Abstract
Dexamethasone-induced Ras-related protein 1 (Rasd1) is a member of the Ras superfamily of monomeric G proteins that have a regulatory function in signal transduction. Rasd1, also known as Dexras1 or AGS1, is rapidly induced by dexamethasone (Dex). While prior data indicates that Rasd1 is highly expressed in the pituitary and that the gene may function in regulation of corticotroph activity, its exact cellular localization in this tissue has not been delineated. Nor has it been determined which endocrine pituitary cell type(s) are responsive to Dex-induced expression of Rasd1. We hypothesized that Rasd1 is primarily localized in corticotrophs and furthermore, that its expression in these cells would be upregulated in response to exogenous Dex administration. Rasd1 expression in each pituitary cell type both under basal conditions and 1-hour post Dex treatment were examined in adult male mice. While a proportion of all endocrine pituitary cell types expressed Rasd1, a majority of corticotrophs and thyrotrophs expressed Rasd1 under basal condition. In vehicle treated animals, approximately 50–60% of corticotrophs and thyrotrophs cells expressed Rasd1 while the gene was detected in only 15–30% of lactotrophs, somatotrophs, and gonadotrophs. In Dex treated animals, Rasd1 expression was significantly increased in corticotrophs, somatotrophs, lactotrophs, and gonadotrophs but not thyrotrophs. In Dex treated animals, Rasd1 was detected in 80–95% of gonadotrophs and corticotrophs. In contrast, Dex treatment increased Rasd1 expression to a lesser extent (55–60%) in somatotrophs and lactotrophs. Corticotrophs of the pars intermedia, which lack glucocorticoid receptors, failed to display increased Rasd1 expression in Dex treated animals. Rasd1 is highly expressed in corticotrophs under basal conditions and is further increased after Dex treatment, further supporting its role in glucocorticoid negative feedback. In addition, the presence and Dex-induced expression of Rasd1 in endocrine pituitary cell types, other than corticotrophs, may implicate Rasd1 in novel pituitary functions.
All organisms strive to maintain a state of homeostasis, which is constantly challenged by a variety of internal and external stressors. Physiological systems have evolved to counterbalance the disruptions caused by stressors (Herman et al., Citation2016). The cells of the pituitary are crucially integrated into the regulation of both basal activity and the response to stress. One of the central stress-related mechanisms is the release of adrenocorticotropic hormone (ACTH) from corticotrophs of the pituitary upon the perception of a stressor through the activation of the hypothalamus-pituitary-adrenal (HPA) axis. Release of ACTH leads to adrenocortical activation and a subsequent rise in glucocorticoid concentrations. Glucocorticoids target cells throughout the body, and make immediate energy available to the organism to overcome acutely stressful situations and to restore homeostasis. It is extremely maladaptive for an organism if pituitary cells fail to respond to stimulation initiated by a stressor or fail to restore homeostasis after the stress response has been initiated. A failure to maintain homeostasis results in excessive or inadequate basal activity and responsiveness to stress associated with a wide variety of health consequences including stress-related mental conditions such as major depressive disorder (Chrousos, Citation2009).
Rasd1, also known as Dexras1 or AGS1, was discovered in 1998 as a gene rapidly induced in mouse corticotroph AtT-20 cells in response to glucocorticoids (Kemppainen & Behrend, Citation1998). The goal of that study was to identify genes/proteins in corticotroph cells whose expression was induced within the early or intermediate time domain (10 min − 3 h) of glucocorticoid negative feedback and could therefore potentially affect ACTH secretion (Shipston, Citation1995). Feedback in this time domain is dependent on transcription/translation and therefore requires synthesis of a protein(s) (Dayanithi & Antoni, Citation1989; Shipston, Citation1995). Although several proteins have been proposed to fulfill such a role (Shipston, Citation1995), the exact mechanism mediating early glucocorticoid negative feedback at the corticotroph has yet to be clearly defined.
Since its discovery, Rasd1 has been found to be expressed in numerous tissues and its possible role(s) in cellular function has been the subject of numerous reports. The gene has been implicated in a wide range of cell functions, including, but not limited to circadian rhythms (Takahashi et al., Citation2003), cancers (Lian et al., Citation2020; Munagala et al., Citation2013; O’Neill et al., Citation2015; Tian et al., Citation2017), brain NMDA–neurotoxicity (Cheah et al., Citation2006), uterine remodeling (Kim et al., Citation2017), and adipogenesis/obesity/osteogenesis (Cha et al., Citation2013; Kim et al., Citation2016; Seok et al., Citation2020). In addition, there is support for Rasd1’s role in the regulation of ACTH release and corticotroph physiology. Tissue screening using RNA expression analysis shows that Rasd1 is highly expressed in pituitary (https://www.proteinatlas.org/ENSG00000108551-Rasd1/tissue). Similarly, we found that the gene was highly expressed in human pituitary by screening a dot blot containing RNA obtained from a wide variety of tissues (Kemppainen et al., Citation2003). Expression of a constitutively active mutant form of Rasd1 inhibited peptide hormone secretion from transfected AtT-20 cells (Graham et al., Citation2001). In addition, Rasd1 knockout mice displayed increased plasma levels of ACTH compared with wild type mice (Seok et al., Citation2020), and a mutation in Rasd1 has been identified in USP8 positive corticotroph tumors from patients with Cushing’s disease (Uzilov et al., Citation2017).
While prior data clearly indicates that Rasd1 is highly expressed in the pituitary and that the gene may function in regulation of corticotrophs, its exact cellular localization in this tissue has not been delineated. Nor has it been determined which endocrine pituitary cell type(s) are responsive to glucocorticoid-induced expression of Rasd1. We therefore undertook the present experiment to co-localize Rasd1 expression in each endocrine pituitary cell type both under basal (vehicle treated) conditions and in response to dexamethasone (Dex). We hypothesized that Rasd1 is primarily localized in corticotrophs and furthermore, that its expression in these cells would be upregulated in response to exogenous Dex administration.
Materials and methods
Animals
Ten male mice (C57BL/6) approximately two months of age, were received from Envigo (Prattville, AL, USA) and were maintained in an AAALAC approved facility at the Auburn University College of Veterinary Medicine (Auburn, AL). All experimental protocols were approved by the Auburn University Institutional Animal Care and Use Committee (Protocol #2019-3593). Animals were communally housed (5 mice/cage) and maintained on a (14/10) photoperiod with ad libitum access to food and water. All animals were given a period of 72 h to allow habituation to the environment prior to beginning the experiment.
Dexamethasone treatment
All animals were habituated to handling, weighing, and intraperitoneal (IP) injections (sterile 1X PBS, 0.25 cc) for 4 days prior to experimental day. On experimental day, animals were weighed then given an IP injection with either vehicle (controls, sterile 1X PBS, 0.25 cc) or Dex (10 ug/mouse, 0.25 cc, 1DEX027, Covetrus, Portland, ME) and returned to their cages. One hour later, mice were euthanized by CO2 inhalation. Pituitaries were collected, flash frozen in 2-methyl-butane and embedded in optimum cutting temperature medium (Tissue-Tek, 25608-930). Pituitaries were sectioned by cryostat at a thickness of 20 µm. Slides were stored at −80 °C until analysis.
RNAscope fluorescent multiplex assay
Protocol was followed according to guidelines provided by Advanced Cell Diagnostics (ACD, Newark, CA). RNAscope fluorescent multiplex assay can use up to a 3-channel system, allowing for the use of three distinct probes, along with DAPI. Based on the five different endocrine cell types of interest, three separate runs were performed (Rasd1-POMC-GH/Rasd1-LH-PRL/Rasd1-TSH). All probes were created using mouse sequences and are as follows: Mm-Rasd1 584651; Mm-GH 445361; Mm-POMC 314081; Mm-Lhb 478401; Mm-PRL 445371; and Mm-TSHb 445381. Slides were removed from −80 freezer and fixed using a 10% neutral buffered formalin at 4 °C for 15 min. Once fixed, tissues followed a series of dehydration (50%, 70%, 100%, 100% EtOH) before beginning RNAscope. A hydrophobic barrier was created around each tissue using an Immedge pen (H-4000, Vector Labs, Burlingame, CA). Protease IV was applied to every tissue and incubated in the HybEZ oven (321720, ACD) for 30 min at 40 °C. Slides were then rinsed with three series of PBS washes before probe hybridization. Probes were applied directly to tissues for 2 hours at 40 °C. Both negative and positive controls were run to ensure viability of the assay. After probe hybridization, slides were washed in 1X wash buffer, incubated in amplification buffers with rinses in the 1X wash buffer between reagents. Tissues were then counterstained with DAPI and coverslipped (0100-35, Fluoromount-G, Southern Biotech, Birmingham, AL) for imaging and analysis. ACD provided universal negative control probe targeting the dihydrodipicolinate reductase (dapB) gene from Bacillus subtilis strain SMY, a soil bacterium to assess nonspecific labeling. Positive control probes were directed toward POLR2A and PPIB to confirm preservation of sample RNA (Supplemental Figure 1).
Imaging and image analysis
Images were acquired using a Nikon A1plus confocal microscope fitted with filter settings for the specific wavelengths used, with a pinhole setting of Airy unit (0.16 μm/px for 40× and 0.11 μm/px for 60× images), resulting in optical slice thickness of 3.6 μm for 40× images and 2.3 μm for 60× images. Images were obtained in sequential acquisition avoiding overlapping spectra. Image capture and thresholds were optimized for each wavelength and held constant throughout study. All post acquisition processing and analysis was performed with NIS-elements 4.6 software (Nikon, Melville, NY). For each animal, a region (300 μm2) of three different pituitary sections of either the pars distalis (PD) or the pars intermedia (PI) were analyzed. A region of interest (ROI) encompassing each positive cell (POMC, GH, LH, PRL, or TSH) was defined. The regions of interests encompassed the center and bilateral regions of the PD. Any cell (ROI) which contained a single puncta/pixel of Rasd1 was considered positive for Rasd1. A Z-Stack was performed on each position to verify that image analysis was restricted to a monolayer of pituitary cells. RNAscope signal is revealed as a punctate staining, with little or no background, and previous studies have demonstrated that each punctum corresponds to one molecule of the intended target mRNA (Wang et al., Citation2012, Citation2014). Thus, quantification of the number of puncta per cell offers a direct measurement of expression of a certain target. As stated above, a single Rasd1 puncta or pixel was used to determine percent of each particular endocrine pituitary cell type positive for Rasd1. However, for highly expressed genes, puncta can oftentimes overlap and fuze, resulting in a difficulty in the accurate numeric quantification. A solution to this issue is to quantify the mean fluorescent intensity (MFI) of a signal within a cell (Chan et al., Citation2018). To determine MFI measurements, the above ROI was placed over every pituitary cell positive for POMC, GH, LH, PRL, or TSH inside a 300 μm2 region of three different pituitary sections of either the PD or PI. The MFI of Rasd1 for each cell was averaged.
Statistical analysis
Percent of POMC, GH, LH, PRL, or TSH cells positive for Rasd1 and MFI of Rasd1 present in each cell type was assessed by Student t test. Two-way ANOVAs followed by individual Bonferroni paired comparisons were performed to evaluate percent Rasd1 positive cells and MFI across cell types and treatment. The level of statistical significance was set at p ≤ 0.05 for all statistical tests. All values were reported as the mean ± SEM. Prism 8 for Mac was used for all analyses (GraphPad Software, Inc., La Jolla, CA).
Results
Rasd1 expression was identified by individual or grouped punctate and granular fluorescence primarily localized to the nuclei or just adjacent. The vast majority of Rasd1 expressing cells were identified in the PD portion of the pituitary gland compared to other pituitary sections. Rasd1 expression was only just above detection in the PI and undetectable in the pars nervosa (posterior pituitary). Overall, Rasd1 expression in the PD was higher in Dex-treated animals than control animals (control = 44.4 ± 7.2 vs Dex = 274.2 ± 24.1 MFI, F(1,3)=11.1, p < 0.0001). There was no difference in Rasd1 MFI in the PI between control (17.68 ± 2.3) and Dex (18.8 ± 4.3) treated animals (F(1,3)=11.1, p < 0.0001).
Pituitary cells expressing one of the five pituitary hormone mRNAs investigated (GH, POMC, LHb, PRL and TSHb), were found throughout the PD. Expression of all hormone mRNA were primarily observed filling the area surrounding the nuclei of pituitary cells, suggesting rough endoplasmic reticulum localization. Overall, the highest proportion of cells expressed PRL or GH followed by POMC, LHb, then TSHb. Dex treatment did not affect the apparent proportion of different cell types or intensity of expression of the mRNA for each pituitary hormone (data not shown). To varying degrees, Rasd1 expression was identified in every endocrine pituitary cell type under control conditions (). In controls, approximately 50–60% of POMC and TSHb expressing cells also expressed Rasd1 while the gene was detected in only 15–30% of lactotrophs, somatotrophs, and gonadotrophs. With the exception of thyrotrophs, Dex treatment resulted in a significantly increased expression of Rasd1 in all other cell types (, F(4,30)=49.08; p < 0.0001). In Dex-treated animals, Rasd1 was detected in 80–95% of gonadotrophs and corticotrophs. In contrast, Dex treatment increased Rasd1 expression to a lesser extent (55–60%) in somatotrophs and lactotrophs. Along with the proportion of cells expressing Rasd1, mean fluorescent intensity (MFI) was used to quantify the expression level of Rasd1 mRNA in each endocrine pituitary cell type (). In Dex-treated animals, Rasd1 MFI was significantly increased above that in controls in all endocrine pituitary cell types, with the exception of thyrotrophs (, F(4,30)=5.9; p = 0.0012). While present in all cell types, the magnitude of Rasd1 expression in GH, LHb, PRL and TSHb expressing cells were relatively low when compared to POMC expressing cells ( and ). Of note, corticotroph Rasd1 MFI levels were significantly elevated above all other cell types in the Dex-treated animals ().
Figure 1. Dexamethasone treatment increases Rasd1 expression in multiple endocrine pituitary cell types. Histograms depicting (A) mean percentage and (B) mean fluorescence intensity (MFI) of Rasd1 expression in POMC, GH, LH, PRL, and TSH expressing cells. Data presented as the mean ± SEM. Asterisks signify difference from control group (p < 0.05).
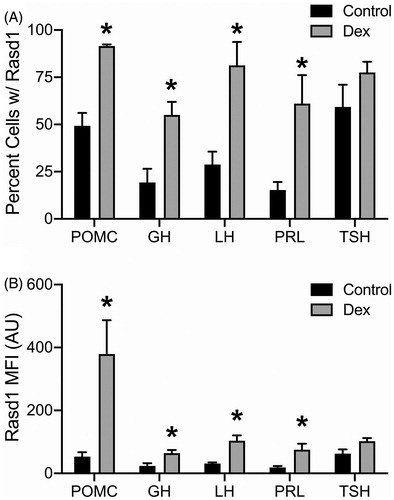
Figure 2. Rasd1 expression in multiple endocrine pituitary cell types. Representative photomicrographs of Rasd1 in somatotrophs (GH), gonadotrophs (LH), lactotrophs (PRL), or thyrotrophs (TSH) in control and dexamethasone (Dex) treated animals. Insets depict 60× magnification images with DAPI nuclear counterstain. scale bars = 10 μm.
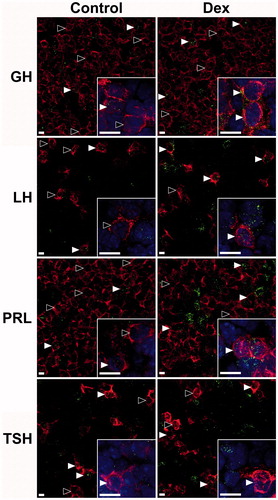
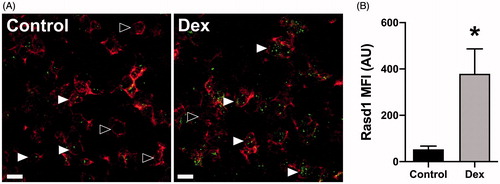
Figure 3. Dexamethasone treatment increases Rasd1 expression in POMC expressing cells. (A) Representative photomicrographs of dual ISH for POMC and Rasd1 in the pituitary of mice treated with vehicle (Control) or dexamethasone (Dex), scale bar = 20 μm. (B) Histogram depicting the mean fluorescent intensity (MFI) of Rasd1 expression in POMC cells. Data presented as the mean ± SEM. Asterisk signifies difference from control group (p < 0.05).
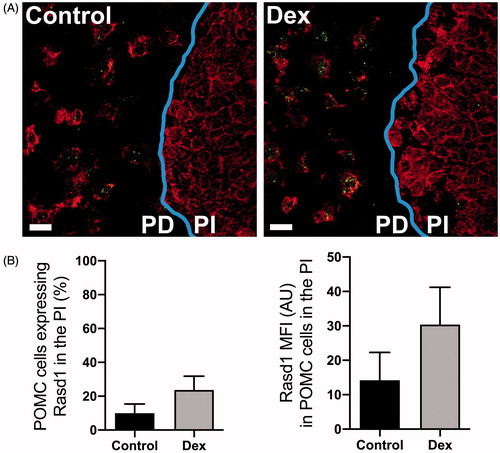
Only POMC expressing cells were identified in the PI and only a small portion of cells contained detectable Rasd1 mRNA (15–30%; ). There were no differences between control and Dex-treated groups in regards to percentage of PI corticotrophs expressing Rasd1 (t(8)=1.41, p = 0.20). Similarly, the Rasd1 MFI relative expression levels were relatively low in corticotrophs of the PI and was not significantly increased following Dex treatment (t(8)=1.14, p = 0.29).
Figure 4. Dexamethasone treatment does not increase Rasd1 expression in POMC cells of the pars intermedia of the pituitary. (A) Representative photomicrographs of dual ISH for POMC and Rasd1 at the border of the pars distalis (PD) and the pars intermedia (PI) of the pituitary of mice treated with vehicle (Control) or dexamethasone (DEX), scale bar = 20 μm. (B) Histograms depicting the mean fluorescent intensity (MFI) of Rasd1 expression in POMC cells of the pars intermedia (PI). Data presented as the mean ± SEM.
Discussion
The major finding of this study is that Rasd1 is expressed in all hormone producing cells in the PD. The findings support our hypothesis that Rasd1 is expressed in PD corticotrophs and furthermore, its expression is increased in these cells in response to Dex. Previous studies have demonstrated that Rasd1 is highly expressed in the pituitary and that Dex treatment can increase Rasd1 expression in this tissue (Brogan et al., Citation2001; Kemppainen et al., Citation2003). However, the specific endocrine pituitary cell type(s) expressing Rasd1 under resting conditions and following Dex treatment have not been determined. The current findings demonstrate that approximately 50% of mouse PD corticotrophs express Rasd1 under basal, non-stressed conditions. However, the proportion of PD corticotrophs showing Rasd1 expression increases to over 90% 1-h after Dex treatment. Somewhat surprisingly, Dex treatment induces Rasd1 in 3 of 4 other PD cell types.
Our results localized Rasd1 to all PD endocrine cell types and, with the exception of thyrotrophs, Dex treatment resulted in a rapid induction in the gene’s expression. There was however, a large difference in the intensity of Dex-induced increase in Rasd1 expression as measured using MFI. The gene was markedly responsive in corticotrophs as compared with all other cell types examined. The proportion and degree to which Dex-induces Rasd1 roughly correlates to the relative expression of glucocorticoid receptors (GR; NR3C1) in endocrine pituitary cell subtypes. Dex has a high avidity and affinity to bind to GR in the pituitary (De Kloet et al., Citation1975; McEwen, Citation1979). Prior co-localization studies have reported variable degrees of GR localization to different cells types in the PD (Kononen et al., Citation1993; Ozawa et al., Citation1999). GR was co-localized to corticotrophs and somatotrophs to a high extent (>70%) in both reports along with significant co-localization (>50%) to thyrotrophs. However, divergent results were found concerning GR in gonadotrophs and lactotrophs. It should also be noted that the single timepoint and dose of Dex used in the current work were specifically chosen to be consistent with our previous studies of Rasd1 and for maximum corticotroph activation. Further work is needed to determine Rasd1’s specific regulation in the other endocrine cells of the pituitary.
The finding that Rasd1 is expressed in all PD endocrine cell types, together with its induction in response to a glucocorticoid raises the question as to whether Rasd1 plays a role in mediating glucocorticoid effects on hormone secretion or synthesis in these cells. It has been well documented that glucocorticoids influence hormone secretion, both positively and negatively, from endocrine cells in the PD. Glucocorticoids have been shown to inhibit GnRH-stimulated secretion of LH (Rivier & Rivest, Citation1991; Suter et al., Citation1988) but both positive and negative effects of these steroids have been reported related to secretion of GH (Vakili & Cattini, Citation2012). In this latter report, Rasd1 was implicated as a potential mediator of the glucocorticoid stimulatory effect on GH secretion. GH and PRL cells are functionally interconvertible and lacto-somatotrophs that store and release GH and PRL have been regarded as an intermediate stage with the capacity to phenotypically switch between mature cell types without cell division; termed transdifferentiation (Frawley & Boockfor, Citation1991). Neonatal Dex treatment increases the proportion of somatotrophs to lactotrophs. It is not known if Rasd1 plays a role in lacto-somatotroph transdifferentiation (Manojlovic-Stojanoski et al., Citation2007). Suppression of TSH secretion by glucocorticoids is well documented (Pamenter & Hedge, Citation1980). This phenomenon may be mediated through glucocorticoid effect on hypothalamic thyrotropin-releasing hormone (TRH) release (Brabant et al., Citation1987). This view is supported by the finding that TRH-immunoreactive neurons in the paraventricular nucleus contain GR (Cintra et al., Citation1990). Dex treatment causes no consistent change in pituitary levels of TSH-alpha and beta mRNA (Ahlquist et al., Citation1989; Gurr et al., Citation1986). Glucocorticoids may also act directly on the TSH cells of the pituitary by changing their sensitivity to TRH (Pamenter & Hedge, Citation1980). Dex treatment has been shown to blunt TRH stimulation of TSH secretion in vivo, however, the change in TRH responsivity does not require tyrosine phosphorylation or change in TSH mRNA levels (John et al., Citation2003). These data indicate that extra pituitary activities are involved in the mechanism underlying the reduced TSH response to TRH induced by glucocorticoid treatment (Coiro et al., Citation2000; Schwinn et al., Citation1976). Our study found that while Rasd1 is expressed in a high percentage of thyrotrophs under basal conditions, Dex treatment did not result in a significant increase in its expression in these cells.
Rasd1 has been shown to influence cell signaling and signal transduction at multiple sites and pathways, thus supporting the notion that this protein may mediate Dex effects on pituitary hormones (Blumer et al., Citation2005; Thapliyal et al., Citation2014). Rasd1 was identified as a receptor independent activator of G-protein signaling (AGS1) in a functional screen involving the yeast pheromone pathway that facilitated GTP exchange on heterotrimeric Gα (Cismowski et al., Citation1999). Examples of findings from other studies demonstrated that Rasd1 acts as an inhibitor of adenylyl cyclase (AC) activity (Graham et al., Citation2004; Harrison & He, Citation2011; Nguyen & Watts, Citation2005, Citation2006); activates transient receptor potential canonical 4 (TRPC4) ion channels (Wie et al., Citation2015), and inhibits protein kinase C delta activity (Nguyen & Watts, Citation2006). The multiple potential effects of Rasd1 on intracellular signaling therefore support its role in regulation of PD hormone synthesis and/or secretion.
Approximately half of corticotrophs in the PD expressed Rasd1 under non-stressed conditions. In Dex-treated animals, over 90% of corticotrophs of the PD were identified as Rasd1 positive. This supports Rasd1’s potential role in regulation of corticotroph secretion of ACTH. Mice lacking Rasd1 present with higher plasma ACTH concentrations (Seok et al., Citation2020), and a mutation in Rasd1 has been identified in patients with Cushing’s disease (Uzilov et al., Citation2017). In contrast, Rasd1 was also expressed in a relatively small percentage of POMC-expressing corticotrophs of the PI. In addition, Dex treatment did not induce expression of the Rasd1 in these cells, which is not surprising given that the PI is one of the few tissues that is devoid (or has very low levels) of GR expression. Thus, our data indicate that while Rasd1 is strongly expressed in POMC-expressing corticotrophs of the PD, its expression does not extend to corticotrophs of the PI. In addition, Rasd1 does not appear to uniformly coexist in cells expressing GR, as we did not identify Rasd1 in the pars nervosa, a tissue previously shown to express this nuclear receptor (Antakly & Eisen, Citation1984; Rees et al., Citation1977).
The fact that Rasd1 is expressed in all endocrine cells of the PD also raises the question as to whether other hormones, besides glucocorticoids, might influence its expression. In a previous study from our lab, Rasd1 in whole pituitaries collected from mice was rapidly induced in response to treatment with corticosterone, Dex and estradiol (E2), but not in response to injections of testosterone, progesterone or aldosterone (Brogan et al., Citation2001). This finding has been more recently confirmed in rat pituitary cells in culture, where treatment with E2 increased Rasd1 expression as well (Wang et al., Citation2017). Further work reported in the latter study suggested that lactotrophs or somatotrophs may be the cell(s) responsible for Rasd1 induction, as Rasd1 expression was similarly induced in cultured mammosomatotropes GH4C1 cells. It would be interesting to determine if Rasd1 in the PD was responsive to thyroid hormones. Rasd1 shows a close structural homology to another Ras family GTPase Rhes (Ras homolog enriched in striatum), with both proteins containing an extended C-terminus along with transcriptional induction in response to hormones (Falk et al., Citation1999; Napolitano et al., Citation2018). Rhes has a more restricted tissue distribution than Rasd1 (Rhes is enriched in corpus striatum) and its expression is responsive to thyroid hormones. Expression of Rhes in rat striatum was not however affected by Dex treatment (Falk et al., Citation1999).
In summary, the gene for the small GTPase Rasd1 is expressed in all endocrine cell types in the mouse PD. Furthermore, with the exception of thyrotrophs, the gene is rapidly induced in endocrine pituitary cells of the PD in response to the glucocorticoid Dex. Rasd1 expression was relatively low in the corticotrophs of the PI and was not increased in Dex-treated animals. As anticipated, Rasd1 is most profoundly expressed in PD POMC-producing corticotrophs. The fact that Rasd1 is expressed widely in the PD renders it a possible mediator of glucocorticoid action on these cells.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Chad D. Foradori
CF and RK conceived the study. CF, LM, and C-CH performed the study and the laboratory analysis. CF and RK wrote the first draft. All authors contributed to the revisions.
Laci Mackay
CF and RK conceived the study. CF, LM, and C-CH performed the study and the laboratory analysis. CF and RK wrote the first draft. All authors contributed to the revisions.
Chen-Che J. Huang
CF and RK conceived the study. CF, LM, and C-CH performed the study and the laboratory analysis. CF and RK wrote the first draft. All authors contributed to the revisions.
Robert J. Kemppainen
CF and RK conceived the study. CF, LM, and C-CH performed the study and the laboratory analysis. CF and RK wrote the first draft. All authors contributed to the revisions.
References
- Ahlquist, J. A., Franklyn, J. A., Ramsden, D. B., & Sheppard, M. C. (1989). The influence of dexamethasone on serum thyrotrophin and thyrotrophin synthesis in the rat. Molecular and Cellular Endocrinology, 64(1), 55–61. https://doi.org/10.1016/0303-7207(89)90064-6
- Antakly, T., & Eisen, H. J. (1984). Immunocytochemical localization of glucocorticoid receptor in target cells. Endocrinology, 115(5), 1984–1989. https://doi.org/10.1210/endo-115-5-1984
- Blumer, J. B., Cismowski, M. J., Sato, M., & Lanier, S. M. (2005). AGS proteins: receptor-independent activators of G-protein signaling. Trends in Pharmacological Sciences, 26(9), 470–476. https://doi.org/10.1016/j.tips.2005.07.003
- Brabant, G., Brabant, A., Ranft, U., Ocran, K., Kohrle, J., Hesch, R. D., & von Zur Muhlen, A. (1987). Circadian and pulsatile thyrotropin secretion in euthyroid man under the influence of thyroid hormone and glucocorticoid administration. The Journal of Clinical Endocrinology and Metabolism, 65(1), 83–88. https://doi.org/10.1210/jcem-65-1-83
- Brogan, M. D., Behrend, E. N., & Kemppainen, R. J. (2001). Regulation of Dexras1 expression by endogenous steroids. Neuroendocrinology, 74(4), 244–250. https://doi.org/10.1159/000054691
- Cha, J. Y., Kim, H. J., Yu, J. H., Xu, J., Kim, D., Paul, B. D., Choi, H., Kim, S., Lee, Y. J., Ho, G. P., Rao, F., Snyder, S. H., & Kim, J. W. (2013). Dexras1 mediates glucocorticoid-associated adipogenesis and diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America, 110(51), 20575–20580. https://doi.org/10.1073/pnas.1320454110
- Chan, S., Filezac de L'Etang, A., Rangell, L., Caplazi, P., Lowe, J. B., & Romeo, V. (2018). A method for manual and automated multiplex RNAscope in situ hybridization and immunocytochemistry on cytospin samples. PLoS One, 13(11), e0207619. https://doi.org/10.1371/journal.pone.0207619
- Cheah, J. H., Kim, S. F., Hester, L. D., Clancy, K. W., Patterson, S. E., 3rd, Papadopoulos, V., & Snyder, S. H. (2006). NMDA receptor-nitric oxide transmission mediates neuronal iron homeostasis via the GTPase Dexras1. Neuron, 51(4), 431–440. https://doi.org/10.1016/j.neuron.2006.07.011
- Chrousos, G. P. (2009). Stress and disorders of the stress system. Nature Reviews. Endocrinology, 5(7), 374–381. https://doi.org/10.1038/nrendo.2009.106
- Cintra, A., Fuxe, K., Wikstrom, A. C., Visser, T., & Gustafsson, J. A. (1990). Evidence for thyrotropin-releasing hormone and glucocorticoid receptor-immunoreactive neurons in various preoptic and hypothalamic nuclei of the male rat. Brain Research, 506(1), 139–144. https://doi.org/10.1016/0006-8993(90)91210-8
- Cismowski, M. J., Takesono, A., Ma, C., Lizano, J. S., Xie, X., Fuernkranz, H., Lanier, S. M., & Duzic, E. (1999). Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nature Biotechnology, 17(9), 878–883. https://doi.org/10.1038/12867
- Coiro, V., Volpi, R., Cataldo, S., Capretti, L., Caffarri, G., Pilla, S., & Chiodera, P. (2000). Dopaminergic and cholinergic involvement in the inhibitory effect of dexamethasone on the TSH response to TRH. Journal of Investigative Medicine : The Official Publication of the American Federation for Clinical Research, 48(2), 133–136.
- Dayanithi, G., & Antoni, F. A. (1989). Rapid as well as delayed inhibitory effects of glucocorticoid hormones on pituitary adrenocorticotropic hormone release are mediated by type II glucocorticoid receptors and require newly synthesized messenger ribonucleic acid as well as protein. Endocrinology, 125(1), 308–313. https://doi.org/10.1210/endo-125-1-308
- De Kloet, R., Wallach, G., & McEwen, B. S. (1975). Differences in corticosterone and dexamethasone binding to rat brain and pituitary. Endocrinology, 96(3), 598–609. https://doi.org/10.1210/endo-96-3-598
- Falk, J. D., Vargiu, P., Foye, P. E., Usui, H., Perez, J., Danielson, P. E., Lerner, D. L., Bernal, J., & Sutcliffe, J. G. (1999). Rhes: A striatal-specific Ras homolog related to Dexras1. Journal of Neuroscience Research, 57(6), 782–788. https://doi.org/10.1002/(SICI)1097-4547(19990915)57:6<782::AID-JNR3>3.0.CO;2-9
- Frawley, L. S., & Boockfor, F. R. (1991). Mammosomatotropes: Presence and functions in normal and neoplastic pituitary tissue. Endocrine Reviews, 12(4), 337–355. https://doi.org/10.1210/edrv-12-4-337
- Graham, T. E., Key, T. A., Kilpatrick, K., & Dorin, R. I. (2001). Dexras1/AGS-1, a steroid hormone-induced guanosine triphosphate-binding protein, inhibits 3',5'-cyclic adenosine monophosphate-stimulated secretion in AtT-20 corticotroph cells. Endocrinology, 142(6), 2631–2640. https://doi.org/10.1210/endo.142.6.8209
- Graham, T. E., Qiao, Z., & Dorin, R. I. (2004). Dexras1 inhibits adenylyl cyclase. Biochemical and Biophysical Research Communications, 316(2), 307–312. https://doi.org/10.1016/j.bbrc.2004.02.049
- Gurr, J. A., Vrontakis, M. E., Athanasian, E. A., Wagner, C. R., & Kourides, I. A. (1986). Hormonal regulation of thyrotropin alpha and beta subunit mRNAs. Hormone and Metabolic Research = Hormon- und Stoffwechselforschung = Hormones et metabolisme, 18(6), 382–385. https://doi.org/10.1055/s-2007-1012322
- Harrison, L. M., & He, Y. (2011). Rhes and AGS1/Dexras1 affect signaling by dopamine D1 receptors through adenylyl cyclase. Journal of Neuroscience Research, 89(6), 874–882. https://doi.org/10.1002/jnr.22604
- Herman, J. P., McKlveen, J. M., Ghosal, S., Kopp, B., Wulsin, A., Makinson, R., Scheimann, J., & Myers, B. (2016). Regulation of the hypothalamic-pituitary-adrenocortical stress response. Comprehensive Physiology, 6(2), 603–621. https://doi.org/10.1002/cphy.c150015
- John, C. D., Christian, H. C., Morris, J. F., Flower, R. J., Solito, E., & Buckingham, J. C. (2003). Kinase-dependent regulation of the secretion of thyrotrophin and luteinizing hormone by glucocorticoids and annexin 1 peptides. Journal of Neuroendocrinology, 15(10), 946–957. https://doi.org/10.1046/j.1365-2826.2003.01081.x
- Kemppainen, R. J., Cox, E., Behrend, E. N., Brogan, M. D., & Ammons, J. M. (2003). Identification of a glucocorticoid response element in the 3'-flanking region of the human Dexras1 gene. Biochimica Et Biophysica Acta, 1627(2-3), 85–89. https://doi.org/10.1016/s0167-4781(03)00079-4
- Kemppainen, R. J., & Behrend, E. N. (1998). Dexamethasone rapidly induces a novel ras superfamily member-related gene in AtT-20 cells. The Journal of Biological Chemistry, 273(6), 3129–3131. https://doi.org/10.1074/jbc.273.6.3129
- Kim, H. J., Cha, J. Y., Seok, J. W., Choi, Y., Yoon, B. K., Choi, H., Yu, J. H., Song, S. J., Kim, A., Lee, H., Kim, D., Han, J. Y., & J. W. Kim. (2016). Dexras1 links glucocorticoids to insulin-like growth factor-1 signaling in adipogenesis. Scientific Reports, 6, 28648. https://doi.org/10.1038/srep28648
- Kim, H. R., Cho, K. S., Kim, E., Lee, O. H., Yoon, H., Lee, S., Moon, S., Park, M., Hong, K., Na, Y., Shin, J. E., Kwon, H., Song, H., Choi, D. H., & Choi, Y. (2017). Rapid expression of RASD1 is regulated by estrogen receptor-dependent intracellular signaling pathway in the mouse uterus. Molecular and Cellular Endocrinology, 446, 32–39. https://doi.org/10.1016/j.mce.2017.02.006
- Kononen, J., Honkaniemi, J., Gustafsson, J. A., & Pelto-Huikko, M. (1993). Glucocorticoid receptor colocalization with pituitary hormones in the rat pituitary gland. Molecular and Cellular Endocrinology, 93(1), 97–103. https://doi.org/10.1016/0303-7207(93)90144-9
- Lian, Y., Yan, C., Lian, Y., Yang, R., Chen, Q., Ma, D., Lian, W., Liu, J., Luo, C., Ren, J., & Xu, H. (2020). Long intergenic non-protein-coding RNA 01446 facilitates the proliferation and metastasis of gastric cancer cells through interacting with the histone lysine-specific demethylase LSD1. Cell Death & Disease, 11(7), 522. https://doi.org/10.1038/s41419-020-2729-0
- Manojlovic-Stojanoski, M., Nestorovic, N., Negic, N., Filipovic, B., Sosic-Jurjevic, B., Sekulic, M., & Milosevic, V. (2007). Influence of maternal dexamethasone treatment on morphometric characteristics of pituitary GH cells and body weight in near-term rat fetuses. Folia Histochem Cytobiol, 45, 51–56.
- McEwen, B. S. (1979). Influences of adrenocortical hormones on pituitary and brain function. Monographs on Endocrinology, 12, 467–492. https://doi.org/10.1007/978-3-642-81265-1_25
- Munagala, R., Aqil, F., Vadhanam, M. V., & Gupta, R. C. (2013). MicroRNA ‘signature’ during estrogen-mediated mammary carcinogenesis and its reversal by ellagic acid intervention. Cancer Letters, 339(2), 175–184. https://doi.org/10.1016/j.canlet.2013.06.012
- Napolitano, F., D'Angelo, L., de Girolamo, P., Avallone, L., de Lange, P., & Usiello, A. (2018). The thyroid hormone-target gene Rhes a novel crossroad for neurological and psychiatric disorders: New insights from animal models. Neuroscience, 384, 419–428. https://doi.org/10.1016/j.neuroscience.2018.05.027
- Nguyen, C. H., & Watts, V. J. (2005). Dexras1 blocks receptor-mediated heterologous sensitization of adenylyl cyclase 1. Biochemical and Biophysical Research Communications, 332(3), 913–920. https://doi.org/10.1016/j.bbrc.2005.05.041
- Nguyen, C. H., & Watts, V. J. (2006). Dexamethasone-induced Ras protein 1 negatively regulates protein kinase C delta: implications for adenylyl cyclase 2 signaling. Molecular Pharmacology, 69(5), 1763–1771. https://doi.org/10.1124/mol.105.019133
- O'Neill, D., Jones, D., Wade, M., Grey, J., Nakjang, S., Guo, W., Cork, D., Davies, B. R., Wedge, S. R., Robson, C. N., & Gaughan, L. (2015). Development and exploitation of a novel mutant androgen receptor modelling strategy to identify new targets for advanced prostate cancer therapy. Oncotarget, 6(28), 26029–26040. https://doi.org/10.18632/oncotarget.4347
- Ozawa, H., Ito, T., Ochiai, I., & Kawata, M. (1999). Cellular localization and distribution of glucocorticoid receptor immunoreactivity and the expression of glucocorticoid receptor messenger RNA in rat pituitary gland. A combined double immunohistochemistry study and in situ hybridization histochemical analysis. Cell and Tissue Research, 295(2), 207–214. https://doi.org/10.1007/s004410051226
- Pamenter, R. W., & Hedge, G. A. (1980). Inhibition of thyrotropin secretion by physiological levels of corticosterone. Endocrinology, 106(1), 162–166. https://doi.org/10.1210/endo-106-1-162
- Rees, H. D., Stumpf, W. E., Sar, M., & Petrusz, P. (1977). Autoradiographic studies of 3H-dexamethasone uptake by immunocytochemically characterized cells of the rat pituitary. Cell and Tissue Research, 182(3), 347–356. https://doi.org/10.1007/BF00219770
- Rivier, C., & Rivest, S. (1991). Effect of stress on the activity of the hypothalamic-pituitary-gonadal axis: peripheral and central mechanisms. Biology of Reproduction, 45(4), 523–532. https://doi.org/10.1095/biolreprod45.4.523
- Schwinn, G., von Zur Muhlen, A., & Warnecke, U. (1976). Effects of dexamethasone on thyrotrophin and prolactin plasma levels in rats. Acta Endocrinologica, 82(3), 486–491. https://doi.org/10.1530/acta.0.0820486
- Seok, J. W., Kim, D., Yoon, B. K., Lee, Y., Kim, H. J., Hwang, N., Fang, S., Kim, H. J., & Kim, J. W. (2020). Dexras1 plays a pivotal role in maintaining the equilibrium between adipogenesis and osteogenesis. Metabolism, 108, 154250. https://doi.org/10.1016/j.metabol.2020.154250
- Shipston, M. J. (1995). Mechanism(s) of early glucocorticoid inhibition of adrenocorticotropin secretion from anterior pituitary corticotropes. Trends in Endocrinology and Metabolism, 6(8), 261–266. https://doi.org/10.1016/1043-2760(95)00149-2
- Suter, D. E., Schwartz, N. B., & Ringstrom, S. J. (1988). Dual role of glucocorticoids in regulation of pituitary content and secretion of gonadotropins. The American Journal of Physiology, 254(5 Pt 1), E595–E600. https://doi.org/10.1152/ajpendo.1988.254.5.E595
- Takahashi, H., Umeda, N., Tsutsumi, Y., Fukumura, R., Ohkaze, H., Sujino, M., van der Horst, G., Yasui, A., Inouye, S. T., Fujimori, A., Ohhata, T., Araki, R., & Abe, M. (2003). Mouse dexamethasone-induced RAS protein 1 gene is expressed in a circadian rhythmic manner in the suprachiasmatic nucleus. Molecular Brain Research, 110(1), 1–6. https://doi.org/10.1016/S0169-328X(02)00543-0
- Thapliyal, A., Verma, R., & Kumar, N. (2014). Small G proteins Dexras1 and RHES and their role in pathophysiological processes. International Journal of Cell Biology, 2014, 308535. https://doi.org/10.1155/2014/308535
- Tian, J., Wang, Y., Zhang, X., Ren, Q., Li, R., Huang, Y., Lu, H., & Chen, J. (2017). Calycosin inhibits the in vitro and in vivo growth of breast cancer cells through WDR7-7-GPR30 Signaling. Journal of Experimental & Clinical Cancer Research : CR, 36(1), 153. https://doi.org/10.1186/s13046-017-0625-y
- Uzilov, A. V., Cheesman, K. C., Fink, M. Y., Newman, L. C., Pandya, C., Lalazar, Y., Hefti, M., Fowkes, M., Deikus, G., Lau, C. Y., Moe, A. S., Kinoshita, Y., Kasai, Y., Zweig, M., Gupta, A., Starcevic, D., Mahajan, M., Schadt, E. E., Post, K. D., & Geer, M. J. (2017). Identification of a novel RASD1 somatic mutation in a USP8-mutated corticotroph adenoma. Molecular Case Studies, 3(3), a001602. https://doi.org/10.1101/mcs.a001602
- Vakili, H., & Cattini, P. A. (2012). The hidden but positive role for glucocorticoids in the regulation of growth hormone-producing cells. Molecular and Cellular Endocrinology, 363(1–2), 1–9. https://doi.org/10.1016/j.mce.2012.08.001
- Wang, F., Flanagan, J., Su, N., Wang, L. C., Bui, S., Nielson, A., Wu, X., Vo, H. T., Ma, X. J., & Luo, Y. (2012). RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. The Journal of Molecular Diagnostics : JMD, 14(1), 22–29. https://doi.org/10.1016/j.jmoldx.2011.08.002
- Wang, H., Wang, M. X., Su, N., Wang, L. C., Wu, X., Bui, S., Nielsen, A., Vo, H. T., Nguyen, N., Luo, Y., & Ma, X. J. (2014). RNAscope for in situ detection of transcriptionally active human papillomavirus in head and neck squamous cell carcinoma. Journal of Visualized Experiments, (85):51426.
- Wang, L., Mitsui, T., Ishida, M., Izawa, M., & Arita, J. (2017). Rasd1 is an estrogen-responsive immediate early gene and modulates expression of late genes in rat anterior pituitary cells. Endocrine Journal, 64(11), 1063–1071. https://doi.org/10.1507/endocrj.EJ17-0148
- Wie, J., Kim, J., Ha, K., Zhang, Y. H., Jeon, J. H., & So, I. (2015). Dexamethasone activates transient receptor potential canonical 4 (TRPC4) channels via Rasd1 small GTPase pathway. Pflugers Archiv : European Journal of Physiology, 467(10), 2081–2091. https://doi.org/10.1007/s00424-014-1666-0
