ABSTRACT
Objective: The objective of our review is to highlight the significance of microsatellite hypervariation in diagnostics of hematologic malignancies.
Methods: For the past few decades, extensive experiments in cancer research have explored all the possible pathways and a number of deleterious mutations that either make the tumor suppressor genes (TSGs) dysfunctional or cause the proto-oncogenes to behave abnormally by changing the cellular phenotype hence rendering disease. To prevent the deleterious effects of mutations and to protect the genomic integrity, our system possesses multiple repair mechanisms. DNA Mismatch Repair (MMR) and Direct Reversal of Damage (DRD) are two repair mechanisms which help in removal of faulty base pairs and alkyl adduct formation respectively to avoid long term effects of toxicity, tumorigenesis and mutagenesis. There are nine major MMR genes – MutS homolog (MSH2, MSH3, MSH4, MSH5, MSH6), MutL homolog (MLH1, MLH3), human post-meiotic segregation genes (PMS1, PMS2), and three major damage reversal genes – O6-methylguanine-DNA-methyltransferase (MGMT), ABH2 and DEPC1.
Results: Any malfunction in DNA repair machinery can cause microsatellite instability (MSI), a form of genomic abnormality with hyper mutable repeats that is directly associated with cancer. Microsatellites are short, repetitive sequences, non-randomly distributed and localized in 3'-UTR (Untranslated Region), introns, coding regions and promoters. Besides MSI, evidence on promoter hypermethylation of selected repair genes also points toward a prominent reason for cancer initiation and progression.
Conclusion: The presence of specific microsatellite marker hyper-mutability and consistent promoter hypermethylation in leukemia or lymphoma can be considered as a part of routine diagnostic test in clinical laboratories.
KEYWORDS:
1. Introduction
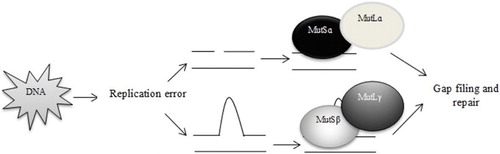
Cancer, the disease of the genes and environment, develops through a complex genetic pathway and the complexity of tumor development varies in different cancers [Citation1]. In colon, lung, gastric, breast cancers and other solid tumors, the molecular complexity of the tumor development is difficult to decipher only using cytogenetic techniques, whereas hematologic malignancies are known to arise mostly due to chromosomal rearrangements [Citation2]. Our body possesses a number of ‘Repair Systems’ that detects the DNA damage, halts the replication process and corrects the error, thus eliminating any flaws in the genome and blocking mutations in somatic cells [Citation3]. Mismatch repair (MMR) machinery is one of the most significant DNA repair mechanisms that is involved in the rectification of base-pair alterations, insertion–deletion loops and hetero-duplexes instigated during replication and recombination. Several repair genes (MSH2, MSH3, MSH6, MLH1, MLH3, PMS1 and PMS2) are involved in this process and each of them is multi-functional in nature. These genes work by forming heterodimers and, depending on the type of lesion a heterodimer is formed – binding of MSH2 gene with MSH6 (MutSɑ) or MSH3 (MutSβ) and MLH1 with either of PMS1 (MutLβ), PMS2 (MutLɑ) or MLH3 (MutLγ). The interaction between mismatch recognition complexes (MutS/MutL) and other proteins such as helicase, proliferating cell nuclear antigen, replication protein A, exonuclease 1 is very much essential to bring about refinement of the errors [Citation4] ().
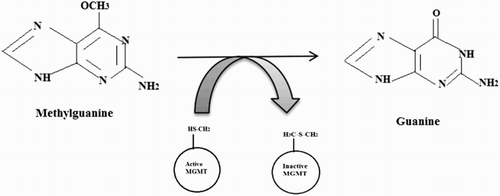
Introduction of an alkyl group and accumulation of DNA-alkyl adducts are considered to be carcinogenic. For instance, an addition of a methyl group to the guanine nucleotide results in complementary base pairing to thymine instead of cytosine causing transition. Direct Reversal of Damage system includes MGMT, a damage reversal enzyme, which transfers the methyl group from O6-methylguanine to the cysteine residue in the enzyme’s active site and thereby eliminating the adduct [Citation5] ().
Microsatellites are simple repeat sequences, prone to hyper variability and thus predisposed to accumulation of mutational errors which are fundamentally caused due to the defects in DNA repair machinery. Promoter hypermethylation of CpG islands causes transcriptional silencing of repair genes and interestingly associated with the development of microsatellite instability (MSI). For example – in absence of MGMT, MMR genes plays the primary role to correct alkylated lession, making chromatin overloaded and resulting an early onset of MSI. Alternatively, DNA-damaging agents are also capable of modifying the repair gene expression; this is another reason for occurrence of MSI [Citation6,Citation7]. Recent studies revealed microsatellite hypervariation due to epigenetic dysregulation of selected DNA repair genes, such as MGMT and MLH1, in primary lymphoid and myeloid tumors, even though this is considered to be a rare event [Citation8,Citation9]. It is possible to introduce MSI testing in the field of diagnostics in blood cancers which are otherwise routinely conducted for tumors of digestive tract. However, based on early reported data accompanied by our pilot study [Citation10], MSIs in hematologic malignancies are yet to be assessed and can be considered diagnostically relevant for diversifying effects of mutations in variable regions.
In this review, we discuss the microsatellite instabilities and epigenetic dysregulation of DNA repair genes and the future prospects of targeting MSI as an effective diagnostic strategy in unexplained cases of hematologic malignancies.
2. Cytogenetic biomarkers in hematologic malignancies
Since the discovery of trisomy 21 in 1959, clinicians are following cytogenetic tests to date to determine the varying effect of non-curable diseases, including malignancies [Citation11]. In hematology, a conventional cytogenetic analysis helps in identifying structural and/or numerical anomalies in chromosomes, which is formally known as karyotyping. Fluorescent in situ hybridization is another technique which requires bone marrow or peripheral blood samples, but inadequate number of DNA probes and cost limit its performance and use. Spectral karyotyping, even though not enlisted in regular practice, uses unique probes/dyes to paint the 24 pairs of chromosomes. Studies have shown that there are many cases where conventional cytogenetics has missed out certain subtle micro anomalies. In malignancies, chromosomal imbalances cannot be detected using array comparative genomic hybridization, but it is likely to be used for tumor classification and progression. A day-to-day use of cytogenetic markers is more assimilated into the diagnostic, prognostic and therapeutic workgroup of hematologic malignancies rather than any other field of oncology. However, it eventually omits out certain molecular level alterations and thus fails to understand the origin and progression of disease which is important for further establishment of novel therapeutic targets [Citation12]. Thus, it is best to enhance the arena of diagnosis in hematologic malignancies to ponder over certain unaccountable cases.
comprises of cytogenetic markers that are used in routine diagnosis and monitoring of hematologic cancers [Citation12,Citation13].
Table 1. List of cytogenetics markers in lymphoid and myeloid malignancies.
3. Microsatellite instability in hematologic malignancies – a different point of view
MSI was originally termed as RER+ (replication error-positive phenotype, because any temporary detachment of the newly synthesized strand or the template strand followed by incorrect re-annealing often leads to unpaired repeat units in daughter or parental strand [Citation14–16]. This phenomenon explains both contraction and expansion of microsatellite region. The abnormality is seen as the presence of extra alleles in cancer patients which results from a loss or a gain of repeats due to polymerase slippage. Unlike hereditary nonpolyposis colorectal cancer (HNPCC) that has Bethesda marker guidelines; there are no selective markers to check for MSI in hematologic malignancies. Although the genetic background behind MSI differs in various cancers, involvement of mono- or dinucleotide markers in hematologic field is still elusive. The study of MSI in leukemia was first reported by Wada et al. [Citation17], wherein they found frequent alteration in microsatellite region using dinucleotide repeat markers, suggesting that MSI is involved in molecular evolution of chronic myeloid leukemia (CML). There have been studies on various cell lines of leukemia and lymphoma which appeared as MMR deficient. In 1997, researchers discovered mutation and inactivation of MLH1 gene in human leukemia cell lines by northern and western blot analysis. The cell lines KCL22, CEM and P300HK showed lack of MLH1 gene expression, suggesting that any disruption in MMR machinery can play a pivotal role in the development of human lymphocytic leukemia [Citation16]. Inoue et al. [Citation18] analyzed MSI in T cell acute lymphocytic leukemia (T-ALL) cell lines where repair genes MSH3 and MSH6 were detected to be less frequently mutated. In another experiment, biochemical analysis showed MSI in CCRF-CEM, CCRF-HSB-2, NALM-6, NAMALWA, MOLT4 and MOLT14 cell lines. These cell lines were seen to have MutSɑ and MutLɑ deficiency directing toward a deletion/point mutation in MSH2, MLH1 and PMS2 genes. For the first time, this study established a MSI-positive Burkitt’s lymphoma cell line NAMALWA, capable of correcting base–base alterations and/or insertion–deletion loops with high efficacy despite faulty DNA MMR system [Citation19]. Studies on patient samples showed an increased frequency of loss of expression in MSH2 and MLH1 genes by immunohistochemical staining, indicating the presence of MSI in chronic B cell lymphocytic leukemia [Citation20]. All of the above experiments show the dynamism of cancers and compel us to consider MSI as a part of hematologic malignancies. However, there are certain cases where researchers failed to locate MSIs in hematologic patient samples with additional cytogenetic abnormalities [Citation21–23]. Owing to less investigation of MSI-related primary leukemia and lymphoma as compared to other tumors, it is very much difficult to interpret the underlying association. Can MSI lead to an acute transformation in primary malignancies with or without cytogenetic rearrangements? Is there any mechanism that causes defective repair system, which in turn leads to MSI in primary blood cancers? What if we can establish a set of microsatellite markers to study MSI in detail like Bethesda markers? Probably, these questions can be answered through an extensive research to set up a benchmark in diagnostics.
4. Promoter hypermethylation – an initiator of microsatellite instability in hematologic malignancies?
The transfer of methyl group from S-adenosyl-l-methionine to cytosine residue (5-C position) is mediated by DNA methyltransferases. This epigenetic event is required to maintain the overall genomic integrity by transcriptional silencing of imprinted genes, transposable elements and genes located on inactive X chromosome [Citation24]. Scientists have recognized both gene hypomethylation and hypermethylation in multiple cancers over times. Altered methylation status of a gene is known to trigger tumorigenesis by modifying genomic integrity and transcriptional gene silencing along with a closed chromatin structure [Citation25–27]. There are fewer researches on DNA repair genes that provided an insight on the occurrence of MSIs in T-cell leukemia and lymphoma as a result of promoter hypermethylation [Citation28–30]. In MSI-positive, aged acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) patients, promoter hypermethylation of both MLH1 and MSH2 genes reflected on the fact that epimutation can contribute to the development of AML in elderly patients [Citation31]. Fülöp et al. [Citation32] designed a study and proved that Richter’s transformation, a rare complication of chronic lymphocytic leukemia (CLL), is associated with MSI and promoter hypermethylation of MLH1 gene. An abnormal expression of MSH2 and PMS1 genes was established in adult T-cell leukemia (ATL) by methylation-specific Polymerase Chain Reaction (PCR) which showed promoter methylation of PMS1 gene proposing significance of defective MMR system in the development and progression of ATL [Citation33]. In CML patients, abnormal promoter methylation was investigated in multiple TSGs to name a few – p14, 15, 16, Rb, MLH1, MSH2, adenomatous polyposis coli, MGMT, fragile histidine triad. In this study, MGMT showed second highest methylation rate, but none of these patients displayed hypermethylation of MLH1 and MSH2 genes suggesting that MMR genes may not be involved in the progression of CML to blast crisis phase [Citation34]. One of the distinguished features in hematologic malignancy is the formation of fusion protein, but how these cluster of proteins are associated with methylation machinery to achieve transcriptional silencing? Are we clear about all the methylation-specific targets and their molecular mechanism that drive pathogenesis of hematologic cancers in different ways? Are MLH1 and MGMT genes be the only risk factor for leukemic or lymphoid patients? The genes coding MutS homolog – MSH2, MSH6 and MSH3 were seen to be epigenetically inactivated in colorectal, ovarian, gastric and oral squamous cell carcinoma [Citation35–39]. Despite their being interesting candidates, there can also be a possibility that MSH2, MSH3 and MSH6 genes are a weak target for hypermethylation unlike MGMT or MLH1.
5. Diagnostic relevance of microsatellite instability and methylation status of DNA repair genes – are we good to go?
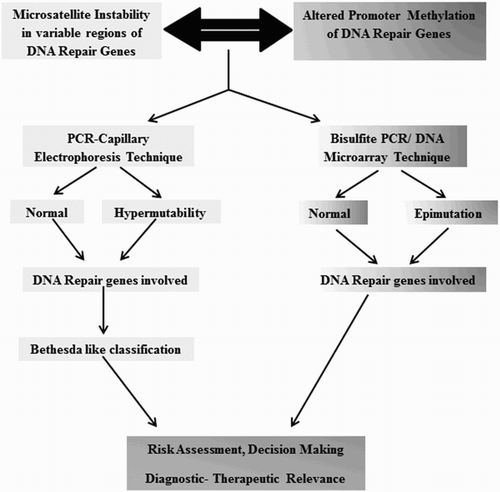
In the diagnosis of primary hematologic malignancies, MSI can be considered a very rare phenomenon until proved by study of various pathways. It may or may not be accompanied by cytogenetic rearrangements. In 1997, National Cancer Institute developed Bethesda guidelines for the detection of specific microsatellite marker mutability in HNPCC patients. According to Bethesda panel of microsatellite markers (BAT25, BAT26, D2S123, D5S346 and D17S250), the MSI tests include three possible results: MSI-H if two or more than two markers out of five show MSI; MSI-L if one of the five markers is MSI positive and Microsatellite Stable (MSS), indicating the absence of MSI [Citation40]. The diagnostic tests for microsatellite markers include immunohistochemistry (IHC)- or PCR-Fragment analysis-based molecular techniques. IHC methods, though cost-effective, could be inconclusive making it difficult for the clinicians to decide the line of treatment [Citation41]. Researches have shown the occurrence of MSI due to altered methylation status of DNA repair genes and their promoters in various cancers along with hematologic malignancies. There are certain limitations in this area, since the extent and frequency of promoter methylation and degree of gene silencing are still covert in most of these cases.
The primary objective of introducing MSI test in the field of hematologic malignancies is to evaluate the patients stage by stage and thus reach to a relevant treatment. Approximately 2–4% of cases, wherein the patients are not showing any conventional chromosomal rearrangements or unresponsive toward drugs could be tested for specific MSI markers. The future direction of research through this article is to develop important MSI makers by studying various cell lines and patient samples. Once the markers show mutability in the primary cases of myelo- or lympho-proliferative conditions, the clinician can diverge from the original diagnostic tests and conventional line of treatment in handful of unaccountable cases. Research showing convincible association between microsatellite hypervariability and promoter hypermethylation, causing tumors of the hematopoietic tissues, is currently deficient. Identifying MSI with a panel of microsatellite markers specific for leukemia/lymphomas would definitely be beneficial for a systematic treatment and clinical management of patients. Thus, this approach has the potential to serve for diagnostic, prognostic and prediction of responses in cancer therapies ().
The major arena for methylation assay is in differential diagnosis where conventional tests turn out to be difficult to conclude [Citation42]. Immune therapy response for DNA repair deficiency caused by mutation and methylation recently opened up a new vision in MSI-positive colorectal tumor [Citation43–46], but whether the same is true for blood-related cancers needs to be elucidated. Diagnostically screening of biologically related family members for MSI markers and epimutation can also help us to evaluate the possible clause in hematologic malignancies.
6. Conclusion
An increased knowledge on DNA repair system, MSI and methylation with respect to their biological pathways is essential to understand the core mechanism of tumor development and to advance current diagnostic approaches. However, not all the patients suffering from hematopoietic malignancies can be positive for mutation in microsatellite regions or altered methylation. Most of them are diagnosed with cytogenetic defects and treated accordingly, but there are handful cases where patients do not show any cytogenetic anomalies by routine assays, but are symptomatic of cancer and hence succumb to mortality. In these circumstances, a proposed panel of microsatellite markers or promoter methylation assays for DNA repair genes can serve as a favorable factor enabling what could be the patient’s response toward drug or chemotherapy and at the same time providing a better survival. We should, therefore, conduct basic research so that precise markers could be evaluated and implemented in diagnostics.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Trupti N. Patel http://orcid.org/0000-0002-7941-602X
References
- Sattler M, Griffin JD. Molecular mechanisms of transformation by the BCR-ABL oncogene. Semin Hematol. 2003;40:4–10. doi: 10.1053/shem.2003.50034
- Garcia-Manero G, Faderl S, O’Brien S, et al. Chronic myelogenous leukemia: a review and update of therapeutic strategies. Cancer. 2003;98:437–457. doi: 10.1002/cncr.11520
- Boland CR, Goel A. The silence of the genes: matching mismatch repair defects with tumors. Cancer. 2003;98:2091–2094. doi: 10.1002/cncr.11769
- Shilpa V, Lakshmi K. Molecular mechanisms of mismatch repair genes in cancer – A brief review. J Proteom Genom. 2014;1:101–108.
- Chikan NA, Shabir N, Shaffi S, et al. N-nitrosodimethylamine in the Kashmiri diet and possible roles in the high incidence of gastrointestinal cancers. Asian Pac J Cancer Prev. 2012;13:1077–1079. doi: 10.7314/APJCP.2012.13.3.1077
- Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res. 1993;285:61–67. doi: 10.1016/0027-5107(93)90052-H
- Hlle SE, Shabashev S, Eckert KA. Tumor-specific microsatellite instability: do distinct mechanisms underlie the MSI-L and EMAST phenotypes? Mutat Res. 2013;743–744:67–77. doi: 10.1016/j.mrfmmm.2012.11.003
- Boultwood J, Wainscoat JS. Gene silencing by DNA methylation in haematological malignancies. Br J Haematol. 2007;138:3–11. doi: 10.1111/j.1365-2141.2007.06604.x
- Maletzki C, Schmidt F, Dirks WG, et al. Frameshift-derived neoantigens constitute immunotherapeutic targets for patients with microsatellite-instable haematologic malignancies, frameshift peptides for treating MSI+ blood cancer. Eur J Cancer. 2013;49:2587–2595. doi: 10.1016/j.ejca.2013.02.035
- Patel T, Chakraborty M, Bhattacharya P. Microsatellite instability in chronic myeloid leukemia using D17S261 and D3S643 markers- a pilot study in western India. Clin Lymphoma Myeloma Leuk. 2016;16:S54–S55. doi: 10.1016/j.clml.2016.07.079
- Put N, Michaux L, Vandenberghe P. The cytogenetic and molecular diagnosis of haematological malignancies: an overview of current techniques. Belg J Hematol. 2014;5:3–11.
- Hussaini M. Biomarkers in haematological malignancies: a review of molecular testing in haematopathology. Cancer Control. 2015;22:158–166. doi: 10.1177/107327481502200206
- Atlas of genetics and cytogenetics in oncology and haematology. Available from: http://www.atlasgeneticsoncology.org/ [accessed 1.3.2017].
- Huntly BJ, Bench A, Green AR. Double jeopardy from a single translocation: deletions of the derivative chromosome 9 in chronic myeloid leukemia. Blood. 2003;102:1160–1168. doi: 10.1182/blood-2003-01-0123
- Claij N, te Riele H. Microsatellite instability in human cancer: a prognostic marker for chemotherapy? Exp Cell Res. 1999;246:1–10. doi: 10.1006/excr.1998.4299
- Hangaishi A, Ogawa S, Mitani K, et al. Mutations and loss of expression of a mismatch repair gene hMLH1 in leukemia and lymphoma cell lines. Blood. 1997;89:1740–1747.
- Wada C, Shionoya S, Fujino Y, et al. Genomic instability of microsatellite repeats and its association with the evolution of chronic myelogenous leukemia. Blood. 1994;83:3449–3456.
- Inoue K, Kohno T, Takakura S, et al. Frequent microsatellite instability and BAX mutations in T cell acute lymphoblastic leukemia cell lines. Leuk Res. 2000;24:255–262. doi: 10.1016/S0145-2126(99)00182-4
- Gu L, Cline-Brown B, Zhang F, et al. Mismatch repair deficiency in haematological malignancies with microsatellite instability. Oncogene. 2002;21:5758–5764. doi: 10.1038/sj.onc.1205695
- Niv E, Bomstein Y, Bernheim J, et al. Microsatellite instability in gastric-MALT lymphoma. Mod Pathol. 2004;17:1407–1413. doi: 10.1038/modpathol.3800207
- Mori N, Takeuchi S, Tasaka T, et al. Absence of microsatellite instability during the progression of chronic myeloid leukemia. Leukemia. 1997;11:151–152. doi: 10.1038/sj.leu.2400524
- Rimsza LM, Kopecky KJ, Ruschulte J, et al. Microsatellite instability is not a defining genetic feature of acute myeloid leukemogenesis in adults: results of a retrospective study of 132 patients and review of the literature. Leukemia. 2000;14:1044–1051. doi: 10.1038/sj.leu.2401699
- Sercan HO, Sercan ZY, Kizildag S, et al. Microsatellite instability is a rare phenomenon in transition from chronic to blastic phase chronic myeloid leukemia. Turk J Cancer. 2001;31:63–71.
- Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1
- Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:900–900. doi: 10.1038/nrg962
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075
- Seedhouse CH, Das-Gupta EP, Russell NH. Methylation of the hMLH1 promoter and its association with microsatellite instability in acute myeloid leukemia. Leukemia. 2003;17:83–88. doi: 10.1038/sj.leu.2402747
- Matsushita M, Takeuchi S, Yang Y, et al. Methylation of the MLH1 gene in hematological malignancies. Oncol Rep. 2005;14:191–194.
- Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901
- Sheikhha MH, Tobal K, Liu Yin JA. High level of microsatellite instability but not hypermethylation of mismatch repair genes in therapy-related and secondary acute myeloid leukaemia and myelodysplastic syndrome. Br J Haematol. 2002;117:359–365. doi: 10.1046/j.1365-2141.2002.03458.x
- Fülöp Z, Csernus B, Timar B, et al. Microsatellite instability and hMLH1 promoter hypermethylation in Richter’s transformation of chronic lymphocytic leukemia. Leukemia. 2003;17:411–415. doi: 10.1038/sj.leu.2402792
- Morimoto H, Tsukada J, Kominato Y, et al. Reduced expression of human mismatch repair genes in adult T-cell leukemia. Am J Hematol. 2005;78:100–107. doi: 10.1002/ajh.20259
- Uehara E, Takeuchi S, Yang Y, et al. Aberrant methylation in promoter-associated CpG islands of multiple genes in chronic myelogenous leukemia blast crisis. Oncol Lett. 2012;3:190–192. doi: 10.3892/ol.2011.419
- Nagasaka T, Rhees J, Kloor M, et al. Somatic hypermethylation of MSH2 is a frequent event in Lynch syndrome colorectal cancers. Cancer Res. 2010;70:3098–3108. doi: 10.1158/0008-5472.CAN-09-3290
- Czerninski R, Krichevsky S, Ashhab Y, et al. Promoter hypermethylation of mismatch repair genes, hMLH1 and hMSH2 in oral squamous cell carcinoma. Oral Dis. 2009;15:206–213. doi: 10.1111/j.1601-0825.2008.01510.x
- Zhang H, Zhang S, Cui J, et al. Expression and promoter methylation status of mismatch repair gene hMLH1 and hMSH2 in epithelial ovarian cancer. Aust N Z J Obstet Gynaecol. 2008;48:505–509. doi: 10.1111/j.1479-828X.2008.00892.x
- Titze S, Peters H, Wahrisch S, et al. Differential MSH2 promoter methylation in blood cells of neurofibromatosis type 1 (NF1) patients. Eur J Hum Genet. 2010;18:81–87. doi: 10.1038/ejhg.2009.129
- Kim HG, Lee S, Kim DY, et al. Aberrant methylation of DNA mismatch repair genes in elderly patients with sporadic gastric carcinoma: a comparison with younger patients. J Surg Oncol. 2010;101:28–35. doi: 10.1002/jso.21432
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257.
- Matos LL, Trufelli DC, Matos MG, et al. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights. 2010;5:9–20. doi: 10.4137/BMI.S2185
- Delpu Y, Cordelier P, Cho WC, et al. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14:15029–15058. doi: 10.3390/ijms140715029
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863
- Gao D, Herman JG, Guo M. The clinical value of aberrant epigenetic changes of DNA damage repair genes in human. Oncotarget. 2016;7:37331–37346.
- Jiang D, Hong Q, Shen Y, et al. The diagnostic value of DNA methylation in leukemia: a systematic review and meta-analysis. PLoS One. 2014;9:1–7.
- Palmieri G, Colombino M, Cossu A, et al. Genetic instability and increased mutational load: which diagnostic tool best direct patients with cancer to immunotherapy? J Transl Med. 2017;15:2189. doi: 10.1186/s12967-017-1119-6