 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The corticotropin-releasing hormone (CRH) is a neuropeptide mediating stress responses. CRH exerts effects via the hypothalamus pituitary adrenal axis as well as immediate effects on the sympathetic-adrenal-medullary system. Genetic variants of the CRH promoter were previously found to be associated with altered CRH promoter activity and physiological reactions. Functional characterization of three CRH promoter haplotypes have been performed in vitro using a reporter gene assay under different stimulation conditions. Furthermore, 232 healthy subjects were genotyped and the influence of CRH haplotypes on basal parameters such as post-awakening cortisol and blood pressure as well as on stress reactivity measured after socially evaluated cold pressor test (SeCPT) was investigated. In vitro, CRH haplotype 2 showed the highest promoter activity under baseline conditions and after forskolin stimulation compared with other haplotypes. Forskolin treatment resulted in a two fold increase of haplotype 2 promoter activity compared with the baseline condition. Cell line-dependent promoter activation was found after hydrocortisone treatment. In vivo, CRH haplotype 2 carriers showed significant higher baseline blood pressure (p = .002) and blood pressure after SeCPT (p < .001), but did not differ in cortisol levels. This study provides converging evidence for the importance of CRH promoter variants on physiological stress response parameters.
Introduction
The physiological stress system includes the hypothalamic-pituitary-adrenal (HPA) axis and the sympathetic-adrenal-medullary system (SAM) (Eysenck, Citation2004). Parameters representing these systems such as cortisol, blood pressure or heart rate define the physiological body reaction in response to a stressor. The corticotropin-releasing hormone (CRH), a 41-amino acid peptide, regulates both systems at different anatomical levels. CRH, a non-hypophyseotropic hormone responding immediately to a stressor, influences the cardiovascular system via neuronal signalling (Kovács, Citation2013; Kronenberg, Polonsky, Larsen, & Melmed, Citation2007). Furthermore, CRH, operating as a hypophyseotropic hormone, also copes with stressors by initiating the endocrine cascade of the HPA axis (Kalogeras et al., Citation1996; Kovács, Citation2013; Kronenberg et al., Citation2007). These different functional pathways depend on the different CRH receptors involved: CRHR1/2, CRH-binding protein (CRHBP), or steroid receptor coactivators (SRC-1a/e) (Kovács, Citation2013; Kronenberg et al., Citation2007). Further, susceptibility to situational and individual influences (Kudielka, Hellhammer, & Wüst, Citation2009), and the contribution of genetic variances needs consideration.
The human CRH gene contains two exons encoding the CRH pre-prohormone (Arbiser, Morton, Bruns, & Majzoub, Citation1988). Two single nucleotide polymorphisms (SNPs), rs3176921 (T/C) and rs5030875 (T/G), located in the 5'-regulatory region of the CRH locus, have been described, assigned to three haplotypes A1B1 (haplotype 1), A2B2 (haplotype 2), and A2B1 (haplotype 3) (Baerwald et al., Citation1999; Baerwald, Panayi, & Lanchbury, Citation1996) and used in a few number of association studies (Baerwald et al., Citation1996, Citation1999; Wagner et al., Citation2006). The minor G allele of rs5030875 was associated with higher stress-induced HPA axis function in patients with rheumatoid arthritis (Malysheva et al., Citation2011) but not with cortisol secretion or sensitivity in healthy men (Rosmond, Chagnon, Bouchard, & Bjorntorp, Citation2001). The minor C allele of rs3176921 was associated with higher verbal intelligence and cognition values in healthy adults (Strohner, Citation2011), but not investigated with respect to stress. So far, only one study analyzed SNPs of CRH with respect to blood pressure, heart rate or cortisol response following stress. The rare allele of rs5030875 showed a significant interaction with a functional SNP (ThtIIII) of the glucocorticoid receptor gene (GR, NR3C1), but no effects on diurnal saliva cortisol variation or dexamethasone suppression (Rosmond et al., Citation2001). Haplotypes of CRH have not yet been investigated in a cohort tested in a standardized laboratory stress test.
The aim of the present study was to characterize CRH promoter haplotypes using a luciferase reporter gene assay in vitro. Following previous findings on CRH promoter functionality (Rosmond et al., Citation2001; Wagner et al., Citation2006) forskolin and hydrocortisone were considered as stimuli for better comparison between in vitro and in vivo markers of stress reactivity. For this purpose, an association study with CRH haplotypes, post-awakening cortisol, and parameters of a laboratory-induced stress response in 232 healthy subjects (cf., Li-Tempel, Larra, Sandt, et al., Citation2016; Li-Tempel, Larra, Winnikes, et al., Citation2016) was performed. We assumed that CRH haplotypes modulate both, promoter activity and in vivo stress responses. Haplotypes resulting in a strong promoter activation are hypothesized to show association with physiological parameters and/or stress reactivity.
Material and methods
Functional characterization of CRH promoter SNPs in vitro
Construction of hCRH reporter plasmids
To analyze the potentially altered promoter activity of the human CRH gene, 3651 bp corresponding to the human CRH promoter region were amplified with PCR using genomic DNA as template and sub-cloned in a sense orientation (antisense was used as negative control) upstream of luc+ into the luciferase reporter vector pGL3-basic (Promega Corporation, Mannheim, Germany) between XmaI and Eco53KI (NEB, R0180 and R0116, Frankfurt a. M., Germany). The 3651 bp DNA fragments were confirmed by sequencing and contain a set of four polymorphisms of which two were examined more closely. The SNP rs3176921 (T/C) at position -684 bp co-segregate with the SNPs rs5030877 and rs5030876 resulting in two alleles (A1/A2) and was selected for further investigations. The biallelic polymorphism rs5030875 (T/G) at position -3371 bp correspond to alleles B1/B2. All positions refer to the transcription start site of the CRH gene. Finally, three different haplotypes could be assigned, haplotype 1 (T-T), haplotype 2 (G-C), and haplotype 3 (T-C) (). Haplotype 1 corresponds to haplotype A1B1, haplotype 2 to A2B2, and haplotype 3 to A2B1 in the study of Wagner and colleagues (2006). Detailed information on plasmid construction is available in Supporting Information Table S1 and Supporting Information Figure S1.
Figure 1. A: Schematic representation of pGl3 constructs containing 3651 bp of the human CRH promoter sequence. The single nucleotide polymorphisms (SNPs) rs503875 (T/G) and rs3176921 (T/C) in the CRH promoter are highlighted in boxes. Combination of both SNPs results in three haplotypes. The arrow represents the transcription start site. B: Location of studied CRH SNPs in the promoter region and diagrammatic representation of the CRH promoter showing the sequential occurrence of response elements that are thought to have a functional role regulating gene expression. (The location in the figure is not in relative correspondence to the actual sequence size).
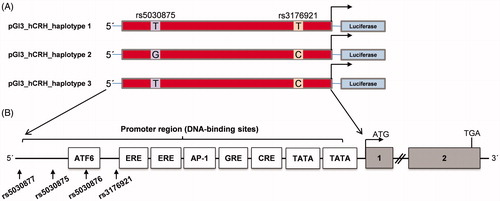
Table 1. Pairwise comparison of promoter activity between three CRH promoter haplotypes in two cell lines under basal and stimulation condition.
Cell culture, transfection, stimulation and luciferase assay
The mouse pituitary corticotroph tumor-derived cell line AtT-20/D16v-F2 (AtT-20) was kindly donated by Christoph Baerwald (Department of Rheumatology, University Hospital, Leipzig, Germany). The AtT-20 cells were grown under standard culture conditions in Dulbecco’s Modified Eagle’s Medium (DMEM, 4.5 g D-glucose/ml without L-glutamine phenol red, Lonza, Basel, Switzerland), supplemented with 10% Fetal Bovine Serum (FBS, Lonza), 2 mM L-glutamine, and 0.1 mg/mL penicillin/streptomycin (Pen/Strep, Sigma, Steinheim, Germany). The human glioblastoma–astrocytoma U373MG cells (ECACC 89081403) were grown in Eagle’s Minimal Essential Medium (EMEM, Lonza), supplemented with 10% FBS (Lonza), 2 mM L-glutamine (Lonza), and 1% NEAA (Lonza). Both cells were maintained in the in 37 °C incubator containing 5% CO2.
AtT-20 Cells and U373MG were transfected using Lipofectamin 3000 transfection reagent (InvitrogenTM, life technologies, California, United States) following the manufactures instruction. Briefly, cells were passaged at 80% confluency and seeded in a density of 3 × 105 cells/mL in 6-well plates. After 24 h of incubation, 3 µg DNA (2.98 µg Firefly luciferase constructs and 0.02 µg pGL4.74 ([HRLUC/TK], Renilla Luciferase; Promega) were added to the cell culture. Since the regulation of CRH gene expression might be affected by the transcription factors (TFs), cAMP response element-binding protein (CREB) and GR, we determined the promoter activity in response to forskolin and hydrocortisone. Both stimulants (in contrast to 8-Br-cAMP and dexamethasone) were selected due to their better physiological compatibility to in vivo studies. Time course and doses below were chosen after reviewing previous studies (Guardiola-Diaz, Kolinske, Gates, & Seasholtz, Citation1996; Reisine, Rougon, & Barbet, Citation1986). The ratio of the dosage conversion of dexamethasone to hydrocortisone was 25 and calculated using an online tool (http://clincalc.com/Corticosteroids/). About 24 h after transfection, 50 µM forskolin (Sigma) or 2.7 µM hydrocortisone (Sigma) was added and cells were incubated for another 24 h. Cells were collected and subjected to passive lysis. Luciferase activity was detected by spectrometry (Berthold, Bad Wildbad, Germany) using the Dual-Luciferase Reporter Assay (DRL) system (Promega), in which Renilla luciferase activity was used to normalize the transfection efficiency. The intensity of the measurement varied between 500 and 5000 light units and the background signal of 6–8 light units was subtracted, respectively. Promoter activities are presented as relative light units (RLUs), calculated as the quotient of the individual Firefly luciferase driven value (promoter activity) divided by the corresponding Renilla luciferase driven value (transfection efficiency control). As controls, pGL3-control, empty pGL3-basic vectors, and antisense direction constructs were used.
Genetic association study in vivo
Participants
About 232 participants were recruited from the University of Trier as previously reported (Larra et al., Citation2014; Li-Tempel, Larra, Sandt, et al., Citation2016; Li-Tempel, Larra, Winnikes, et al., Citation2016). Briefly, 218 healthy men and women (115 females, mean age years =23 ± 2.8; BMI 22.4 ± 0.25) completed the experimental protocol. Participants with an increased objective or subjective sensitivity to cold, any indication of circulatory disturbances, cardiovascular problems, smoking more than five cigarettes per day or alcohol abuse were excluded. Participants had to be medication free, contraceptives (except ethinylestradiol and drospirenone containing ones such as Yasmin (Bayer-Schering Pharma, Leverkusen, Germany) were allowed at the time of testing as studies showed no influence of contraceptives on the cortisol awakening response (Wüst et al., Citation2000). As previously reported, caffeinated and alcoholic drinks, physical exercise, and meals were not permitted in the 3 h immediately preceding the experimental visit. The protocol was approved by the ethics committee of the state’s medical association (Landesärztekammer Rheinland-Pfalz) and in accordance with the declaration of Helsinki. All participants gave written informed consent.
DNA extraction, genotyping, and haplotype construction
DNA was extracted from 10 mL EDTA blood following a standard NaCl salting out method according to the protocol of Miller (Miller, Dykes, & Polesky, Citation1988). The CRH SNPs rs3176921 and rs5030875 were amplified using standard PCR method. For allelic discrimination, the amplified PCR products of rs3176921 and rs5030875 were subsequently digested with the restriction enzyme BsmAI or XmnI, respectively. Detailed information about primer sequences and PCR conditions are available in Supporting Information (Supporting Information Table S2).
Table 2. The association of haplotypes of the CRH promoter with the cortisol level, blood pressure and heart rate under basal and stress condition.
Assessment of post-awakening cortisol
Participants were asked to collect at five times saliva samples using Salivettes (Sarstedt, Nümbrecht, Germany) at home; immediately after awakening and 15, 30, 45, and 60 min thereafter on two consecutive weekdays for the assessment of post-awakening cortisol. They were instructed to abstain from food, drinks other than water and brushing their teeth before completion of saliva sampling. Furthermore and for reasons of compliance, they were told to pay special attention to keep the timing right to ensure the reliability of their data. Salivary cortisol was analyzed as previously reported (Li-Tempel, Larra, Winnikes, et al., Citation2016), in correspondence with consensus guidelines for the assessment of post-awakening cortisol (Stalder et al., Citation2016).
Socially evaluated cold pressor test (SeCPT)
The SeCPT was performed as previously reported (Lass-Hennemann, Kuehl, Schulz, Oitzl, & Schachinger, Citation2011; Schwabe, Haddad, & Schachinger, Citation2008). Briefly, participants assigned to the SeCPT group were asked to completely immerse their hand in ice-cold (2–3 °C) water. Participants assigned to the control group were asked to completely immerse their hand in isothermic (35–37 °C) water. Participants in the SeCPT group were under the social surveillance of an experimenter; their perceptions of social evaluation, uncertainty, and lack of control were enhanced by warning them that the procedure may be painful, not communicating the duration of immersion during the test, and informing them that their performance would be recorded for subsequent facial expression analysis. Participants assigned to the control group were not under social surveillance and no video camera was present. All participants were asked to remove their hand from the water after 3 min. Heart rate (HR) and systolic, diastolic blood pressure (SBP, DBP) were measured throughout the experiment using the Dinamap System (Critikon; Tampa, Florida) with the cuff placed on the right upper arm. Assessments at three times were used to calculate the recovery period: 10, 25, and 55 min after SeCPT finished. Cortisol, measured in salivary samples collected at five time points (one time pre SeCPT and four times post SeCPT), served as an indicator for HPA-axis reactivity.
Statistical analysis
Group comparisons in the in vitro studies were performed using an analysis of variance (ANOVA). Both SNPs were tested for Hardy-Weinberg equilibrium (HWE). Haploview 4.2 (Barrett, Fry, Maller, & Daly, Citation2005) was used to assess inter-marker linkage disequilibrium scores (LD scores), expressed as D′ and r2. Individual haplotypes were reconstructed using the program PHASE, version 2.1 [(Stephens, Smith, & Donnelly, Citation2001) http://stephenslab.uchicago.edu/software.html#phase], which uses an algorithm based on coalescence-based Bayesian haplotype inference for predicting haplotypes from genotype data, combining modelling strategy with computational strategies. Post-awakening cortisol levels assessed on two consecutive days were averaged in order to enhance the reliability of the measure (Hellhammer et al., Citation2007). The area under the curve with respect to ground (AUCg) and increase (AUCi) was calculated as previously reported (Pruessner, Kirschbaum, Meinlschmid, & Hellhammer, Citation2003). Briefly, the formula for five times measures for AUCg = and AUCi =
(
denoting the individual measurement, and
the total number of measurements). AUCg and AUCi served as dependent variables in the subsequent analyses. Cardiovascular data (HR, SBP, DBP) were reduced by extracting the mean increase of dependent variables from baseline to the peak after the water task and mean decrease from the peak to the recovery period. These are referred to as, for example, “SBP peak”, “SBP increase,” and “SBP decrease.” Baseline was considered as average value of three measurements before water task. Recovery period was calculated using the three measurements after the water task. Results were considered statistical significant with a p-value <.05. The threshold of significance was set at p < .05, *, **, and *** represent p < .05, p < .01, and p < .001, respectively. Statistical analyses were performed using SPSS 20.0. Analyzed data are indicated as mean ± SEM.
Results
Functional characterization of CRH haplotypes in vitro
Baseline condition
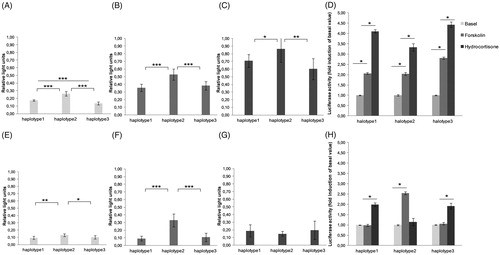
The luciferase activity for three CRH promoter constructs under baseline and stimulated conditions in two cell lines (AtT-20 and U373MG) is shown in . In both cell lines, the promoter activity was highest for haplotype 2. In detail, comparison of transcriptional activity between the haplotypes measured in the AtT-20 cell line revealed significantly higher transcriptional activity of haplotype 2 compared with haplotype 1 [t(16) = 8.85, p < .001)], and haplotype 1 showed significantly higher transcriptional activity than haplotype 3 under unstimulated conditions (basal) [t(16) = 5.07, p < .001]. In the U373MG cell line, haplotype 2 showed significantly higher transcriptional activity compared with haplotype 1 [t(16) = 3.867, p = .001], as well as compared with haplotype 3 [t(16) = 2.572, p = .020], whereas haplotype 1 and haplotype 3 did not differ significantly [t(16) = 0.785, p = .444].
Figure 2. Activity of different CRH promoter alleles (haplotypes 1, 2, and 3) measured by luciferase activity in the AtT-20 cell line (A–D) and the U373MG cell line (E–H). Cells were treated with forskolin (50 µM) and hydrocortisone (2.7 µM) for 24 h before subjected to dual luciferase reporter assay. (A) AtT-20 cells in control condition. (B) AtT-20 cells under forskolin stimulation. (C) AtT-20 cells under hydrocortisone stimulation. (E) U373MG cells in the control condition. (F) U373MG cells under forskolin stimulation. (G) U373MG cells under hydrocortisone stimulation. Fold induction of luciferase activity of three haplotypes with respect to the control condition in AtT-20 (D) and U373MG cells (H). The results are represented as the mean ± SEM of three independent experiments performed in triplicate. (*p < .05; **p < .01; ***p < .001).
Stimulation condition
We used a three (haplotype: 1, 2, 3) × three (condition: basal, forskolin, hydrocortisone) ANOVA to examine the promoter activity in AtT-20 cells. In addition to significant main effects of haplotype, F(2, 16) = 309.88, p < .001, and of condition, F(2, 16) = 92.48, p < .001, the interaction was significant, F(4, 32) = 4.01, p = .010. Transcriptional activity increased for all three haplotypes constructs after stimulation with forskolin or hydrocortisone (). A detailed overview about pairwise comparisons of all significant results is given in . Comparison of transcriptional activity between the haplotypes in the AtT-20 cell line revealed a significantly higher transcriptional activity of haplotype 2 compared with haplotype 1 as well as compared with haplotype 3 if stimulated with forskolin or hydrocortisone whereas haplotype 1 and haplotype 3 did not differ significantly (see ).
Another three (haplotype) × three (condition) ANOVA examined promoter activity in the U373MG cell line. In addition to significant main effects of haplotype, F(2, 10) = 19.13, p < .001, and of condition, F(2, 10) = 20.72, p < .001, the interaction was significant, F(4, 20) = 8.35, p = .010. Forskolin increased transcriptional activity for haplotype 2 but not haplotypes 1 and 3 (). In contrast, hydrocortisone increased the promoter activity for haplotypes 1 and 3 but not haplotype 2 (). Comparison of transcriptional activity between the haplotypes revealed that under stimulation with forskolin, haplotype 2 showed significantly higher transcriptional activity compared with haplotype 1 as well as compared with haplotype 3 whereas haplotype 1 and haplotype 3 did not differ significantly. Under hydrocortisone stimulation, there were no significant differences between haplotypes. All pairwise comparisons can be found in .
Comparison of forskolin and hydrocortisone stimulation relative to baseline is depicted in . In AtT-20 cells, treatment with hydrocortisone resulted in a higher increase in luciferase activity compared with forskolin. This effect was found for all three CRH haplotypes (). However, the fold increase to baseline varied with 4.424 ± 0.138 times (p < .001) in haplotype 3, 4.107 ± 0.848 times (p < .001) in haplotype 2, and 3.326 ± 0.174 times (p < .001) in haplotype 1. In contrast, this pattern did not occur in U373MG cells ().
Genetic association study in vivo
Sample characteristics
A relatively homogenous cohort of 232 undergraduate students with minimal lifestyle differences, determined from exclusion and inclusion criteria, were recruited, and data from 218 (103 males and 115 females) were analyzed (Supporting Information Table S3).
Two CRH promoter polymorphisms, rs5030875 and rs3176921, were analyzed and an overview of the genetic data is given in the supplements (Supporting Information Table S4). In line with the literature, both CRH SNPs were in high linkage disequilibrium (D′ = 1, r2 = .48). The haplotype structure was successfully created with PHASE. CRH haplotype 1 (T-T) had a frequency of 89.7% and consisted of the major alleles of each SNP. Haplotype 2 (G-C) with a frequency of 5.3% contained the minor alleles of each SNP. Haplotype 3 (T-C) showed a frequency of 5.0%. The haplotype frequencies were similar to that previously reported (Wagner et al., Citation2006). For the CRH genotypes and haplotypes, there were no differences in the distributions with respected to age and gender.
Basal condition
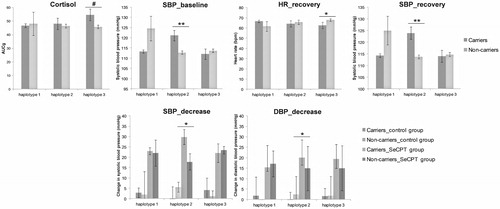
Post-awakening cortisol is used as an indicator for the basal state of the HPA axis. The average value of three repeated measures of blood pressure and heart rate before the SeCPT experiment served as cardiovascular parameters for the basal sympathetic-adrenal-medullary system (SAM) state. To evaluate the influences of CRH haplotypes on post-awakening cortisol expressed as AUCi and AUCg as well as baseline blood pressure and heart rate, a series of one-way ANOVAs were performed (). Haplotype 2 had a significant effect on baseline SBP [F(1, 216) = 9.696, p = .002]. Carriers of haplotype 2 had higher SBP compare to non-carriers (mean 121.1 ± 2.56; mean 112.732 ± 0.818). In addition, an association of haplotype 3 and AUCg approached significance [F(1, 215) = 3.883, p = .05] with higher AUCg in carriers of haplotype 3 compared with non-carriers (mean 54.715 ± 4.253; mean 45.92 ± 1.355). Statistically significant results are depicted in .
Figure 3. Mean values corresponding to ANOVA results: Asterisks indicate a significant main effect of haplotype (carriers, non-carriers) in the upper section or a significant interaction of a haplotype (carriers, non-carriers) and the socially evaluated cold pressure (SeCPT) manipulation in the lower section. Baseline data were considered as average value of three measurements before water task. Recovery was calculated using the three measurements after the water task. Decrease referred to the dependent variables from the peak to the recovery period. The results are represented as the mean ± SEM. (*p < .05; **p < .001, #p = .05).
Stress condition
Five repeated measures of cortisol levels during the SeCPT served as an indicator for the HPA-axis reactivity. Cardiovascular data (HR, SBP, DBP) as well as AUCg and AUCi calculated from five cortisol measures were used as the dependent variable in separate between-participants ANOVAs with the two factors SeCPT group and haplotypes (). There was a significant main effect of haplotype 2 on the “SBP recovery,” with higher SBP for carriers [F(1, 212) = 13.429, p < .001] compared with non-carriers (mean 123.912 ± 2.64; mean 113.777 ± 0.825). Moreover, “SBP decrease” was influenced by a significant interaction of haplotype 2 and SeCPT group [F(1, 209) = 4.860, p = .029]. Carriers of haplotype 2 showed a higher “SBP decrease” in the SeCPT group whereas in the control group carriers of that haplotype had a smaller “SBP decrease.” Similar, “DBP decrease” was also influenced by a significant interaction of haplotype 2 and SeCPT group [F(1, 212) = 6.831, p = .010]. In addition, there was a significant main effect on the “HR recovery” with lower HR for haplotype 3 carriers [F (1, 212) = 13.429, p = .036] compared with non-carriers (62.641 ± 3.065; 67.831 ± 1.354).
Discussion
Three CRH haplotypes constituted of the two SNPs rs5030875 and rs3176921 were investigated in vitro and in vivo regarding their possible influence on stress regulation. In the first part of the study, we characterized the haplotypes in two cell lines with respect to their baseline activity as well as their activation after stimulation with forskolin and hydrocortisone. Haplotype 2 (G-C), which contains the minor alleles of rs5030875 and rs3176921, showed the highest transcriptional activity in both cell lines under baseline conditions and after treatment with forskolin. The stimulation with hydrocortisone alone, however, increased the promoter activity of all three haplotypes in AtT-20 cells but not in U373MG. In our study, we choose hydrocortisone as it can, rather than dexamethasone, activate both types of glucocorticoid receptors. Further, tissue-specific actions of ligand-activated glucocorticoid receptors might take place on the CRH promoter in a cell-type-specific way (King & Nicholson, Citation2007). In addition and in contrast to other studies, our promoter construct contains not only the proximal 600–900 bp of the CRH promoter but also the distal promoter, which harbors more potential transcription factor binding sites that might indirectly enhance and inhibit the regulation via GCs (Vamvakopoulos & Chrousos, Citation1993). In the second part, we found that under baseline conditions carriers of haplotype 2 showed an effect on the SAM system, represented by higher blood pressure compared with non-carriers, but did not differ in cortisol levels. After stress exposure, haplotype 2 was associated with different blood pressure indices but again not with any HPA-related measure. In addition, we observed a trend for carriers of haplotype 3 to possess higher post-awakening cortisol compared with non-carriers.
In our study, we worked with AtT-20 cells, a widely used cell line in the studies focusing on CRH (King & Nicholson, Citation2007), and U373 cells as a human cell line to have two possible models for the CRH action. It is known that CRH expression is highly tissue specific, with highest expression levels found in placenta followed by brain, testis, and thyroid (Fagerberg et al., Citation2014). Interestingly, only one CRH promoter appears to be responsible for this tissue-specific regulation of CRH expression. For example, in pituitary tumor-derived AtT-20 cells cAMP produces a high level of promoter activity through the cAMP response element (Jensen et al., Citation2009), and the caudal type homeobox protein response element (CDXARE), whereas in placental cells the CRH promoter had a low intrinsic basal activity and cAMP caused a modest increase in activity because CRE acts in isolation (King & Nicholson, Citation2007). Therefore, AtT-20 and U373MG cells might differ in their capacity to mediate signals to the CRH promoter, which could explain differences observed in our experiments. Functional consequences of the investigated haplotypes are supposed to vary in different cell types and seem to depend on specific TFs. These differences can be related to the actual TFs present, splice variants, cofactors, post-translational modifications, differences in activation pathways, or sequence variations. As such, our findings might inspire further research on the biological mechanisms and need additional evaluation.
Previously and similarly to our study, a significant increase in the promoter activity after stimulation with 8-Br-cAMP has been reported (Wagner et al., Citation2006). However, and in contrast to our findings, haplotype 1 (A1B1) resulted in the highest luciferase activity after 36 h. This difference might be attributed to the longer stimulation time and/or to the use of different stimulation reagents. Both 8-Br-cAMP and forskolin, are related to cAMP activation but serve different molecular mechanisms. In our study, we used forskolin, an activator of the adenylate cyclase that increases intracellular levels of cAMP and its availability for different TFs. Moreover, a previous study suggested that the effects of forskolin on the AtT-20 cell may be due to mechanisms occurring in normal pituitary cells, too (Guild, Itoh, Kebabian, Luini, & Reisine, Citation1986). In comparison, Wagner and colleagues treated cells with 8-Br-cAMP, a brominated derivate of cAMP, which is long-acting on the activation of cAMP-dependent protein kinases. Due to the stimulation of intracellular cAMP production and possible recruitment of specific cAMP-dependent TFs (Kapatos, Stegenga, & Hirayama, Citation2000; Wagner et al., Citation2006), forskolin might lead to the slightly different activation profile observed in our settings.
We focused on two SNPs that were previously shown to impact mainly cAMP-dependent transcription factor binding. Rs3176921 changes the consensus sequence of the binding box of nuclear transcription factor Y (NF-Y) from CCAAT to CCACT, and therewith disrupts NF-Y binding (Wagner et al., Citation2006) as well as significantly affects the expression of genes involved in complex pathways (Chassanidis et al., Citation2009; Chen, Mo, Li, Zeng, & Xu, Citation2007; Dolfini, Gatta, & Mantovani, Citation2012). Further evidence is accumulating that alteration of the NF-Y structure is not the direct cause of any specific disease. Rather the efficiency of DNA-binding might play a role on the development of several pathological conditions, resulting either in altered proteins, or altered gene expression patterns (Dolfini et al., Citation2012). Further, rs5030875 is in a high linkage disequilibrium with the SNP rs5030876, which results in the exchange of a nucleotide within the binding site of the transcription factor activating transcription factor 6 (ATF6). ATF6 is a member of the human ARF/CREB (cAMP response element binding protein) family. ATF6 participates in two independent signaling pathways, both leading to transcriptional activation in the nucleus (Yoshida, Haze, Yanagi, Yura, & Mori, Citation1998), interaction with cAMP responsive elements (Fawcett, Martindale, Guyton, Hai, & Holbrook, Citation1999), and finally differences in the CRH promoter activity (Wagner et al., Citation2006).
The pivotal role of CRH in integrating various aspects of the stress response by regulating the HPA axis is well understood. In particular, stress-induced hypothalamic CRH results in the release of ACTH, which increases blood pressure secondary to secretion of glucocorticoids from the adrenal cortex. In addition to these indirect effects, CRH also exerts direct effects on the cardiovascular system, which can be centrally and peripherally. Therefore, several pathways have been discussed (Yang et al., Citation2010). First, stress-induced hypothalamic CRH enhances contractility of vasculature and increases blood pressure and heart rate via β-endorphin-induced catecholamine release from the adrenal medulla. Second, central CRH modulates autonomic nervous system activity via the dorsal motor nucleus of the vagus, decreasing heart rate, and increasing heart rate variability due to enhanced sympatho-vagal antagonism. Third, peripheral CRH mediates subsequent decrease in blood pressure and relaxation of vasculature, while direct action on CRHR2 in the heart enhanced myocyte contractility (Yang et al., Citation2010). Complementary to our in vitro findings, we found independent of the stress exposure higher blood pressure in carriers of haplotype 2 under baseline conditions. This might reflect the direct rather than the indirect effects of CRH on the cardiovascular system, probably due to a haplotype dependent increase of CRH expression. We suggest that more CRH binds to the CRH receptors but did not have a significant impact on cortisol levels in our cohort. In contrast, the observed trend for higher post-awakening cortisol (calculated as AUCg) and lower heart rate recovery of carriers of haplotype 3 indicates genetic impacts on the HPA that are unrelated to blood pressure. This might reflect a protective aspect of higher AUCg and only moderate changes under stress. Although the present findings do to not permit to explain the exact pathways of the observed effects, they demonstrate the putative importance of the examined SNPs in stress responses and a susceptibility to situational and individual influences. We suggest considering and testing the various ways of CRH affecting the cardiovascular system (directly or indirectly) when trying to understand the association between stress responses and genetic variations.
For the interpretation of our study results, some limitations need to be taken into account. To control for false positive results of promoter activity only due to the inserted DNA sequences, the hCRH promoter region was also used in an antisense orientation. The promoter activity of our constructs was tested in two tumor cells derived from pituitary or glioblastom but not in peripheral cells. However, as central CRH signalling is involved in HPA axis regulation as well as direct regulation of cardiovascular processes, the investigation of those cells seemed a plausible choice. In our study, we detected positive glucocorticoid regulation in AtT-20 cells, which is a relatively unique finding. Many other studies do not report any detectable change with glucocorticoids alone but use 8BrcAMP in combination with dexamethasone. The increase in promotor activity with hydrocortisone in our settings might be in part explained by the absence of cAMP-mediated pathways and a possible interaction of ligand-activated glucocorticoid receptors. Further experiments using the combination of forskolin and hydrocortisone together with the use of respective antagonist of each of the glucocorticoid receptors are warranted to shed light on the complex molecular mechanisms of the CRH regulation. Even with a sample size of over 200 participants, the low-frequency (MAF 5%–10%) of many genetic variants are outside the reach of sufficient statistical power for association studies. Therefore, some challenges continue to exist in both the discovery and interpretation of findings from genetic association studies. Given the sample size of N = 218 in our study, the statistical power to detect a small-to-medium-size of ηp2 = 0.04 was 1 – β = 0.76 for analyses comparing three haplotype groups. Although this power can be considered as adequate for the present purpose of our investigation, the significance of the reported associations still has to be interpreted carefully because smaller effects might have remained undetected. In particular, a careful interpretation of the association results related to haplotype 2 and 3 is warranted due to the small sample size. In addition, to control for the relatively small number of donors, we aimed at a high homogeneity of the young, infrequent-smoking, undergraduate student population reducing confounding socioeconomic factors. Hence, our results should not be generalized to other than a Caucasian undergraduate student population. For future studies, it might be necessary to increase the samples size and population diversity. Perhaps, the reported associations change with age or differ across ethnicities. If available, a focus on low-frequency alleles might be considered in particular.
Conclusions
The present study links the functional inspection in vitro and the genetic association of CRH promoter SNPs in vivo to physiological stress parameters. In vitro assays demonstrated differences in promoter activity depending on the haplotype. Haplotype 2 revealed the highest transcriptional activity under baseline conditions and after treatment with forskolin. The molecular mechanism underlying these differences might be affected by the SNPs that change DNA sequences and therewith important regulatory elements. In vivo, individuals with haplotype 2 had a higher dynamic response in systolic blood pressure under baseline and after stress exposure compared with non-carriers. Taken together, we conclude that these CRH haplotypes contribute to individual differences in the neuroendocrine response when coping with stress. In addition, our results suggest that direct and indirect effects of CRH differ regarding the individual susceptibility to situational stimuli and genetic variance.
SM_Figure_1_Legend_resubmission.docx
Download MS Word (13.5 KB)SM_Figure_1_EWM1.tif
Download TIFF Image (1.6 MB)SM_Table_4_resub.docx
Download MS Word (13.5 KB)SM-Table_3_resub.docx
Download MS Word (14.2 KB)SM-Table_2_final.docx
Download MS Word (14 KB)SM-Table_1_final.docx
Download MS Word (14.3 KB)Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
References
- Arbiser, J.L., Morton, C.C., Bruns, G.A.P., & Majzoub, J.A. (1988). Human corticotropin releasing hormone gene is located on the long arm of chromosome 8. Cytogenetic and Genome Research, 47, 113–116. http://www.karger.com/DOI/10.1159/000132525
- Baerwald, C.G.O., Mok, C.C., Fife, M.S., Tikly, M., Lau, C.S., Wordsworth, B.P., … Lanchbury, J.S. (1999). Distribution of corticotropin-releasing hormone promoter polymorphism in different ethnic groups: Evidence for natural selection in human populations. Immunogenetics, 49, 894–899. doi:10.1007/s002510050570
- Baerwald, C.G.O., Panayi, G.S., & Lanchbury, J.S. (1996). A new XmnI polymorphism in the regulatory region of the corticotropin releasing hormone gene. Human Genetics, 97, 697–698. doi:10.1007/s004390050121
- Barrett, J.C., Fry, B., Maller, J., & Daly, M.J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. doi:10.1093/bioinformatics/bth457
- Chassanidis, C., Kalamaras, A., Phylactides, M., Pourfarzad, F., Likousi, S., Maroulis, V., … Kollia, P. (2009). The Hellenic type of nondeletional hereditary persistence of fetal hemoglobin results from a novel mutation (g.-109G > T) in the HBG2 gene promoter. Annals of Hematology, 88, 549–555. doi:10.1007/s00277-008-0643-0
- Chen, X.-W., Mo, Q.-H., Li, Q., Zeng, R., & Xu, X.-M. (2007). A novel mutation of −73(A→T) in the CCAAT box of the β-globin gene identified in a patient with the mild β-thalassemia intermedia. Annals of Hematology, 86, 653–657. doi:10.1007/s00277-007-0312-8
- Dolfini, D., Gatta, R., & Mantovani, R. (2012). NF-Y and the transcriptional activation of CCAAT promoters. Critical Reviews in Biochemistry and Molecular Biology, 47, 29–49. doi:10.3109/10409238.2011.628970
- Eysenck, M.W. 2004. Psychology: An international perspective (pp. 165). London: Taylor & Francis.
- Fagerberg, L., Hallström, B.M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., … Uhlén, M. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular and Cellular Proteomics, 13, 397–406. doi:10.1074/mcp.M113.035600
- Fawcett, T.W., Martindale, J.L., Guyton, K.Z., Hai, T., & Holbrook, N.J. (1999). Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochemical Journal, 339, 135–141. doi:10.1042/bj3390135
- Guardiola-Diaz, H., Kolinske, J., Gates, L., & Seasholtz, A. (1996). Negative glucorticoid regulation of cyclic adenosine 3', 5'- monophosphate-stimulated corticotropin-releasing hormone-reporter expression in AtT-20 cells. Mol Endocrinol, 10, 317–329. doi:10.1210/mend.10.3.8833660
- Guild, S., Itoh, Y., Kebabian, J.W., Luini, A., & Reisine, T. (1986). Forskolin enhances basal and potassium-evoked hormone release from normal and malignant pituitary tissue: The role of calcium. Endocrinology, 118, 268–279. doi:10.1210/endo-118-1-268
- Hellhammer, J., Fries, E., Schweisthal, O.W., Schlotz, W., Stone, A.A., & Hagemann, D. (2007). Several daily measurements are necessary to reliably assess the cortisol rise after awakening: State- and trait components. Psychoneuroendocrinology, 32, 80–86. doi:10.1016/j.psyneuen.2006.10.005
- Jensen, L.J., Kuhn, M., Stark, M., Chaffron, S., Creevey, C., Muller, J., … von Mering, C. (2009). STRING 8 –a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Research, 37, D412–D416. doi:10.1093/nar/gkn760
- Kalogeras, K.T., Nieman, L.K., Friedman, T.C., Doppman, J.L., Cutler, G.B., Chrousos, G.P., … Yanovski, J.A. (1996). Inferior petrosal sinus sampling in healthy subjects reveals a unilateral corticotropin-releasing hormone-induced arginine vasopressin release associated with ipsilateral adrenocorticotropin secretion. Journal of Clinical Investigation, 97, 2045–2050. doi:10.1172/JCI118640
- Kapatos, G., Stegenga, S.L., & Hirayama, K. (2000). Identification and characterization of basal and cyclic amp response elements in the promoter of the rat gtp cyclohydrolase i gene. The Journal of Biological Chemistry, 275, 5947–5957. doi:10.1074/jbc.275.8.5947
- King, B.R., & Nicholson, R.C. (2007). Advances in understanding corticotrophin-releasing hormone gene expression. Frontiers in Bioscience, 12, 581–590. Retrieved from http://europepmc.org/abstract/MED/17127319. https://doi.org/10.2741/2084
- Kovács, K.J. (2013). CRH: The link between hormonal-, metabolic- and behavioral responses to stress. Journal of Chemical Neuroanatomy, 54, 25–33. doi:10.1016/j.jchemneu.2013.05.003
- Kronenberg, H.M., Polonsky, K.S., Larsen, P.R., & Melmed, S. 2007. Williams textbook of endocrinology, 12/e (pp. 125–133). New York, NY: Elsevier India.
- Kudielka, B.M., Hellhammer, D.H., & Wüst, S. (2009). Why do we respond so differently? Reviewing determinants of human salivary cortisol responses to challenge. Psychoneuroendocrinology, 34, 2–18. doi:10.1016/j.psyneuen.2008.10.004
- Larra, M.F., Schulz, A., Schilling, T.M., Ferreira De Sá, D.S., Best, D., Kozik, B., & Schächinger, H. (2014). Heart rate response to post-learning stress predicts memory consolidation. Neurobiology of Learning and Memory, 109, 74–81. doi:10.1016/j.nlm.2013.12.004
- Lass-Hennemann, J., Kuehl, L.K., Schulz, A., Oitzl, M.S., & Schachinger, H. (2011). Stress strengthens memory of first impressions of others' positive personality traits. PLoS ONE, 6, e16389. doi:10.1371/journal.pone.0016389
- Li-Tempel, T., Larra, M.F., Sandt, E., Mériaux, S.B., Schote, A.B., Schächinger, H., … Turner, J.D. (2016). The cardiovascular and hypothalamus-pituitary-adrenal axis response to stress is controlled by glucocorticoid receptor sequence variants and promoter methylation. Clinical Epigenetics, 8, 12. doi:10.1186/s13148-016-0180-y
- Li-Tempel, T., Larra, M.F., Winnikes, U., Tempel, T., DeRijk, R.H., Schulz, A., … Schote, A.B. (2016). Polymorphisms of genes related to the hypothalamic-pituitary-adrenal axis influence the cortisol awakening response as well as self-perceived stress. Biological Psychology, 119, 112–121. doi:10.1016/j.biopsycho.2016.07.010
- Malysheva, O., Wagner, U., Wahle, M., Pierer, M., Wagner, U., Stalla, G.K., & Baerwald, C.G.O. (2011). Hypothalamic-pituitary-adrenal axis stress test in patients with early RA: Role of corticotropin-releasing hormone promoter polymorphisms. Annals of the Rheumatic Diseases, 70, 2058–2059. Retrieved from: http://europepmc.org/abstract/MED/21666229. doi:10.1136/ard.2011.151340
- Miller, S.A., Dykes, D.D., & Polesky, H.F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research, 16, 1215. doi:10.1093/nar/16.3.1215
- Pruessner, J.C., Kirschbaum, C., Meinlschmid, G., & Hellhammer, D.H. (2003). Two formulas for computation of the area under the curve represent measures of total hormone concentration versus time-dependent change. Psychoneuroendocrinology, 28, 916–931. doi:10.1016/S0306-4530(02)00108-7
- Reisine, T., Rougon, G., & Barbet, J. (1986). Liposome delivery of cyclic AMP-dependent protein kinase inhibitor into intact cells: Specific blockade of cyclic AMP-mediated adrenocorticotropin release from mouse anterior pituitary tumor cells. The Journal of Cell Biology, 102, 1630–1637. doi:10.1083/jcb.102.5.1630
- Rosmond, R., Chagnon, M., Bouchard, C., & Bjorntorp, P. (2001). A polymorphism in the regulatory region of the corticotropin-releasing hormone gene in relation to cortisol secretion, obesity, and gene-gene interaction. Metabolism: Clinical and Experimental, 50, 1059–1062. Retrieved from http://europepmc.org/abstract/MED/11555839 doi:10.1053/meta.2001.25598
- Schwabe, L., Haddad, L., & Schachinger, H. (2008). HPA axis activation by a socially evaluated cold-pressor test. Psychoneuroendocrinology, 33, 890–895. doi:10.1016/j.psyneuen.2008.03.001
- Stalder, T., Kirschbaum, C., Kudielka, B.M., Adam, E.K., Pruessner, J.C., Wüst, S., … Clow, A. (2016). Assessment of the cortisol awakening response: Expert consensus guidelines. Psychoneuroendocrinology, 63, 414–432. doi:10.1016/j.psyneuen.2015.10.010
- Stephens, M., Smith, N.J., & Donnelly, P. (2001). A New Statistical Method for Haplotype Reconstruction from Population Data. American Journal of Human Genetics, 68, 978–989. Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1275651/. doi:10.1086/319501
- Strohner, C. (2011). Auswirkungen des Polymorphismus rs3176921 im CRH-Gen auf kognitive Phänotypen (Dissertation, University of Munich: Faculty of Medicine). Retrived from https://edoc.ub.uni-muenchen.de/12882/
- Vamvakopoulos, N.C., & Chrousos, G.P. (1993). Structural organization of the 5′ flanking region of the human corticotropin releasing hormone gene. Mitochondrial DNA, 4, 197–206. doi:10.3109/10425179309015632
- Wagner, U., Wahle, M., Moritz, F., Wagner, U., Häntzschel, H., & Baerwald, C.G. (2006). Promoter polymorphisms regulating corticotrophin-releasing hormone transcription in vitro. Hormone and Metabolic Research, 38, 69–75. doi:10.1055/s-2006-925115
- Wüst, S., Wolf, J., Hellhammer, D.H., Federenko, I., Schommer, N., & Kirschbaum, C. (2000). The cortisol awakening response – normal values and confounds. Noise and Health, 2, 79–88. Retrieved from doi: http://europepmc.org/abstract/MED/12689474
- Yang, L.-Z., Tovote, P., Rayner, M., Kockskämper, J., Pieske, B., & Spiess, J. (2010). Corticotropin-releasing factor receptors and urocortins, links between the brain and the heart. European Journal of Pharmacology, 632, 1–6. doi:10.1016/j.ejphar.2010.01.027
- Yoshida, H., Haze, K., Yanagi, H., Yura, T., & Mori, K. (1998). Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins: Involvement of basic leucine zipper transcription factors. Journal of Biological Chemistry, 273, 33741–33749. doi:10.1074/jbc.273.50.33741
