ABSTRACT
Background: The aim of the study is to determine the prognostic relevance of CD200/ CD56 expression in adult acute lymphoblastic leukemia patients.
Methods: The expression of CD200 and CD56 by blast cells was assessed by flow cytometry before the start of chemotherapy in 70 B-ALL patients.
Results: Positive expression of CD200 was detected in forty-six patients (66%) and CD56 was detected in 7 patients (10%) out of 70 patients, respectively. Only three patients (4.3%) had co-expression for CD200+ and CD56+. Splenomegaly and thrombocytopenia were frequently observed more in CD200+ patients. Increased frequency of CD34+ was associated with CD200+and CD56+ patients. The CD200+ and CD56+ subgroups of B-ALL patients had inferior OS and disease free survival compared to CD 200− and CD 56− patients.
Conclusions: CD200+ and/or CD56+ positive expression in B-ALL patients at diagnosis is a poor prognostic biomarker. Identification of CD200+ and CD56+ expression at diagnosis is recommended for a better stratification of adult B-ALL patients.
KEYWORDS:
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous condition and an aggressive cancer, especially in adults and only 20–40% is cured with the current treatment regimens [Citation1]. Adults with ALL tend to have higher risk factors at diagnosis and increasing age which often requires dose reduction [Citation2]. New immunotherapeutic approaches are used to improve the outcome of ALL therapy based mainly on non-chemotherapeutic approaches [Citation3]. In addition, prognostic features play a critical role in directing therapy for ALL, so this area of investigation changes rapidly.
CD200 is a trans-membrane cell surface glycoprotein encoded by a gene residing at chromosome 3q12. It belongs to type-I immunoglobulin superfamily and also known as MOX-2″ [Citation4]. It is related to the B7 family of co-stimulatory receptors, with two extracellular domains, a single trans-membrane region, and cytoplasmic tail without signal motif [Citation5].
CD200-CD200R interactions have a role in modulated cell-mediated immunity with a significant lymphocytic involvement [Citation6], regulation of myeloid differentiation and or activation, fetal loss, transplant rejection [Citation7] and autoimmunity [Citation8]. However, CD200 has a central role in immune tolerance that protects stem cells and other critical tissues from immune damage [Citation7]. CD200 can induce neurogenesis and promote neuronal survival in primary neurons through activation of the fibroblast growth factor receptor (FGFR) [Citation9].
CD200 with its receptor CD200R delivers an inhibitory signal to the target cell in most circumstances [Citation10] and imparts an immunosuppressive signal leading to the inhibition of macrophage function [Citation11], induction of regulatory T cells [Citation12], switching of cytokine profiles from Th1 to Th2 and inhibition of tumor-specific T-cell immunity [Citation13].
CD56 is a neural-cell adhesion molecule (N-CAM) known as a marker of natural killer cells (NK) [Citation14]. It is expressed on a subset of normal T cells and occasionally on blasts in T-cell ALL especially a subgroup of T-ALL patients who did not respond to therapy [Citation15]. NK-cells are phenotypically and functionally very similar to T-cells (cytotoxic T-cells) as it arises from T/NK bi-potential common progenitor [Citation16].
High incidence of CNS disease was found in adult ALL patients who expressed CD56 [Citation17]. The expression of CD56 has been shown to be of prognostic relevance in certain hematologic malignancies as well as it was associated with inferior survival in a small series of CD56+ T-ALL [Citation18].
The aim of the present study was the estimation of the prognostic impact of CD200and CD56 expression in adult B-cell ALL.
Patients and methods
Pretreatment bone marrow and peripheral blood samples were obtained from 70 patients (age range 18–52 years; 40 males and 30 females) at Mansoura University Oncology Center (MUOC). The diagnosis of B-cell ALL was based on the morphologic examination of peripheral blood and bone marrow smears stained with Leishman stain and immunophenotypic analysis using flowcytometry. For all patients included in this study, the full conventional laboratory investigation as well as clinical examination was done. An informed consent was taken from all patients. Complete remission (CR) was considered when bone marrow blast cell counts were <5%.
Chemotherapy protocols
Philadelphia chromosome-negative ALL
In young adults (age 15–39 years) BFM chemotherapy protocol was used [Citation19]. In the older adults (age ≥40 years) GRAALL protocol was used [Citation20]. In patients ≥65 years MRCUK protocol [Citation21].
Philadelphia chromosome-positive ALL
In young adults (age 15–39 years) BFM chemotherapy protocol plus tyrosine kinase inhibitors (TKI) were used. In the older adult (age≥40 years); chemotherapy (hyper-CVAD) plus TKI were used [Citation22]. In patients ≥65 years TKI plus corticosteroids or TKI plus MRCUK chemotherapy protocol were used.
Immuno-phenotyping
Immunophenotypic analyses for ALL patients were performed according to standard techniques on an FACSCanto flow cytometer (BD Bioscience, San Diego, CA, U.S.A.). In order to perform immuno-phenotyping of the blasts a broad panel of fluorochrome conjugated monoclonal antibodies (mAbs) were used including: anti-CD3(PE-HIT3a), CD4(FITC-RPA-T4), CD5(FITC-UCHT2), CD7(FITC-8H8.1), CD8(APC-RPA-T8), CD10(PE-HI10a), CD13(PECY5-Immu103-44), CD14(PE-M5E2), CD19(PE-HIB19), CD20 (PE-2H7), CD22 (PE-HIB22), CD33(FITC-HiM3-4), CD34(PE-581), HLADR (PE-Immu-357) and TDT (FITC-E17-1519). In addition, CD200 (PE-MRC OX-104) and CD56 (FITC-NCAM16.2) mAbs were used. Samples were stained with monoclonal antibodies against cell surface markers as described before using stain – lyse – wash then direct immunofluorescence technique was done [Citation23]. Briefly, 100 μl of BM or PB sample were added to 20 μl of mAb of each CD, and then incubated for 15 minutes at room temperature. Cell suspensions were washed with phosphate buffer saline (PBS) and 2 ml of 1× BD FACS lysing solution. Then incubated for10 minutes at room temperature and centrifuged for 5 minutes at 550 rpm. Cells were washed twice with PBS; the cell pellet was re-suspended in 500 μl PBS. After that, the sample became ready for acquisition and analysis by flow cytometry. When expression of a marker on the lymphatic blasts ≥20%; the case was defined as positive.
BCR-ABL detection
Samples were tested for t (9;22) using real-time quantitative PCR. The assay involved a duplicate real-time amplification reaction for the target and a duplicate real-time amplification reaction for the control. There were two main parts in the test, the amplification reaction was carried out specific for a region of the mRNA of P190 KD, while in the second step, an amplification reaction was carried out specific for a region of the mRNA of ABL using the cDNA produced by the reverse transcription reaction of the RNA extracted from the test samples. System standardization was carried out on Applied Biosystems ABI PRISMTM 7000 series instruments.
Statistical analysis
All statistical analyses were performed using the SPSS software package and Graph Pad Prism (Graph Pad Software, Inc; San Diego, CA, U.S.A.) to analyze p values. Data were statistically described in terms of mean ± SE, mean ± SD and mean with range. Overall survival was calculated from the date of first diagnosis to the date of death from any cause. Whereas, remission duration was calculated from the time of achievement of CR to the time of relapse or death in CR. The OS and disease free survival (DFS) were assessed by the Kaplan–Meier curve. A probability value (p value) less than 0.05 was considered statistically significant.
Results
The clinical and laboratory characteristics according to CD200 and CD56 expression
Positive expression of CD200 () and CD56 was detected in forty-six out of 70 B-ALL cases (66%); and 7/70 (10%), respectively ( and ). Only three cases (4.3%) had co-expression for both CD200+ and CD56+. Comparing the clinical, the laboratory and the cytogenetic findings in CD200 positive vs. CD200 negative ALL patients point out to frequent CNS infiltrations; higher number of patients with CD34+ blasts; lower hemoglobin level; lower platelets count; higher peripheral blast cells count; lower induction remission response, more association with FAB subtypes in CD200+ve subgroup (p = 0.001; 0.001; 0.046; 0.027, 0.053, 0.001, respectively) (). Moreover; the clinical, the laboratory and the cytogenetic findings in CD56 positive vs. CD56 negative B-ALL patients showed significantly higher CNS infiltrations; higher number of patients with CD34+ blasts; lower platelet counts; lower induction remission response rates in CD56+ve subgroup (p = 0.001;0.001; 0.024; 0.018, respectively) ().
Figure 1. Shows the expression of CD200 and CD56 in adult B-ALL case. The patient is positive for CD200 but negative for CD56.
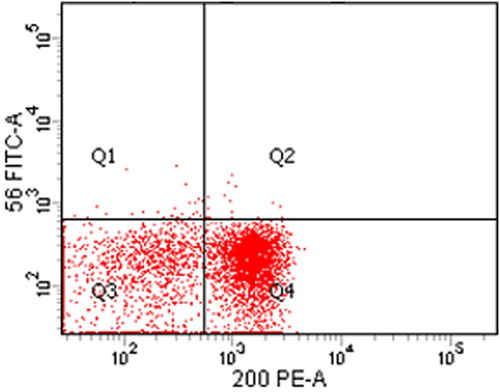
Table 1. Hematological and clinical data in CD200+ B-ALL patients vs. CD200− B-ALL patients.
Table 2. Hematological and bio-chemical data in CD56+ B-ALL vs. CD56− ALL patients.
CD200+/CD56+ subgroup characteristics
CD200+/CD56+ patients were three males and one female. All of them have hepatosplenomegaly and suffer from abdominal pain and very low platelets count. The lowest induction remission response rate was reported in this B-ALL subgroup of patients with co-expression of both CD200+/CD56+ (0%) (data not shown).
Cox regression analysis results
Cox regression analysis was conducted for prediction of OS in adult B-ALL patients using clinical and laboratory data; CD200 and CD56 expressions as covariates. Higher bone marrow blast cells; failure to induction remission response; CD200+, CD56+ were significantly associated with shorter OS in univariate analysis. On the other hand, in multivariate analysis CD200+, CD56+ and failure for induction of remission were considered as independent risk factors for shorter OS. Furthermore; Cox regression analysis was conducted for prediction of DFS in all B-ALL patients, using age, sex, clinical data, laboratory findings, cytogenetics, and CD200 and CD56 expressions as covariates. Positive CD200 and CD56 expression by blast cells, lower serum albumin were associated with shorter DFS in univariate analysis. However, in multivariate analysis, only higher CD200 and CD56 were considered as independent risk factors for shorter DFS in ALL cases ( and ).
Table 3. Cox regression analysis for prediction of OS in adult B-ALL cases.
Table 4. Cox regression analysis for prediction of DFS in adult B-ALL cases.
Survival analysis
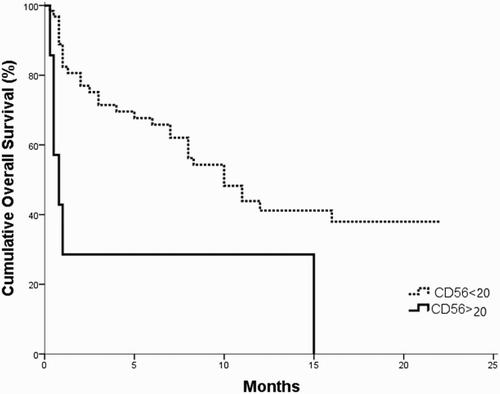
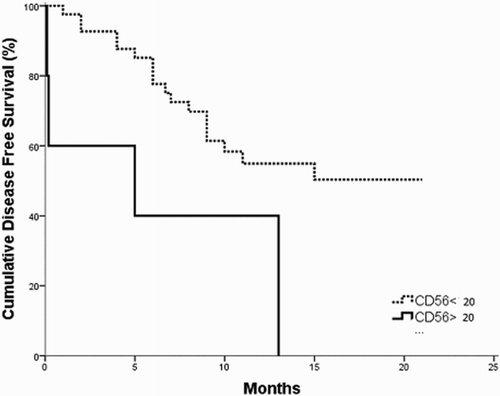
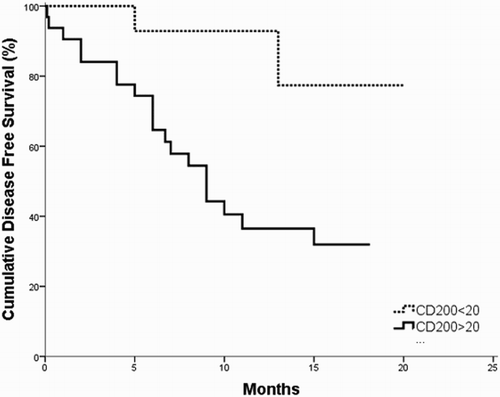
The survival analysis for the investigated B-ALL patients revealed that the OS and DSF times were significantly shorter in CD200+ patients as compared to CD200− patients (p = 0.042; 0.006, respectively) ( and ). Moreover, the OS and DFS time of CD56+ patients subgroup were significantly shorter as compared to CD56− patients (p = 0.003; 0.007, respectively ( and ).
Figure 2. OS in CD200+ vs. CD200− B-ALL patients; the OS is shorter in CD200+ group as compared to CD200− one (p = 0.042).
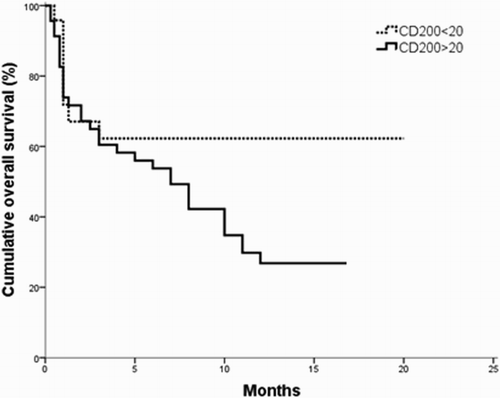
Figure 3. DFS in CD200+ vs. CD200− B-ALL patients; the DFS is shorter in CD200+ group as compared to CD200− one (p = 0.006).
Discussion
In the present study, the CD200+ expression was frequently detected in our cases (66%). Higher figures (95 and 80.3%) were reported by Alapat et al. [Citation24] and Adnan Awad et al. [Citation25], respectively and it was suggested to be used as minimal residual disease markers as well as for targeted therapy. However, Tembhare et al. [Citation26] reported 28.9% among B cell precursor ALL.
In our study, CD56+ positive expression was found in only 10% of cases. This finding is consistent with the results in the previous studies which are 13.9% [Citation27], 9% [Citation28], 8% [Citation17], 3% [Citation29], 5% [Citation30] and 2.2% [Citation31]. CD56 is an isoform of the neural adhesion molecules (NCAM) and has been reported in several myeloproliferative disorders including acute leukemia [Citation32]. A significant reduction of CR and OS duration was found in patients who express CD56 antigen on the membrane of the leukemic blasts [Citation33].
The co-expression of both CD200 and CD56 was detected in 4.3% of cases. CD200+/CD56+ patients were associated with more pain and increased frequency of splenomegaly. This is in accordance with the results of Murakami et al. [Citation34] who explained this by malignant cells infiltration of the liver and spleen leading to hepatosplenomegaly. Pain may be due to the low intensity of the bone marrow in regions adjacent to localized musculoskeletal symptoms as reported by Yoshikawai et al. [Citation35]. High incidence of the previous clinical data in CD200+/CD56+ patients could be explained on the basis that CD200 was a key immunosuppressive molecule [Citation36].
The patients positively expressed CD200+ and CD56+ had the lowest platelet count at diagnosis and higher LDH activity compared to the other studied subgroups. No previous data was listed in this point. This could be attributed to the high frequency of splenomegaly among those patients.
Six patients were positive for BCR-AbL (Ph+) gene t (9;22) (8.6%). Most Ph+ patients in the present study were associated with CD200+ and CD56+ subgroup. This is in agreement with the finding reported by Salami et al. [Citation37] who found (6.6%) of the studied ALL patients had Ph+ ALL. This is also in agreement with the fact that Ph+ B-ALL is an aggressive form of ALL that frequently presents in older children with FAB L2 morphology [Citation38].
Association studies between B-ALL immunophenotypic markers and CD200+/CD56+ positive expression revealed that there is a good association between CD200+ and CD34+ expression. Similar finding was reported by Alapat et al. [Citation24]. This finding point out to the frequency of CD200+/CD56+ positive expression in more immature blast associated B-ALL.
Induction of remission response was the lowest in CD200+ and CD56+ subgroups. This could be explained on the basis that positivity for both CD200 and CD56 define a subgroup of B-ALL patients with immature blast cells which are more resistant to chemotherapy. Tonks et al. [Citation39] reported that CD200 is a marker of LSC activity in acute myeloid leukemia.
OS and DFS studies revealed that CD200+and CD56+ patients have shorter OS and DFS as compared to CD200− and CD56−, respectively. Previous studied listed the effect of these CDs individually. CD200 had a prognostic impact in many similar studies. The CD200 antigen expression in AML may be associated with a poor prognosis [Citation32]. CD200 may have an important role for the diagnosis, prognosis and choice of treatment in CLL. In addition, low expressions of CD200 in chronic lymphocytic leukemia patients may predict shorter time to treatment response [Citation40]. In Cutaneous Squamous Cell Carcinoma (CSCC), CD200 is up-regulated and may be used as a novel biomarker for the prognosis in CSCC that predicts poor prognosis [Citation41]. CD200 signaling may play a key role in regulating macrophage polarization toward anti-inflammatory phenotypes [Citation42].
CD56 expression has been shown to have a prognostic relevance in certain hematologic malignancies [Citation43]. An inferior overall survival of CD56+ T-ALL was reported by Montero et al. [Citation18]. In addition, Dalmazzo et al. [Citation15] estimated that T-cell ALL CD56+ patients expressed cytotoxic molecules at a higher frequency and presented with shorter OS compared to CD56− patients. Moreover, DFS was significantly lower for patients with CD56+ immunophenotype compared with patients with CD56− [Citation44].
CD200 expression on tumor cells inhibits the ability of human lymphocytes to eradicate tumor cells [Citation45]. Blocking CD200 alone was sufficient to recover a significant proportion of NK cell cytolytic activity [Citation13]. In addition, CD56 expression in leukemic cells could allow infiltration through the adhesion of these cells to tissues that also express this marker [Citation46].
Taken together, the data of this study suggest that CD200 and CD56 positive expression may play a fundamental role in ALL survival and has a poor influence because it promotes tumor progression.
Conclusion
CD200+ and/ or CD56+ positive expression in adult B-ALL patients at diagnosis is a poor prognostic biomarker. Identification of CD200+ and CD56+ expression at diagnosis is recommended for a better stratification of adult B-ALL.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Allahyari A, Hashemi SM, Nazemian F, et al. The relationship between risk factors and survival in adult acute lymphoblastic leukemia. Iran J Cancer Prev. 2016;9(4):e50. doi: 10.17795/ijcp-5045
- Paul S, Kantarjian H, Jabbour EJ. Adult acute lymphoblastic leukemia. Mayo Clin Proc. 2016;91(11):1645–1666. doi: 10.1016/j.mayocp.2016.09.010
- Al Ustwani O, Gupta N, Bakhribah H, et al. Clinical updates in adult acute lymphoblastic leukemia. Crit Rev Oncol Hematol. 2016;99:189–199. doi: 10.1016/j.critrevonc.2015.12.007
- Wright GJ, Jones M, Puklavec MJ, et al. The unusual distribution of the neuronal/lymphoid cell surface CD200 (OX2) glycoprotein is conserved in humans. Immunology. 2001;102(2):173–179. doi: 10.1046/j.1365-2567.2001.01163.x
- Barclay AN, Wright GJ, Brooke G, et al. CD200 and membrane protein interactions in the control of myeloid cells. Trends Immunol. 2002;23(6):285–290. doi: 10.1016/S1471-4906(02)02223-8
- Walker DG, Lue LF. Understanding the neurobiology of CD200 and the CD200 receptor: a therapeutic target for controlling inflammation in human brains? Future Neurol. 2013;8(3):321–332. doi: 10.2217/fnl.13.14
- Gorczynski RM. Transplant tolerance modifying antibody to CD200 receptor, but not CD200, alters cytokine production profile from stimulated macrophages. Eur J Immunol. 2001;31(8):2331–2337. doi: 10.1002/1521-4141(200108)31:8<2331::AID-IMMU2331>3.0.CO;2-#
- Gorczynski RM, Chen Z, Lee L, et al. Anti-CD200R ameliorates collagen-induced arthritis in mice. Clin Immunol. 2002;104(3):256–264. doi: 10.1006/clim.2002.5232
- Pankratova S, Bjornsdottir H, Christensen C, et al. Immunomodulator CD200 promotes neurotrophic activity by interacting with and activating the fibroblast growth factor receptor. Mol Neurobiol. 2016;53(1):584–594. doi: 10.1007/s12035-014-9037-6
- Jenmalm MC, Cherwinski H, Bowman EP, et al. Regulation of myeloid cell function through the CD200 receptor. J Immunol. 2006;176(1):191–199. doi: 10.4049/jimmunol.176.1.191
- Hoek RM, Ruuls SR, Murphy CA, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science. 2000;290(5497):1768–1771. doi: 10.1126/science.290.5497.1768
- Gorczynski RM, Lee L, Boudakov I. Augmented induction of CD4+CD25+ Treg using monoclonal antibodies to CD200R. Transplantation. 2005;79(4):488–491.
- Coles SJ, Wang EC, Man S, et al. CD200 expression suppresses natural killer cell function and directly inhibits patient anti-tumor response in acute myeloid leukemia. Leukemia. 2011;25(5):792–799. doi: 10.1038/leu.2011.1
- Farag SS, VanDeusen JB, Fehniger TA, et al. Biology and clinical impact of human natural killer cells. Int J Hematol. 2003;78(1):7–17. doi: 10.1007/BF02983234
- Dalmazzo LF, Jacomo RH, Marinato AF, et al. The presence of CD56/CD16 in T-cell acute lymphoblastic leukaemia correlates with the expression of cytotoxic molecules and is associated with worse response to treatment. Br J Haematol. 2009;144(2):223–229. doi: 10.1111/j.1365-2141.2008.07457.x
- Shibuya A, Nagayoshi K, Nakamura K, et al. Lymphokinerequirement for the generation of natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1995;85(12):3538–3546.
- Ravandi F, Cortes J, Estrov Z, et al. CD56 expression predicts occurrence of CNS disease in acute lymphoblastic leukemia. Leuk Res. 2002;26(7):643–649. doi: 10.1016/S0145-2126(01)00188-6
- Montero I, Rios E, Parody R, et al. CD56 in T-cell acute lymphoblastic leukaemia: a malignant transformation of an early myeloid-lymphoid progenitor? Haematologica. 2003;88(9):E127–E128.
- Nachman J, Sather HN, Gaynon PS, et al. Augmented Berlin-Frankfurt-Munster therapy abrogates the adverse prognostic significance of slow early response to induction chemotherapy for children and adolescents with acute lymphoblastic leukemia and unfavorable presenting features: a report from the Children’s Cancer Group. J Clin Oncol. 1997;15:2222–2230. doi: 10.1200/JCO.1997.15.6.2222
- Huguet F, Leguay T, Raffoux E, et al. Pediatric-inspired therapy in adults with Philadelphia chromosome-negative acute lymphoblastic leukemia: the GRAALL-2003 study. J Clin Oncol. 2009;27(6):911–918. doi: 10.1200/JCO.2008.18.6916
- Chessells JM, Bailey CC, Richards S. MRC UKALL X. The UK protocol for childhood ALL: 1985-1990. The Medical Research Council Working Party on childhood leukaemia. Leukemia. 1992;6(Suppl. 2):157–161.
- Kantarjian HM, O’Brien S, Smith TL, et al. Long-term follow-up results of hyper fractionated cyclophosphamide, vincristine, doxorubicin, and dexamethasone (Hyper-CVAD), a dose-intensive regimen, in adult acute lymphocytic leukemia. Cancer J. 2004;101(12–15):2788–2801. doi: 10.1002/cncr.20668
- Porwit A, Rajab A. Flow cytometry immunophenotyping in integrated diagnostics of patients with newly diagnosed cytopenia: one tube 10-color 14-antibody screening panel and 3-tube extensive panel for detection of MDS-related features. Int J Lab Hematol. 2015;37(Suppl 1):133–143. doi: 10.1111/ijlh.12368
- Alapat D, Coviello-Malle J, Owens R, et al. Diagnostic usefulness and prognostic impact ofCD200 expression in lymphoid malignancies and plasma cell myeloma. Am J Clin Pathol. 2012;137(1):93–100. doi: 10.1309/AJCP59UORCYZEVQO
- Adnan Awad S, Kamel MM, Ayoub MA, et al. Immunophenotypic characterization of cytogenetic subgroups in Egyptian pediatric patients with B-cell acute lymphoblastic leukemia. Clin Lymphoma Myeloma Leuk. 2016;16(Suppl.):S19–S24.e1. doi: 10.1016/j.clml.2016.02.032
- Tembhare PR, Ghogale S, Ghatwai N, et al. Evaluation of new markers for minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia: CD73 and CD86 are the most relevant new markers to increase the efficacy of MRD 2016; 00B: 000-000. Cytometry B Clin Cytom. 2016;96:2691. doi:10.1002/cyto.b.21486. [Epub ahead of print].
- Fischer L, Gokbuget N, Schwartz S, et al. CD56 expression in T-cell acute lymphoblastic leukemia is associated with non-thymic phenotype and resistance to induction therapy but no inferior survival after risk-adapted therapy. Haematologica. 2009;94(2):224–229. doi: 10.3324/haematol.13543
- Abdulateef NA, Ismail MM, Aljedani H. Clinical significance of co-expression of aberrant antigens in acute leukemia: a retrospective cohort study in Makah Al Mukaramah, Saudi Arabia. Asian Pac J Cancer Prev. 2014;15(1):221–227. doi: 10.7314/APJCP.2014.15.1.221
- Paietta E, Neuberg D, Richards S, et al. Rare adult acute lymphocytic leukemia with CD56 expression in the ECOG experience shows unexpected phenotypic and genotypic heterogeneity. Am J Hematol. 2001;66(3):189–196. doi: 10.1002/1096-8652(200103)66:3<189::AID-AJH1043>3.0.CO;2-A
- Seegmiller AC, Kroft SH, Karandikar NJ, et al. Characterization of immunophenotypic aberrancies in 200 cases of B acute lymphoblastic leukemia. Am J Clin Pathol. 2009;132(6):940–949. doi: 10.1309/AJCP8G5RMTWUEMUU
- Hussein S, Gill KZ, Sireci AN, et al. Aberrant T-cell antigen expression in B lymphoblastic leukaemia. Br J Haematol. 2011;155(4):449–456. doi: 10.1111/j.1365-2141.2011.08870.x
- Damiani D, Tiribelli M, Raspadori D, et al. Clinical impact of CD200 expression in patients with acute myeloid leukemia and correlation with other molecular prognostic factors. Oncotarget. 2015;6(30):30212–30221. doi: 10.18632/oncotarget.4901
- Raspadori D, Damiani D, Lenoci M, et al. CD56 antigenic expression in acute myeloid leukemia identifies patients with poor clinical prognosis. Leukemia. 2001;15:1161–1164. doi: 10.1038/sj.leu.2402174
- Murakami J, Shimizu Y. Hepatic manifestations in hematological disorders. Int J Hepatol. 2013;2013:1–13, Article ID 484903, 13 pages. doi: 10.1155/2013/484903
- Yoshikawai T, Tanizawa A, Suzuki K, et al. The usefulness of T1-weighted magnetic resonance images for diagnosis of acute leukemia manifesting musculoskeletal symptoms prior to appearance of peripheral blood abnormalities. Case Rep Pediatr. 2016:2016. Article ID 2802596, 6 pages. doi: 10.1155/2016/2802596
- Cox CV, Diamanti P, Hazell M, et al. CD200 may be a potential target for therapy in standard risk childhood ALL. Blood. 2014;124(21):4787–4478.
- Salami K, Alkayed K, Halalsheh H, et al. Hemopoietic stem cell transplant versus chemotherapy plus tyrosine kinase inhibitor in the treatment of pediatric Philadelphia chromosome positive acute lymphoblastic leukemia. Hemat Oncol Stem Cell Ther. 2013;6(1):34–41. doi: 10.1016/j.hemonc.2013.03.001
- Crist W, Carroll A, Shuster J, et al. Philadelphia chromosome positive childhood acute lymphoblastic leukemia: clinical and cytogenetic characteristics and treatment outcome. A Pediatric Oncology Group study. Blood. 1990;76(3):489–494.
- Tonks A, Hills R, White P, et al. CD200 as a prognostic factor in acute myeloid leukaemia. Leukemia. 2007;21:566–568. doi: 10.1038/sj.leu.2404559
- Maio Y, Fani L, Wu Y, et al. Low expression of CD200 predicts shorter time to tratment in chronic lymphocytic lrukrmia. Oncotarget. 2016;7(12):13551–13562.
- Li L, Tian Y, Shi C, et al. Over-expression of CD200 predicts poor prognosis in cutaneous squamous cell carcinoma. Med Sci Monit. 2016;22:1079–1084. doi: 10.12659/MSM.895245
- Hayakawa K, Wang X, Lo EH. CD200 increases alternatively activated macrophages through cAMP-response element binding protein – C/EBP-beta signaling. J Neurochem. 2016;136(5):900–906. doi: 10.1111/jnc.13492
- Suzuki R, Murata M, Kami M, et al. Prognostic significance of CD7(+)CD56(+) phenotype and chromosome 5 abnormalities for acute myeloid leukemia M0. Int J Hematol. 2003;77(5):482–489. doi: 10.1007/BF02986617
- Hu W, Wang X, Yang R, et al. Expression of CD56 is a risk factor for acute lymphocytic leukemia with central nervous system involvement in adults. Hematology. 2016;22:1–7.
- Kretz-Rommel A, Qin F, Dakappagari N, Ravey EP, McWhirter J, Oltean D, et al. CD200 expression on tumor cells suppresses antitumor immunity: new approaches to cancer immunotherapy. J Immunol. 2007;178:5595–5605. doi: 10.4049/jimmunol.178.9.5595
- Zocchi MR, Vidal M, Poggi A. Involvement of CD56/N-CAM molecule in the adhesion of human solid tumor cell lines to endothelial cells. Exp Cell Res. 1993;204(1):130–135. doi: 10.1006/excr.1993.1017