresolution Thermal Hyperspectral and Digital Color Data: Outcome of the 2014 IEEE GRSS Data Fusion Contest.” IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing 8 (6): 2984–2996.10.1109/JSTARS.2015.2420582
- Loncan, L., L. B. D. Almeida, J. M. Bioucas-Dias, X. Briottet, J. Chanussot, N. Dobigeon, S. Fabre, et al. 2015. “Hyperspectral Pansharpening: A Review.” IEEE Geoscience and Remote Sensing Magazine 3 (3): 27–46.10.1109/MGRS.2015.2440094
- Mahlein, A. K., U. Steiner, H. W. Dehne, and E. C. Oerke. 2010. “Spectral Signatures of Sugar Beet Leaves for the Detection and Differentiation of Diseases.” Precision Agriculture 11 (4): 413–431.10.1007/s11119-010-9180-7
- Mookambiga, A., and V. Gomathi. 2016. “Comprehensive Review on Fusion Techniques for Spatial Information Enhancement in Hyperspectral Imagery.” Multidimensional Systems and Signal Processing 27 (4): 863–889.10.1007/s11045-016-0415-2
- Ochoa, D., J. Cevallos, G. Vargas, R. Criollo, D. Romero, R. Castro, and O. Bayona. 2016. “Hyperspectral Imaging System for Disease Scanning on Banana Plants.” In Sensing for Agriculture and Food Quality and Safety VIII, edited by M. S. Kim, K. Chao, and B. A. Chin, Proc. of SPIE Vol. 9864: 98640 M.
- Paris, S., and F. Durand. 2009. “A Fast Approximation of the Bilateral Filter Using a Signal Processing Approach.” International Journal of Computer Vision 81 (1): 24–52.10.1007/s11263-007-0110-8
- Rumpf, T., A. K. Mahlein, U. Steiner, E. C. Oerke, H. W. Dehne, and L. Plümer. 2010. “Early Detection and Classification of Plant Diseases with Support Vector Machines Based on Hyperspectral Reflectance.” Computers and Electronics in Agriculture 74 (1): 91–99.10.1016/j.compag.2010.06.009
- Scharr, H., H. Dee, A. P. French, and S. A. Tsaftaris. 2016. “Special Issue on Computer Vision and Image Analysis in Plant Phenotyping.” Machine Vision and Applications 27 (5): 607–609.10.1007/s00138-016-0787-1
- Soille, P. 2003. Morphological Image Analysis: Principles and Applications. 2nd ed. Secaucus, NJ: Springer-Verlag, New York, Inc. ISBN 3540429883.
- Steger, C. 1998. “An Unbiased Detector of Curvilinear Structures.” IEEE Transactions on Pattern Analysis and Machine Intelligence 20 (2): 113–125.10.1109/34.659930
- Tomasi, C., and R. Manduchi. 1998. “Bilateral Filtering for Gray and Color Images.” Proceedings of the Sixth International Conference on Computer Vision 839–846.
- Triest, D., and M. Hendrickx. 2016. “Postharvest Disease of Banana Caused by Fusarium Musae: A Public Health Concern?” PLOS Pathogens 12 (11): 1–5.

![Figure 5. Morphological profiles for a disk shaped SE of sizes [1, 2, 4].](/cms/asset/8b54c3c3-3375-4bce-906e-11856b04e5c5/tgsi_a_1399673_f0005_oc.gif)