ABSTRACT
Objectives: Imatinib, a breakpoint cluster region-Abelson murine leukemia tyrosine kinase inhibitor, has revolutionized the treatment of chronic myelogenous leukemia (CML). However, the development of multidrug resistance (MDR) limits the clinical application of imatinib. In this study, we aimed to investigate the mechanisms of long noncoding RNA (lncRNA) HOTAIR in CML resistance to imatinib.
Methods: Thirty-four CML patients were divided into multidrug resistance protein 1 (MRP1)-low and MRP1-high groups according to the median expression. Real-time PCR (qPCR) was used to detect the expression of lncRNA HOTAIR in CML patients, and MTT assay and flow cytometry assay were employed to detect the biological function of silencing lncRNA HOTAIR on the cell survival rate and apoptotic rate. An imatinib-resistant human CML cell line K562 (K562-R) was established, and western blot was used to detect the impact of lncRNA HOTAIR on the activation of PI3K/Akt signaling pathway.
Results: Our results showed that lncRNA HOTAIR was greatly upregulated in the MRP1-high patients as well as in the K562-imatinib-resistant cells compared with control. Knockdown of HOTAIR expression downregulated the MRP1 expression levels in the K562-imatinib cells and resulted in higher sensitivity to the imatinib treatment. In addition, the activation of PI3K/Akt was greatly attenuated when HOTAIR was knocked down in K562-imatinib cells.
Discussions: These data suggest that the knockdown of HOTAIR may play a crucial role in improving acquired resistance to imatinib in CML K562-R cells via PI3K/Akt pathway.
Conclusions: LncRNA HOTAIR modulates CML cell MDR in a PI3K/Akt-dependent way.
Introduction
Acquired multidrug resistance (MDR) and disease relapse are often regarded as the causes of the failure of chemotherapeutic drug treatments in patients diagnosed with malignant neoplasm, including leukemia [Citation1,Citation2]. Numerous reports have focused on the mechanisms of MDR of tumor cells, for example the expression of drug-transporting pumps, changes in the targets of anticancer drugs, decrease of drug activity, as well as changes in apoptosis regulatory pathways that contribute to MDR [Citation3]. Imatinib has revolutionized the treatment of chronic myelogenous leukemia (CML). Indeed, 95% of CML patients in chronic phase achieve remission with imatinib [Citation4,Citation5]. However, many patients with accelerated phase or blast crisis develop resistance to imatinib [Citation5,Citation6]. The proposed mechanisms underlying imatinib resistance in CML include impaired drug binding due to breakpoint cluster region-Abelson murine leukemia (BCR-ABL) kinase domain point mutations, amplification of the gene, and overexpression of the multidrug resistance protein 1 (MRP1) gene in tumor cells [Citation7,Citation8]. Although the phenomenon of acquired resistance to imatinib has been observed in CML, little is known about the effects of imatinib on the development of leukemia drug-resistance.
Long noncoding RNAs (lncRNAs) are a heterogeneous class of RNAs that are generally defined as nonprotein-coding transcripts longer than 200 nucleotides [Citation9]. lncRNA, which was considered as only transcriptional ‘noise’ in the past decades, can participate in various critical biological processes, such as chromatin remodeling, gene transcription, and protein transport and trafficking [Citation10,Citation11]. HOTAIR (HOX Antisense Intergenic RNA) is a recently discovered 2.2 kb long lncRNA that is located at the antisense strand of the HOXC gene locus in chromosome 12, flanked by HOXC11 and HOXC12 [Citation12]. A growing body of literature has revealed that lncRNA HOTAIR is highly expressed in variety of cancers, including breast tumor [Citation13], hepatocellular carcinoma [Citation14], pancreatic cancer [Citation15], non-small cell lung cancer [Citation16], colorectal cancer [Citation17], and gastrointestinal stromal tumor [Citation18]. LncRNA HOTAIR expression levels are correlated with tumor metastases and loss of lncRNA HOTAIR has been linked with the decrease in cancer invasiveness [Citation13]. Although lncRNAs play multiple roles in myeloid leukemia oncogenesis [Citation10,Citation19,Citation20], little is known in the acquired resistance to imatinib in CML.
PI3K/Akt signaling network is crucial to widely divergent physiological processes that include cell cycle progression, differentiation, transcription, translation, and apoptosis [Citation21]. Recent studies illustrated that aberrant activation of PI3K/Akt pathway contributed to the drug resistance of different types of leukemia [Citation22]. The PI3K inhibitor LY294002 may have therapeutic potential when combined with doxorubicin in the treatment of MRP1-mediated drug resistance [Citation23]. However, whether lncRNA HOTAIR plays any role in PI3K/AKT-dependent drug resistance of CML is never investigated. In this study, we investigated the expression level of lncRNA HOTAIR in CML patients and analyzed its function in acquired resistance to imatinib in CML. In a cell model obtained by passing of K562 cells in progressively increasing doses of imatinib, we detected the expression and function of lncRNA HOTAIR in imatinib-resistant K562 cells (K562-R), and we describe here a novel expressional profile of lncRNA HOTAIR in peripheral blood mononuclear cells (PBMCs) of CML patients, as well as MRP1-positive CML K562 cells. In addition, we also elucidate that the regulatory mechanism of HOTAIR mediates MDR in CML cell lines via PI3K/Akt pathway.
Materials and methods
Samples from leukemia patients
Thirty-four CML patients comprising 16 males and 18 females with age ranging from 22 to 75 years (median age of 41.7 years) and four healthy donors (aging from 18 to 45 years with a median age of 35.8 years) were included in this study. All the samples were obtained from October 2011 to December 2013 at the Department of Hematology in the Affiliated Hospital of Weifang Medical University, and the Department of Hematology and Blood and Marrow Transplantation in the Tianjin Medical University Cancer Institute and Hospital, and the written informed consent was obtained from all participants. Both the study and the contents of the written consent were approved by the institutional ethics committees of Weifang Medical University and Tianjin Medical University. Bone marrow was collected from patients by the bone marrow puncture at diagnosis. The diagnosis of multidrug-resistant CML patients was based on the expression level of MRP1 via real-time PCR. This study was approved by the ethics committee of the Affiliated Hospital of Weifang Medical University.
Cell culture and generation of imatinib-resistant cell lines
Human chronic myelogenous leukemia K562 cells bought from Zhongyuan Biotech (Beijing, China) were cultured in a complete RPMI 1640 medium with 10% fetal bovine serum (Biological Industries, Beit HaEmek, Israel) and 1% penicillin/ streptomycin at 37°C in a humidified atmosphere containing 5% CO2. A stable imatinib (Sigma-Aldrich, St. Louis, MO, USA)-resistant cell line variant (K562-R) was established from K562 by the continuous exposure of the cells to increasing concentrations up to 2.0 μM. Subsequently, K562-R cells were cultured in the presence of 2.0 μM of imatinib to maintain the drug-resistant phenotype.
RNA interference
Small interfering RNAs (siRNAs) for the lncRNA HOTAIR, and a scramble control (Scramble) were purchased from Gene-Pharma (Gene-Pharma, Shanghai, China). The target sequences for HOTAIR siRNAs were as follows: si-HOTAIR1, sense: 5′-GGACUUUGCACUCUAAAUAUU-3′ antisense: 3′-UUCCUGAAACGUGAGAUUUAU-5′, si-HOTAIR2, sense: 5′-GCCCAAUUUAAGAAUUACAUU-3′ antisense: 3′-UUCGGGUUAAAUUCUUAAUGU-5′ and si-HOTAIR3, sense: 5′-GCACAGAGCAACUCUAUAAUU-3′ antisense: 3′-UUCGUGUCUCGUUGAGAUAUU-5′. Scramble siRNA was as follows: sense: 5′-ACUCUAAAUAGGACUUUGCUU-3′ antisense: 3′-UUAGAUUUAUCCUGAAACGUG-5′. K562 cells (1 × 105/well) were plated in 24-well plates overnight separately. Cells were then transfected with 20pM Scramble or 20pM siRNA against lncRNA HOTAIR (siRNA-1, siRNA-2 or siRNA-3) for 48 hours using an Lipofectamine 2000 transfection reagent from Invitrogen (Thermo Fisher, Shanghai, China) according to the manufacturer’s protocol.
In vitro drug cytotoxic assay
MTT assay was used to measure the drug resistance. 1 × 104 cells per well were plated in a 96-well plate and incubated with different concentrations of imatinib (Sigma-Aldrich, St. Louis, MO, USA) for 48 hours, respectively. Then cells were treated with 100 μl MTT (0.5 mg/ml, Sigma). After 4 hours of incubation at 37°C in 5% CO2, 100 μl DMSO from Gibco (Thermo Fisher, Shanghai, China) was pipetted to solubilize the formazan product for 30 minutes at room temperature. The spectrometric absorbance was measured at 490 nm by a microplate reader (Model 680; Bio-Rad, Hercules, CA, USA). Each test was repeated three times. The drug resistance was estimated by comparing the IC50 values (drug concentration that inhibits cell growth by 50%) from growth inhibition curves.
Real-time PCR
Total RNA was isolated and quality-checked as previously described [Citation24]. First strand cDNAs were synthesized using the iScript™ cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA). Real-time quantitative PCR was performed using the iTaq™ SYBR® Green Supermix with ROX (Bio-Rad Laboratories, Hercules, CA, USA) on a Mastercycler® ep realplex thermal cycler (Eppendorf, Shanghai, China), with quantitation by the relative standard curve method. Relative expression levels were calculated on the basis of the threshold cycle for amplification. The primers for different genes were as follows: HOTAIR: sense, 5′-AAACAGAGTCCGTTCAGTGTCA-3′, antisense, 5′-ATTCTTAAATTGGGCTGGGTC-3′; GAPDH: sense, 5′-AATGGACAACTGGTCGTGGAC-3′, antisense, 5′-CCCTCCAGGGGATCTGTTTG-3′; MRP1, sense, 5′-ATGCGAATGAGGAGGTGGAG-3′, antisense, 5′-CAGGCAGGCAGTGACAAACA-3′. ABL1, sense, 5′-GGAGAGCTGCAGAGCACAGA-3′, antisense, 5′-CAATGGAGACACGGCAGGCT-3′.
Western blot analysis
Total cell protein was electrophoresed in 10% SDS-PAGE gel and blotted onto a polyvinylidene difluoride membrane. After being blocked with 5% powdered skim milk for 2 hours in phosphate buffered saline containing 0.1% Tween 20 (PBST), the membranes were incubated with antibody overnight at 4°C (anti-MRP1, Cat. No.sc-365635, 1:500 diluted, Santa Cruz Biotech, Santa Cruz, CA, USA; anti-PI3K p85, Cat. No. 4292, anti-PI3K p110a Cat. No. 4228, anti-Akt, Cat. No. 4685, anti-p-Akt, Cat. No. 4060, all 1:1000 diluted, Cell Signaling Technology, MA, USA), and then incubated with secondary antibody anti-rabbit/mouse-HRP (1:2000 diluted, Santa Cruz Biotech, Santa Cruz, Dallas, TX, USA). GAPDH antibody (1:200 diluted, Santa Cruz Biotech, Santa Cruz, Dallas, TX, USA) was used as a control. All bands were detected using ECL Western blotting kit (Amersham Biosciences, Amersham, UK), according to the manufacturer’s instruction.
Annexin V/propidium iodide staining assay
Apoptosis was analyzed using a flow cytometer with FITC conjugated-Annexin V/propidium iodide (PI) double staining to detect membrane events. In brief, after treatment with imatinib, whole cells were collected in the HEPES buffer (10 mM HEPES at pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2). Subsequently, cells were stained with FITC conjugated-Annexin V (2.5 μg/ml) and PI (2 ng/ml) for 20 minutes at room temperature, followed by the analysis on a flow cytometer (Becton Dickinson, CA, USA) using the CellQuest software (Becton Dickinson, CA, USA). The sum of early apoptosis (annexin V+/PI−) and late apoptosis (annexin V+/PI+) is presented as total apoptosis.
Statistical analysis
Results are shown as mean ± standard deviation (SD). Statistical significance of differences among different designed groups was calculated using a two-tailed unpaired Student’s t-test. Pearson correlation analysis was used to estimate the relationship between the expression level of lncRNA HOTAIR and MRP1. A p value of less than 0.05 was considered to be statistically significant. All experiments were repeated at least three times except other explanation. In the figures, * and # indicate statistical significance at p < 0.05 and p < 0.01, respectively. The statistical calculations were performed with GraphPad Prism 6.0 for Windows (GraphPad Software, La Jolla, CA, USA).
Results
Expression of MRP1 and HOTAIR in CML patients
Because the increased expression of MRP1 is involved in cancer cell resistance to chemotherapeutic agents [Citation25], we first analyzed the expression of MRP1 and lncRNA HOTAIR in CML patients. Significant difference was observed in the expression level of lncRNA HOTAIR and MRP1, and positive correlation was found between the expression level of lncRNA HOTAIR and MRP1 ((a)). The 34 samples were divided into two groups according to the median expression of MRP1, in which 17 samples belonged to MRP1 high expression group and 17 samples belonged to MRP1 low expression group ((b)). To identify the altered mRNA expression level of lncRNA HOTAIR in CML patients, the PBMC isolated from both MRP1 high expression and MRP1 low expression CML patients and health donors were analyzed by real-time PCR. We found that the expression of lncRNA HOTAIR was significantly upregulated in all CML patients compared with health donors, in which the MRP1 high expression group showed higher expression of lncRNA HOTAIR compared with the MRP1 low expression group ((c)), which indicated that HOTAIR might play important roles in CML tumor pathological grade and prognosis.
Figure 1. Expressions of MRP1 and lncRNA HOTAIR in CML patients. (a) The expression of MRP1 and lncRNA HOTAIR were both detected via real-time PCR in CML patients (n = 34), and the correlation between the expression level of lncRNA HOTAIR and MRP1 was analyzed using the Pearson correlation analysis. (b) The 34 samples were divided into two groups according to the median expression of MRP1 gene. (c) lncRNA HOTAIR expression was detected in CML patients including healthy donors (n = 4), and the MRP1 low expression group (n = 17) and MRP1 high expression group (n = 17) were classified according to the median number. Relative lncRNA HOTAIR expression was determined using the formula 2−ΔΔCt with GAPDH as an internal control.
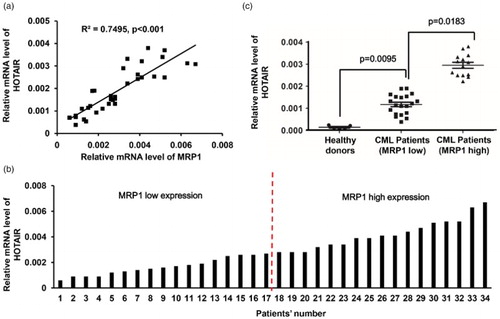
Expression of MRP1 and lncRNA HOTAIR in K562 and K562-R cell lines
To investigate the mechanisms of cellular resistance to imatinib in CML, we first established an imatinib-resistant cell line using CML K562 cells. These cells were made resistant to imatinib by stepwise selection in imatinib. The expression of MRP1 was detected in parental K562 cells (K562) and the imatinib-resistant K562 cells (K562-R), in which we found that MRP1 was significantly upregulated in K562-R cells ((a)). The analysis of drug sensitivity using the MTT assay indicated that K562-R cells were more resistant to imatinib than parental K562 cells, because the IC50 was ∼250 nM in the parental K562 cells, but it went up to ∼10 μM in the K562-R cells ((b)). In addition, we analyzed the expression of lncRNA HOTAIR in K562 cells and K562-R cells by real-time PCR, and the result showed that lncRNA HOTAIR was overexpressed by nearly 10-folds in K562-R cells compared to K652 cells ((c)). Taken together, the expression of lncRNA HOTAIR was related to the imatinib resistance of the K562 cells.
Figure 2. Expression of MRP1 and lncRNA HOTAIR in the K562 and K562-R cell lines. (a) Western blot shows the MRP1 protein level in membrane vesicle preparations from K562 and K562-R cells with GAPDH as a loading control. (b) Sensitivity of K562 cells and K562-R cells to increasing doses of imatinib was analyzed using the MTT cytotoxicity assay. Data points are presented as means ± standard deviation of at least three separate experiments. (c) Real-time PCR analysis of mRNA expression levels of lncRNA HOTAIR in the K562 and K562-R cells with GAPDH as an internal control. *p < 0.05; #p < 0.001.
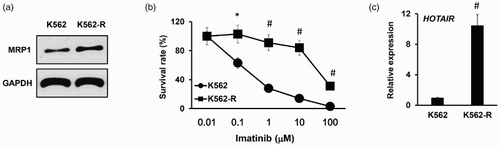
Altered expression of HOTAIR effects on the chemosensitivity of K562 cells
To investigate the functional effects of lncRNA HOTAIR on leukemia cells, we performed loss-of-function analysis of lncRNA HOTAIR. Three individual HOTAIR siRNAs were designed and transfected into the K562-R cells using Lipofectamine 2000 reagent, and real time-PCR was used to examine mRNA level of lncRNA HOTAIR 48 hours after transfection. The result showed that the expression of lncRNA HOTAIR was greatly silenced with siRNA-1 and si-RNA-2, whereas siRNA-3 only slightly downregulated the expression of HOTAIR; therefore, we used only the miRNA-1 and miRNA-2 for further experiments ((a)). To elucidate the direct effect of lncRNA HOTAIR expression on the chemosensitivity of K562-R cells, we silenced lncRNA HOTAIR in the K562-R cells with siRNA-1 and siRNA-2 and then exposed them to increasing concentration of imatinib treatment (up to 100 μM). We found that the cells with lncRNA HOTAIR knockdown using both the siRNA- and siRNA-1 were less resistant to imatinib compared with those transfecting with Scramble control, and the IC50 of K562-R cells with lncRNA HOTAIR knockdown was reversed to ∼500 nM compared with the ∼10 μM in the control cells ((b)). Furthermore, scramble or siRNA-transfected cells exposed to 500nM of imatinib treatment for 24 hours and the apoptosis rate was detected with flow cytometry assay using FITC-Annexin V/PI double staining kit. The result was also consistent with results obtained from MTT assay: more apoptosis was induced in the K562-R cells with HOTAIR knockdown compared with the scramble control ((c)). Since BCR-ABL1 mutation is often correlated with CML drug resistance, we also detected whether knockdown of lncRNA HOTAIR changes the expression of ABL1. As shown in the result, K562-R cells transfected with siRNA-1 did not alter the expression of ABL1 significantly ((d)).
Figure 3. Manipulation of lncRNA HOTAIR expression changes the chemosensitivity of K562 cells to imatinib treatment. (a) Real time-PCR analysis of mRNA expression levels of lncRNA HOTAIR in the K562-R cells transfected with two siRNAs targeting HOTAIR, or Scramble as the control. (b) Sensitivity of the K562-R cells transfected with siRNAs targeting HOTAIR to increasing doses of imatinib was analyzed using the MTT cytotoxicity assay, with scramble as the control. Data points are presented as means ± standard deviation of at least three separate experiments. (c) Apoptosis rate of K562-R cells transfected with siRNAs targeting HOTAIR to increasing doses of imatinib was analyzed using flow cytometry assay, with scramble as the control. (d) Detection of expression of ABL1 gene in the K562 cells and K562-R cells transfected with HOTAIR siRNA-1 using real-time PCR, and GAPDH was used as an internal control. *p < 0.05; #p < 0.001.
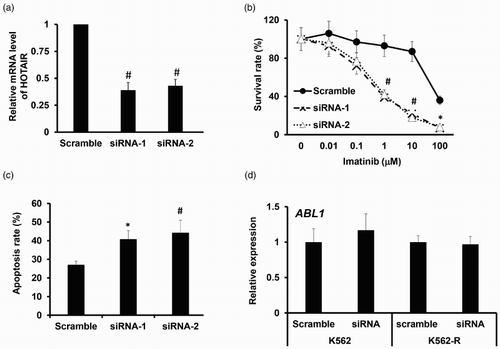
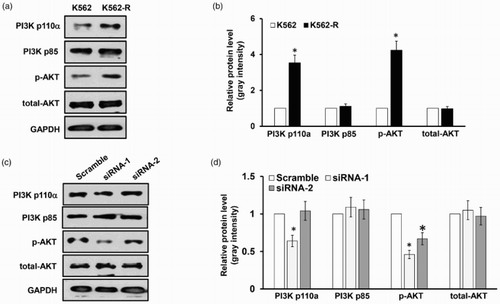
LncRNA HOTAIR mediates the activity of PI3K/Akt signaling pathway
In order to investigate whether lncRNA HOTAIR is correlated with activation of the PI3K/Akt in CML, the levels of the main signal molecules of PI3K/Akt pathway were assessed in K562-R cells. Western blot analysis showed that the levels of PI3 kinase p110α (the catalytic subunit of PI3K) and phosphorylation Akt were significantly upregulated in K562-R cells compared with normal K562 cells ((a,b)). Further on, we investigated whether knockdown of lncRNA HOTAIR affects the activation of the PI3K/Akt in K562-R cells. We found that there was no great change in PI3K p85 (regulatory subunits p85α and p85β) and the total amount of Akt proteins, whereas knockdown of lncRNA HOTAIR in K562 cells significantly decreased protein expression of PI3 kinase p110α and phosphorylation Akt as illustrated ((c,d)). From these data, we conclude that lncRNA HOTAIR was implicated in key steps of imatinib development in CML.
Figure 4. LncRNA HOTAIR mediates the activity of PI3K/Akt signaling pathway. (a). Western blot analysis of PI3K p100a, PI3K p85, p-Akt, total Akt in membrane vesicle preparations from K562 and K562-R cells with GAPDH as a loading control. The quantitative grayscale intensity analysis result is shown in (b). (c). Western blot analysis of PI3K p100a, PI3K p85, p-Akt, total Akt in membrane vesicle preparations from K562-R cells transfected with siRNA-1 and siRNA-2 targeting lncRNA HOTAIR, with GAPDH as a loading control. The quantitative grayscale intensity analysis result is shown in (d). *p < 0.05; #p < 0.001.
Discussion
Despite the significant activity of imatinib in CML treatment, acquired MDR has been observed in some patients, particularly those in advanced stages [Citation26]. MDR in CML continues to be a significant problem. Recently, some progresses have been gained in revealing the mechanism of MDR [Citation27]. Our study continued to investigate the role of lncRNA HOTAIR in mediating MDR in human leukemia cells and its possible mechanisms.
Aberrant expression of lncRNA HOTAIR has been detected in carcinomas of gastric cancer [Citation28], pancreatic cancer [Citation29], lung cancer [Citation30], hepatocelluar carcinoma [Citation31], colon cancer [Citation32], and ovarian cancer [Citation33]. LncRNA HOTAIR is known to be involved in the process of proliferation, invasion, and apoptosis in myeloid leukemia, especially acute myeloid leukemia [Citation10,Citation19,Citation20]. From our results, we postulate that abnormal expression of lncRNA HOTAIR might also involve in the development of MDR possibly in CML. In this study, we detected the expression profiles of lncRNA HOTAIR in the tumor cells of leukemia patients using real-time PCR analysis. The patients with high MRP1 expression showed higher expression levels of lncRNA HOTAIR compared with those with low MRP1 expression group. To note, the PBMC cells isolated from CML patients may also contain cancer stem cell, such as the CD34+ stem/progenitor cells. Since CD34+ stem/progenitor cells contain most of leukemia stem cells (LSCs), and LSC is also a factor for IM resistance, the failure to separate CD34+-enriched cells instead of using PBMCs as the primary CML sample is a weakness of this study. To investigate the cellular-resistant mechanisms to imatinib in CML, we established an imatinib-resistant cell line through stepwise drug selection of a CML cell line (K562-R). Our results suggested that the K562 cells with imatinib resistance were characterized with higher expression levels of lncRNA HOTAIR compared to their parental cells. Western blot results showed that MRP1 was overexpressed in K562-R cells compared with K562 cells; additionally, the lncRNA HOTAIR mRNA level was significantly elevated in the resistant K562 cells. These results suggest that overexpression of lncRNA HOTAIR might confer resistance to imatinib in K562 imatinib cells.
Recently, Yan et al. showed that the expression levels of lncRNA HOTAIR were upregulated in diffuse large B cell lymphoma tumor tissues and cell lines compared with normal tissues and cells; furthermore, knockdown of the lncRNA HOTAIR led to growth inhibition, cell cycle arrest and apoptosis in vitro, possibly through the regulation of phosphoinositide 3 kinase/AKT/nuclear factor κB pathway [Citation34]. Dou et al. showed that when knockdown of lncRNA HOTAIR in CD133(+) colorectal cancer stem cells suppressed cellular proliferation, migration, invasion, colony-forming, in particular, markedly attenuated the tumor growth and lung metastasis in xenograft nude mice [Citation17]. Gao et al. showed that the inhibition of lncRNA HOTAIR in liver cancer cells resulted in the suppression of cell proliferation and invasion in vitro, and lncRNA HOTAIR depletion significantly inhibited the rate of growth of liver cancer cells in vivo [Citation35].
In this study, we found that lncRNA HOTAIR plays a pivotal role in controlling K562-R cell survival, apoptosis and drug resistance functionally. Our studies demonstrated that knockdown of lncRNA HOTAIR could effectively increase the drug sensitivity of K562-R cells to imatinib treatment. Furthermore, we also observed that the apoptosis rate was significantly upregulated in K562-R cells with lncRNA HOTAIR knockdown. This study might provide a better understanding of a new mechanism of lncRNAs-mediated drug resistance in leukemia.
BCR-ABL1 kinase-independent IM resistance, including activation of downstream signaling pathways, such as PI3K, MAPK, or JAK/STAT, play important part in acquired IM resistance. The PI3K/Akt signaling pathway controls the expression and function of many proteins that are necessary for leukemia cell MDR [Citation36,Citation37]. Although increasing evidences showed that PI3K/Akt signaling was frequently activated in CML [Citation21,Citation38]; however, no data have confirmed the correlation of the HOTAIR-mediated PI3K/Akt signaling pathway with MDR to our knowledge. This study demonstrated that the resistant cell line K562-R presented higher PI3K/Akt activity than the sensitive one, which was in accordance with the MDR. Furthermore, decreased expression of lncRNA HOTAIR markedly modulated the activity of PI3K/Akt pathway in CML cell lines. Inhibition of PI3K/AKT signaling through knockdown of lncRNA HOTAIR has been found to render CML cells more sensitive to imatinib treatment. These results indicated that HOTAIR-modulated CML cell MDR was, at least in part, PI3K/Akt-dependent. However, mechanisms of acquired IM resistance include the acquisition of point or compound mutations in the BCR-ABL1 kinase domain and the gene amplification [Citation39]. According to our results, we detected the expression of ABL1 gene expression in the lncRNA HOTAIR knockdown K562-R cells and did not find significant change; therefore, the mechanism of lncRNA HOTAIR on CML IM resistance is BCR-ABL1 kinase-independent. Notably, previous studies demonstrated that lncRNA HOTAIR silences the HOXD locus genes present at chromosome 2 in humans, in trans, via recruitment of various gene silencing machineries [Citation12]. For example, lncRNA HOTAIR interacts with PRC2 (polycomb repressive complex 2) and histone demethylases LSD1/CoREST/REST complexes through its 5′- and 3′-end, respectively [Citation12,Citation13,Citation40]. Notably, the ability to tether two distinct complexes enables RNA-mediated assembly of PRC2 and LSD1 and coordinates targeting of PRC2 and LSD1 to chromatin for coupled histone H3 lysine 27 methylation and lysine 4 demethylation [Citation40]. In future, the detailed molecular biology mechanism of how HOTAIR regulated the PI3K/Akt signaling pathway should be focused on. In summary, we first divided the patient samples to two groups according to the endogenous expression level of MRP1, and we found that the samples overexpressed HOTAIR with MRP1 high expression, and highly expressed HOTAIR is associated with the imatinib resistance in K562 cells. Increased MRP-1 has been unraveled in IM-resistant CML cells, and low-level of MRP-1 may serve as a good marker for prognosis in CML patients. However, to date, our study is the first one to report that MRP-1 level could be a marker or predictor for IM resistance, and more investigation should be done to support this point. Moreover, grouping IM-sensitive and IM-resistant patients according to relapse or refractory during IM treatment should be considered in the future studies. Taken together, the present study provides evidence, for the first time, that lncRNA HOTAIR may play important roles in the imatinib resistance in CML cell lines. However, the mechanisms of imatinib resistance in CML may be multifactorial and certainly more complex than only dysregulated expression of lncRNA HOTAIR; the precise molecular mechanisms behind the involvement of lncRNA HOTAIR in CML require further investigation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes on contributors
Haiying Wang, M.D., is a hematologist of the Affiliated Hospital of Weifang Medical University, Department of Hematology; she is a Ph.D. candidate of Shandong University, major in blood malignances. Her research interests in the mechanisms of lncRNA in lymphoma drug resistance.
Qian Li, M.D., is a physician of the Department of Hematology and Blood and Marrow Transplantation, Tianjin Medical University Cancer Institute and Hospital; she is also a Ph.D. candidate of Chinese Academy of Medical Science and Peking Union Medical College; her research focuses on myeloma and lymphoma refractory and drug resistance.
Shusen Tang, M.D., Surgeon, is Associate Professor of the Affiliated Hospital of Weifang Medical University.
Meifang Li is a master candidate of Weifang Medical University; her major is hematological malignance.
Anhua Feng, M.D., is a physician of the Department of Hematology, the Affiliated Hospital of Weifang Medical University.
Lili Qin, M.D., is a physician of the Department of Hematology, the Affiliated Hospital of Weifang Medical University.
Zhiqiang Liu, M.D., Ph.D., now is full professor of the Department of Pathophysiology, Tianjin Medical University after he came back from the University of Texas MD Anderson Cancer Center. His researches mainly focus on cancer drug resistance, especially in multiple myeloma and lymphoma.
Xin Wang, M.D., is Professor and Director of the Department of Hematology, Shandong Provincial Hospital Affiliated to Shandong University. She is an expert in leukemia, lymphoma, multiple myeloma, myelodysplastic syndromes (MDS) diagnosis and chemotherapy, immunotherapy as well as stem cell transplantation.
ORCID
Meifang Li http://orcid.org/0000-0002-7698-4021
Anhua Feng http://orcid.org/0000-0001-5850-0890
Xin Wang http://orcid.org/0000-0001-8051-1481
Additional information
Funding
References
- Wang H, Wang X, Li Y, et al. The proteasome inhibitor bortezomib reverses P-glycoprotein-mediated leukemia multi-drug resistance through the NF-kappaB pathway. Die Pharmazie. 2012;67(2):187–192.
- Ma H, Cheng L, Hao K, et al. Reversal effect of ST6GAL 1 on multidrug resistance in human leukemia by regulating the PI3K/Akt pathway and the expression of P-gp and MRP1. PLoS One. 2014;9(1):e85113. doi: 10.1371/journal.pone.0085113
- Bosch TM. Pharmacogenomics of drug-metabolizing enzymes and drug transporters in chemotherapy. Methods Mol Biol. 2008;448:63–76. doi: 10.1007/978-1-59745-205-2_5
- Walz C, Sattler M. Novel targeted therapies to overcome imatinib mesylate resistance in chronic myeloid leukemia (CML). Crit Rev Oncol/Hematol. 2006;57(2):145–164. doi: 10.1016/j.critrevonc.2005.06.007
- Spolitu S, Uda S, Deligia S, et al. Multidrug resistance P-glycoprotein dampens SR-BI cholesteryl ester uptake from high density lipoproteins in human leukemia cells. Am J Can Res. 2016;6(3):615–627.
- Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344(14):1038–1042. doi: 10.1056/NEJM200104053441402
- Huang H, Luo Y, Liang Y, et al. CD4 + CD25+ cells in multiple myeloma related renal impairment. Sci Rep. 2015;5:16565. Epub 2015/11/14. doi: 10.1038/srep16565
- Peng XX, Tiwari AK, Wu HC, Chen ZS. Overexpression of P-glycoprotein induces acquired resistance to imatinib in chronic myelogenous leukemia cells. Chin J Cancer. 2012;31(2):110–118. doi: 10.5732/cjc.011.10327
- Deng H, Zhang J, Shi J, et al. Role of long non-coding RNA in tumor drug resistance. Tumour Biol. 2016;37(9):11623–11631. Epub 2016/10/27. doi: 10.1007/s13277-016-5125-8
- Hao S, Shao Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int J Clin Exp Pathol. 2015;8(6):7223–7228.
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975
- Ishibashi M, Kogo R, Shibata K, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29(3):946–950.
- Xie Z, Chen X, Li J, et al. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget. 2016.
- Zhou C, Ye L, Jiang C, et al. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1alpha activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. 2015;36(12):9179–9188. doi: 10.1007/s13277-015-3453-8
- Dou J, Ni Y, He X, et al. Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am J Transl Res. 2016;8(1):98–108.
- Niinuma T, Suzuki H, Nojima M, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Can Res. 2012;72(5):1126–1136. doi: 10.1158/0008-5472.CAN-11-1803
- Wu S, Zheng C, Chen S, et al. Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol Lett. 2015;10(4):2410–2414.
- Xing CY, Hu XQ, Xie FY, et al. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589(15):1981–1987. doi: 10.1016/j.febslet.2015.04.061
- Zhang X, Dong W, Zhou H, et al. Alpha-2,8-sialyltransferase is involved in the development of multidrug resistance via PI3K/Akt pathway in human chronic myeloid leukemia. IUBMB Life. 2015;67(2):77–87. doi: 10.1002/iub.1351
- Tazzari PL, Cappellini A, Ricci F, et al. Multidrug resistance-associated protein 1 expression is under the control of the phosphoinositide 3 kinase/Akt signal transduction network in human acute myelogenous leukemia blasts. Leukemia. 2007;21(3):427–438. doi: 10.1038/sj.leu.2404523
- Abdul-Ghani R, Serra V, Gyorffy B, et al. The PI3K inhibitor LY294002 blocks drug export from resistant colon carcinoma cells overexpressing MRP1. Oncogene. 2006;25(12):1743–1752. doi: 10.1038/sj.onc.1209201
- Liu Z, Xu J, He J, et al. A critical role of autocrine sonic hedgehog signaling in human CD138+ myeloma cell survival and drug resistance. Blood. 2014;124(13):2061–2071. Epub 2014/07/23. doi: 10.1182/blood-2014-03-557298
- Lu JF, Pokharel D, Bebawy M. MRP1 and its role in anticancer drug resistance. Drug Metab Rev. 2015;47(4):406–419. Epub 2015/11/07. doi: 10.3109/03602532.2015.1105253
- Benjamin JE, Stein AS. The role of blinatumomab in patients with relapsed/refractory acute lymphoblastic leukemia. Ther Adv Hematol. 2016;7(3):142–156. doi: 10.1177/2040620716640422
- Deininger MW. Diagnosing and managing advanced chronic myeloid leukemia. American Society of Clinical Oncology educational book/ASCO American Society of Clinical Oncology Meeting. 2015:e381-8.
- Guo LL, Song CH, Wang P, et al. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21(41):11680–11687. doi: 10.3748/wjg.v21.i41.11680
- Kishikawa T, Otsuka M, Ohno M, et al. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol. 2015;21(28):8527–8540. doi: 10.3748/wjg.v21.i28.8527
- Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4
- He Y, Meng XM, Huang C, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Can Lett. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021
- Luo ZF, Zhao D, Li XQ, et al. Clinical significance of HOTAIR expression in colon cancer. World J Gastroenterol. 2016;22(22):5254–5259. doi: 10.3748/wjg.v22.i22.5254
- Wu H, Shang X, Shi Y, et al. Genetic variants of lncRNA HOTAIR and risk of epithelial ovarian cancer among Chinese women. Oncotarget. 2016.
- Yan Y, Han J, Li Z, et al. Elevated RNA expression of long noncoding HOTAIR promotes cell proliferation and predicts a poor prognosis in patients with diffuse large B cell lymphoma. Mol Med Rep. 2016;13(6):5125–5131.
- Gao JZ, Li J, Du JL, et al. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11(3):1791–1798.
- Wang H, Jia XH, Chen JR, et al. Osthole shows the potential to overcome P-glycoprotein-mediated multidrug resistance in human myelogenous leukemia K562/ADM cells by inhibiting the PI3K/Akt signaling pathway. Oncol Rep. 2016;35(6):3659–3668.
- Chen JR, Jia XH, Wang H, et al. Timosaponin A-III reverses multi-drug resistance in human chronic myelogenous leukemia K562/ADM cells via downregulation of MDR1 and MRP1 expression by inhibiting PI3K/Akt signaling pathway. Int J Oncol. 2016;48(5):2063–2070.
- Bertacchini J, Heidari N, Mediani L, et al. Targeting PI3K/AKT/mTOR network for treatment of leukemia. Cell Mol Life Sci: CMLS. 2015;72(12):2337–2347. doi: 10.1007/s00018-015-1867-5
- Berman E, Jhanwar S, Hedvat C, et al. Resistance to imatinib in patients with chronic myelogenous leukemia and the splice variant BCR-ABL1(35INS). Leuk Res. 2016;49:108–112. Epub 2016/09/23. doi: 10.1016/j.leukres.2016.08.006
- Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002
