Abstract
In the past decade a remarkable rebirth of serum/plasma lipoprotein(a) (Lp(a)) as an independent risk factor of cardiovascular disease (CVD) occurred. Updated evidence for a causal continuous association in different ethnic groups between Lp(a) concentrations and cardiovascular outcomes has been published in the latest European Atherosclerosis Society (EAS) Lp(a) consensus statement. Interest in measuring Lp(a) at least once in a person’s lifetime moreover originates from the development of promising new Lp(a) lowering drugs. Accurate and clinically effective Lp(a) tests are of key importance for the timely detection of high-risk individuals and for future evaluation of the therapeutic effects of Lp(a) lowering medication. To this end, it is necessary to improve the performance and standardization of existing Lp(a) tests, as is also noted in the Lp(a) consensus statement. Consequently, a state-of-the-art internationally endorsed reference measurement system (RMS) must be in place that allows for performance evaluation of Lp(a) field tests in order to certify their validity and accuracy. An ELISA-based RMS from Northwest Lipid Research Laboratory (University of Washington, Seattle, USA) has been available since the 1990s. A next-generation apo(a)/Lp(a) RMS is now being developed by a working group from the International Federation of Clinical Chemistry and Laboratory Medicine (IFCC). The envisioned apo(a) RMS is based on the direct measurement of selected proteotypic fragments generated after proteolytic digestion using quantitative protein mass spectrometry (MS). The choice for an MS-based RMS enables selective measurement of the proteotypic peptides and is by design apo(a) isoform insensitive. Clearly, the equimolar conversion of apo(a) into the surrogate peptide measurands is required to obtain accurate Lp(a) results. The completeness of proteolysis under reaction conditions from the candidate reference measurement procedure (RMP) has been demonstrated for the quantifying apo(a) peptides. Currently, the candidate apo(a) RMP is endorsed by the IFCC and recommendations for suitable secondary reference materials have been made in a recent commutability study paper. Ongoing efforts toward a complete apo(a) RMS that is listed by the Joint Committee on Traceability in Laboratory Medicine (JCTLM) are focused on the peptide-based calibration and the establishment of a network of calibration laboratories running the apo(a) RMS in a harmonized way. Once completed, it will be the holy grail for evaluation and certification of Lp(a) field methods.
1. Introduction
The pilgrimage to selective and accurate quantification of lipoprotein(a) (Lp(a)) finds its origin more than half a century ago. It started with the first description of Lp(a) in 1963 by Kåre Berg and coworkers, which boosted research interest in the elusive Lp(a) particle until the 1990s [Citation1]. Yet, a major setback was caused by three negative Ridker studies in 1993, 1995, and 2001. In the Physician Health Study, more than 15,000 physicians aged between 40 and 84 years without a history of myocardial infarction (MI), stroke, or peripheral arterial disease were monitored, and almost 300 individuals experienced cardiovascular events during the study. The median plasma Lp(a) levels from patients and controls were similar (103.0 mg/L vs. 102.5 mg/L), and it was concluded that Lp(a) should no longer be considered a useful cardiovascular disease (CVD) biomarker [Citation2–4]. However, in 2004, Rifai and coworkers demonstrated that the Ridker studies were flawed by the immunoassay-based Lp(a) test used that was inaccurate due to apo(a) size polymorphisms [Citation2–5]. Test result inaccuracy was demonstrated using the International Federation of Clinical Chemistry and Laboratory Medicine (IFCC)-WHO reference material (RM) SRM2B and the internationally endorsed enzyme-linked immunosorbent assay (ELISA)-based reference measurement procedure (RMP) from Marcovina [Citation6]. Reproduction of the Physician Health Study revealed that the association with CVD indeed had been masked by inaccurate Lp(a) test results [Citation7]. The renewed interest in Lp(a) further increased in 2009 when genetic evidence for its causal association with CVD was presented in Mendelian randomized trials [Citation8]. High levels of Lp(a) were associated with an increased risk of MI, stroke, and peripheral arterial disease irrespective of other traditional risk factors like low-density lipoprotein (LDL) [Citation9]. From this, the European Atherosclerosis Society (EAS)/European Society of Cardiology (ESC) guideline on the management of dyslipidemias advocated that Lp(a)-corrected LDL cholesterol (LDL-c) should be assessed at least once in patients with suspected or known high Lp(a). Recently, the 2022 expert EAS consensus on Lp(a) recommends independent measurement of Lp(a) on top of the serum lipid profile in all patients at least once in a lifetime [Citation10].
In this review, the key features of properly defining the “quantity intended to be measured” (i.e. the measurand) at the top of the metrological traceability chain and the necessity of developing apo(a) kringle-independent higher order RMPs to evaluate fitness-for-purpose of contemporary Lp(a) in vitro diagnostic (IVD) kits will be discussed. Over the past 5–10 years, several guidelines and consensus statements, particularly in Northern America, Europe, and Asia, have adopted recommendations to quantify Lp(a) levels in all adults at least once in their lifetime [Citation7–13]. In case there is a family history of cardiac events or CVD risk, it is recommended to determine Lp(a) also in youth. Importantly, the recent EAS Lp(a) consensus statement supports a strong causal association between Lp(a) levels and the risk of developing CVD, which together with the establishment of promising Lp(a) lowering medication such as PCSK9 inhibitors as well as anti-sense oligonucleotides currently under evaluation in phase II and III clinical trials, have led to a renaissance of Lp(a) testing [Citation10]. A summary of the currently available guidelines and consensus statements, including their recommendations for Lp(a) testing, is outlined in .
Table 1. Overview of clinical guidelines and consensus papers and their decision limits and recommendations for Lp(a) testing.
In , relevant milestone publications that reflect on the inter-dependent clinical and analytical performance challenges are mentioned.
Figure 1. Schematic representation of relevant manuscripts reports on the inter-dependencies between analytical and clinical performance during the exploration of the clinical value of Lp(a) as a CVD biomarker.
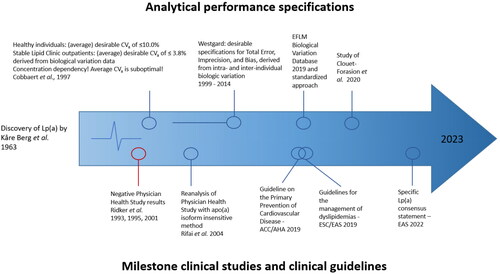
Starting in the early 2000s, evidence for a strong causal association between Lp(a) levels and the risk of developing CVD was reestablished, specifically with MI and aortic valve stenosis [Citation14–17]. In several studies, the role of Lp(a) in cardiovascular-related morbidity and mortality was reported [Citation10,Citation16–18]. Consequently, a new focus on Lp(a) as an additional biomarker of CVD appeared independent of the traditional risk factors such as LDL [Citation19]. As clinical performance and clinical effectiveness of Lp(a) tests are determined by the analytical performance of Lp(a) tests, it is of utmost importance that the Lp(a) tests used in clinical trials are fit-for-clinical-purpose. An aspect of the evaluation of fitness for purpose of Lp(a) field methods is the significant amount of information that can be obtained by the generation and application of data on intra- and inter-individual biological variation. In order to determine desirable analytical goals for Lp(a) testing, Cobbaert and coworkers reported previously on a comprehensive biological variability study in a healthy cohort of Caucasians as compared to stable outpatients from the Lipid Clinic with elevated Lp(a)>300 mg/L [Citation19]. These researchers found an inverse relationship between both biological and analytical coefficients of variation (CVs) and serum Lp(a) concentrations, signifying that desirable analytical performance goals are concentration dependent. This means that average intra-individual biological CVs and analytical performance goals are inadequate for Lp(a) with its 1000-fold concentration differences [Citation20]. Nevertheless, other researchers kept proposing average and conflicting intra-individual biological CVs and analytical performance goals. A relevant conceptual framework for determining analytical performance specifications (APS) from biological variation data was published by the European Federation of Clinical Chemistry and Laboratory Medicine (EFLM) as a second-best approach when clinical outcome data are not available [Citation21,Citation22]. In 2014, Westgard published an updated dataset for APS followed by Ricos and coworkers with a dataset based on EFLM’s standardized approach [Citation23,Citation24]. Clouet-Foraison and Marcovina defined APS for Lp(a) using the fully Biological Variation Data Critical Appraisal Checklist (BIVAC)-compliant protocol. Their data confirmed biological variation estimates of Lp(a) listed in the EFLM database and in reference [Citation19]. They also reinforced concerns regarding the suitability of older APS recommendations for Lp(a) measurements. Given the heterogeneity of Lp(a), more BIVAC-compliant studies on large numbers of individuals of different ethnic groups are desirable.
The EAS recommendation from 2022 to measure Lp(a) in all individuals has consequences for the IVD industry, laboratory specialists, and clinicians, as they have to manufacture, implement, order, interpret, and act on elevated Lp(a) measurement results. The management of patients with raised Lp(a) levels so far includes: (1) reducing overall atherosclerotic risk, (2) controlling dyslipidemia with a desirable non-HDL-cholesterol level of <100 mg/dl (2.5 mmol/l), and (3) consideration of lipoprotein apheresis. Currently, a multitude of immunoassay-based Lp(a) tests are commercially available with between-test result variations as large as 2-fold and inter-method CVs between 16% and 32% [Citation25]. In order to guarantee the accuracy of Lp(a) test results within allowable limits of uncertainty, all elements of the metrological traceability chain have to be in place. A proper calibration hierarchy model according to the International Organization for Standardization (ISO) 17511:2020 forms the basis for anchoring test results from field methods to endorsed RMs and RMPs of higher order. Ideally, a network of calibration laboratories is operational to implement global standardization of internationally endorsed reference measurement systems (RMSs) via certification programs for IVD manufacturers. The conceptual approach for establishing a state-of-the-art RMS for Lp(a) (and other apolipoproteins) has been described recently [Citation26]. Given the large variation in measurement results obtained for Lp(a) when using various tests from different suppliers, implementation of standardization seems of utmost importance [Citation25]. Not only would standardization allow for universal interpretation of measurement results by clinicians, potentially resulting in the delivery of more consistent clinical care, but it would also simultaneously enable meta-analysis of epidemiological research and pharmaceutical trials. In both applications, test trueness and comparability are crucial [Citation27]. In the 1990s, an IFCC-WHO ELISA-based RMS was developed that has been operational for certifying Lp(a) kits from IVD manufacturers and has been the “gold standard” RMP until recently [Citation28,Citation29]. Here the value assignments of Lp(a) were reported in molar concentrations, whereas most commercially available tests report Lp(a) mass concentrations. When reporting molar concentrations, it is assumed that each Lp(a) particle carries one molecule apolipoprotein(a) (apo(a)). Notwithstanding the claimed metrological traceability of IVD manufacturers to the endorsed Lp(a) RM and RMS, Lp(a) mass tests vary between labs from 16.4% to 32.1% at Lp(a) levels of ∼150 to 450 mg/L [Citation25,Citation30,Citation31]. In this review, we elaborate on our hypothesis that the substantial Lp(a) inter-method variation may have a multifactorial origin. As the former RMS no longer exists, a new RMS based on mass spectrometry (MS) and traceability on internationally accepted SI-units is being developed by the IFCC working group on Apolipoproteins by Mass Spectrometry (WG-APO MS) under the auspices of the IFCC Scientific Division.
This review targets IVD manufacturers, laboratory specialists, and clinicians with an interest in the accurate measurement of Lp(a). It puts into context the inter-dependencies between clinical and analytical performance, as well as reemphasizes accuracy and selectivity as the predominant requirements for obtaining valid medical test results. The goals for laboratory medicine and IVD industry should be to understand all determinants of Lp(a) test validity, especially the molecular characterization of the quantity intended to be measured in relation to the measurement principles, technologies, and applied kit design. In addition, adequate implementation of current metrological traceability concepts based on ISO 17511:2020 calibration models; normative ISO standards 15193, 15194, and 15195 for RMs, RMPs, and calibration labs, respectively; and commutable value-assigned matrix-based RMs should be realized. In this review, the currently available Lp(a) field methods are reviewed and discussed with regard to their strengths and potential drawbacks. Determinants of Lp(a) test accuracy are summarized and recommendations for improving Lp(a) accuracy and selectivity are deduced from method comparisons and commutability studies between immunoassays (mass and molar) and MS-based molar apo(a) tests. Finally, we reflect on the impact of the next generation apo(a) RMS for manufacturers and for clinical practice.
2. Metrological traceability within allowable measurement uncertainty for accurate measurement of Lp(a)
The implementation of the metrological traceability chain provides an unbroken relationship between an end result in a medical lab and higher-order RMs, of which the concentration has been defined accurately. Traceability of results from all commercially available IVD tests to the same RMS allows comparisons in time and space, provided the measurements of the same measurand are addressed across measurement procedures [Citation32,Citation33]. In addition, a metrological traceability chain is accompanied by a predefined allowable measurement uncertainty (MU) to avoid traceability toward an “untrue” value. This section explains the concept of metrological traceability and provides insight into the importance of a molecularly defined measurand(s) and the difficulties that are encountered with regard to highly heterogenous apo(a). The definitions and terminology regarding the concept of metrological traceability are explained in detail in the International Vocabulary of Metrology (VIM), and we follow these definitions in the current review [Citation34]. The technical requirements and documentation necessary to achieve metrological traceability of medical laboratory tests are described in ISO 17511 [Citation33]. Within ISO 17511:2020, the concept of metrological traceability is materialized in six calibration hierarchies. The ultimate goal and highest level of traceability of Lp(a) tests is traceability toward a “true” value that is traceable to SI-units [Citation25,Citation35]. Metrological traceability chains of the former and future apo(a) RMS are shown in .
Figure 2. Metrological traceability chains are presented for contemporary and emerging serum apo(a) RMS consisting of an unbroken sequence of calibrators and measurement procedures that are used to relate a measurement result to a reference of higher order. Serum apo(a)/Lp(a) test results are currently traceable to WHO-IFCC secondary reference materials (left), whereas envisioned traceability to SI for serum apo(a) is presented (right). Note that the top of the traceability chain on the right side must be made complete by including the development of peptide-based certified primary reference materials for apo(a) standardization. Figure adapted from Cobbaert et al. [Citation26].
![Figure 2. Metrological traceability chains are presented for contemporary and emerging serum apo(a) RMS consisting of an unbroken sequence of calibrators and measurement procedures that are used to relate a measurement result to a reference of higher order. Serum apo(a)/Lp(a) test results are currently traceable to WHO-IFCC secondary reference materials (left), whereas envisioned traceability to SI for serum apo(a) is presented (right). Note that the top of the traceability chain on the right side must be made complete by including the development of peptide-based certified primary reference materials for apo(a) standardization. Figure adapted from Cobbaert et al. [Citation26].](/cms/asset/39971272-f554-40d9-ab68-45e6798efa92/ilab_a_2199353_f0002_c.jpg)
The amino acid standards are calibrators with verified high purity and well-defined amount of substance, better known as certified primary reference standards. The content (purity and amount) of these primary standards can be determined using a variety of state-of-the-art analytical methods, including mass balance, amino acid analysis, quantitative nuclear magnetic resonance, and elemental analysis [Citation26]. The next order of materials in the apo(a) peptide-based primary RMs, which are generally synthetically produced using regulated production processes. Peptide calibrators are extensively purified to achieve a high level of purity, typically >98%, to ensure reliable results for amino acid analysis. These RMs are created by RM producers such as the Joint Research Center (JRC), World Health Organization (WHO), National Institute of Standards and Technology (NIST), and National Institute for Biological Standards and Control (NIBSC). The values assigned to the peptide-based primary reference materials, including the corresponding uncertainties, are selected based on their suitability for the intended use in the calibration chain. The primary RM is followed by a commutable secondary RM which is matrix-based. The primary and the secondary RM are interconnected via an RMP. The development and validation of higher-order RMPs as well as their application in calibration laboratories should be in compliance with ISO guidelines 15193:2009 and 15195:2018, respectively [Citation36,Citation37]. Once an RMS is in place, IVD manufacturers can then create traceability of their Lp(a) test results to the RMS through either a protocolized certification process using native split samples that cover the Lp(a) measuring range or by purchasing proven commutable value-assigned secondary RMs to directly calibrate the Lp(a) field tests. To ensure that Lp(a) tests are fit-for-clinical-purpose, the total MU should be within the allowable MU of 24% derived from biological variation, as demonstrated in (right). The MU budget consumed by the stakeholders of the apo(a) RMS should as a rule of thumb be a maximum 1/3 to 1/2 of the total allowable MU. Multiple stakeholders are involved and shared responsibility is key for establishing and maintaining all components of the future apo(a) RMS. Regular stakeholders are metrology institutes (such as JRC, NIBSC, “Laboratoire National de Métrologie et d‘Essais” (LNE), NIST), (candidate) calibration laboratories and/or academia, IVD industry, and end users.
Besides the availability of a complete metrological traceability chain, another prerequisite for test accuracy is the unequivocal definition of the measurand [Citation38–40]. This part of the traceability chain has commonly been neglected. From a structural point-of-view, Lp(a) is an LDL particle that includes a single copy of apo(a) in addition to apolipoprotein B100 (apoB100) [Citation28]. Both apolipoproteins are covalently connected through a disulfide bond [Citation41]. Apo(a) is transcribed from the LPA gene and consists of ten kringle IV subunits (KIV1–10), a kringle V subunit, and a peptidase S1 domain [Citation42]. A critically important aspect of Lp(a) is the size polymorphism of KIV2 that is present in the LPA gene, which results in apo(a) molecules of variable length, with KIV2 repeats of 2 to >40 [Citation43]. Notably, the apo(a) size polymorphism is not the only cause of the variable composition of the Lp(a) particle. The total mass of an Lp(a) particle also depends on the lipid/protein ratio, compositions, genetic variations, and the presence of post-translational modifications (PTMs) (e.g. glycosylation) on apo(a) and apoB100 [Citation25,Citation44–46]. Hence, by definition, it is impossible to accurately express Lp(a) in total mass [Citation25].
All tests that aim to measure Lp(a) in fact target apo(a), thus defining this protein as the measurand. Notably, apo(a) may also be present in the circulation as lipid-free apo(a), [Citation47] and higher levels of lipid-free apo(a) have been reported for individuals with renal disease [Citation48]. In addition, other variants of apo(a) occur such as truncated apo(a). LPA has evolved from PLG through duplications, deletions, and conversions [Citation49,Citation50]. A frequent nonsense mutation in this LPA gene causes truncation of apo(a) [Citation51]. Apo(a) has been observed in fragmented form excreted in the urine, in atherosclerotic plaques and in the circulation [Citation52,Citation53]. In atherogenic plaques, cleavage of apo(a) by MMP9 has been reported [Citation54]. Clearly, the variation in Lp(a) composition, as well as the non-constant composition of apo(a) and the presence of free and fragmented apo(a), hampers the unequivocal definition of the measurand [Citation39]. We speculate that Lp(a) particle composition may even vary during lipid-lowering treatment regimes in a person’s lifetime. The effects thereof will be highlighted in Section 5.
Every step in the traceability chain adds a certain MU that must be limited in order to render the Lp(a) test clinically useful (as outlined in ISO 15193:2009). MU of field methods is determined by medical laboratories in accordance with ISO 20194 TS [Citation55], whereas calibration labs determine MU of the RMS according to the Guide to the Expression of Uncertainty in Measurement (GUM) [Citation37]. RMPs, as part of an RMS, should as a rule of thumb require no more than 33% to 50% of the total allowable uncertainty of the measurand [Citation26]. The APS for end users’ tests should be predefined and aligned with recommended clinical performance requirements in guidelines. A collection of criteria that quantify the clinical performance of a test to enable better patient management than current practice is commonly referred to as clinical performance specifications (CPS) [Citation56]. Lord and coworkers defined multiple steps for defining the CPS: (1) define the intended benefits, (2) map current practice, (3) propose test role, (4) link clinical performance requirements to intended benefits, and (5) set minimum acceptable clinical performance levels [Citation56]. A major question that is asked to set the CPS is “which harm-benefit compromise are you willing to make?”; it is, however, not trivial to quantitatively define CPS.
APS are key feature of every medical test because they determine whether the laboratory’s reported variation from the target value is acceptable [Citation57,Citation58]. To ensure that medical tests are fit-for-intended-purpose, a hierarchical consensus system for predefining APS was established at the first strategic EFLM conference in Milan in 2015. In this hierarchy, the first and best APS model is based on results from clinical outcome studies (randomized control trials (RCTs) for biomarkers are rarely available). The second model estimates APS from biological variation studies and the third and least best model is based on the state of the art [Citation21].
Predefining APS for apo(a) is challenging (). There are currently no large studies directly assessing the effects of Lp(a) testing on clinical outcomes. Multiple attempts have therefore been made to derive APS from biological variation data [Citation21]. Initial APS were set based on research conducted more than 25 years ago, using Lp(a) tests that no longer exist. Intra-individual variation (CVi) of 20.8% and within-group variation of 18.1% (CVG) were found. Based on these results, desirable imprecision was set at 10.4%, while the total allowable error (TEa) was set at 24.1% [Citation24]. More recently, Clouet-Foraison and Marcovina used the newly developed BIVAC to define renewed APS, see [Citation59]. The specification for imprecision was reported to be 3.3% in males and 5.3% in females. The bias (BAPS) was not calculated as the CVG for Lp(a) was not defined. We question the validity of average APS [Citation60], as the models used for the calculation of imprecision assume a constant intra-individual variation and do not consider the 1000-fold variation in Lp(a) levels and the concentration dependency with the apo(a) isoforms. Previously, Cobbaert and coworkers demonstrated in the late 1990s that the intra-individual biological CV (CVis) and analytical CV (CVas) for Lp(a) in healthy Caucasians and in stable Caucasian outpatients with hyper-Lp(a)-lipoproteinemia were variable, with CVa’s as high as 8.4% at a low Lp(a) concentration and as low as 1.3% at high Lp(a) concentrations, as shown in [Citation20]. Looking at the approach of Harris and coworkers [Citation61], which states that the maximum allowable analytical imprecision should be ≤ ½ CVi, overall their data corroborate the findings of Clouet-Foraison and coworkers. The KIV2 repeats that are associated with apo(a) make that there is inter-individual variation over the total measurement range. We conclude that APS for Lp(a) tests should be based on biological variation data in a concentration-dependent way [Citation20,Citation62]. Eventually, new BIVAC-compatible studies in specific populations that consider all determinants of accuracy for Lp(a) tests, including the 1000-fold differences in levels, are needed to carefully determine these biological variations.
Figure 3. Confidence intervals (95%) for the subject’s true value of Lp(a) as a percentage of the observed serum Lp(a) value for one, three, and five serial specimens. Figure re-used with permission from Cobbaert et al. [Citation20].
![Figure 3. Confidence intervals (95%) for the subject’s true value of Lp(a) as a percentage of the observed serum Lp(a) value for one, three, and five serial specimens. Figure re-used with permission from Cobbaert et al. [Citation20].](/cms/asset/6ece872d-2e64-4226-993f-2dcafe66d5a4/ilab_a_2199353_f0003_b.jpg)
3. Overview of field Lp(a) tests and their traceability
The various immunoassay-based Lp(a) tests that are commercially available are summarized in . Detection technologies include particle-enhanced immunoturbidimetric assays (PETIAs) [Citation63], dissociation-enhanced lanthanide fluorescence immunoassays (DEIFIAs) [Citation64], immunoradiometric assays (IRMAs), immunonephelometric assays (INAs), and ELISAs [Citation64] (). It is important to note that most high-volume tests apply light scattering methods (immunoturbidimetry or immunonephelometry), which could be affected by variation in Lp(a) particle size or even more by the presence of lipid-free or fragmented apo(a). The end-user measurement procedures summarized in are widely used and commonly referred to as “field methods”. These tests have been used since the pioneering researchers reported on Lp(a) analysis [Citation65]. As was emphasized previously, for a sound interpretation of Lp(a) levels, the nature of the actual measurand is apo(a) which permits selective and molar detection and quantitation [Citation39]. However, immunoassays that are commonly based on polyclonal antibodies inherently lack selectivity. In addition, the type and number of independent product calibrators, their concentration level and range, and their apo(s) isoform composition affect the apo(a) recovery in specimens, and moreover limit the traceability of the product calibrators to SRM2B and the WHO-IFCC ELISA-based RMP. All immunoassays start with an antibody that binds apo(a) on Lp(a) in a mixture of biomolecules (native matrix). The first quantitative method for measuring Lp(a)was based on radial immunodiffusion (RID) of precipitated complex [Citation66]. Around the same time, Albers and coworkers reported the development of a radioimmunoassay (RIA) to determine Lp(a) levels in 1000 individuals [Citation67]. Note that in both of these assays, the measurement of Lp(a) is apo(a) isoform dependent, potentially resulting in biases. The currently available commercial tests demonstrate excellent reproducibility, although these still apply polyclonal antibodies that are kringle IV2-dependent ().
Figure 4. Schematic representation of the currently applied immunoaffinity-based (and LC-MS/MS-based) methods for quantification of Lp(a) levels. Test read-out principles from top to bottom: immunonephelometry (not measured in the direct path), immunoturbidimetry (measured in the direct path), immunoradiometry, enzyme-linked immunosorbent assay, dissociation-enhanced fluorescence immunoassays, and liquid chromatography coupled to mass spectrometry. Recognition of “free” apo(a) indicates if tests are possibly creating bias due to not taking particle size into account. Note that sensitivity and specificity are conceptual, relative to each other, and defined as either “+”, “++”, “+++”. Quantitative protein mass spectrometry is a direct KIV2 repeat of the independent and non-immune-based method. *Whether or not INA or ITA recognize free apo(a) depends on the size of the antibody-antigen complex in the assays. Since this site cannot be predicted the effect on the Raleigh light scatter detection is uncertain.
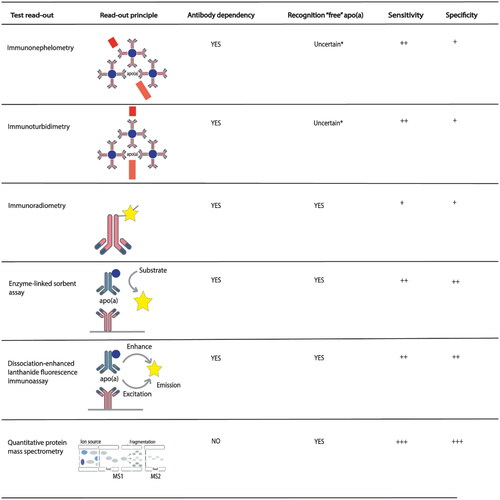
Table 2. Immunoassay-based Lp(a) tests that are commercially available.
Besides the use of kringle-dependent antibodies and readout methods that could be particle size dependent, Lp(a) has been reported in three units or modes: Lp(a) mass (mg/dL), Lp(a) particle number (nmol/L), and more recently, Lp(a) cholesterol (Lp(a)-c, mg/dL) [Citation68]. This latter assay could be important to accurately determine the LDL-c value, as the cholesterol portion of Lp(a) may make up a substantial amount of detected LDL-c when the Lp(a) particle concentration is high [Citation69]. The Lp(a)-c test is not further discussed in this review, as it has another intended use. Initially, the mass composition of the Lp(a) particle, including apo(a), apoB-100, cholesterol, phospholipid, triglyceride, and the carbohydrate content were included in Lp(a) assay results reported in mass units (mg/dL). Since measurement procedures only target apo(a) and no other components of Lp(a), the use of mass composition is incorrect from a metrological perspective. Misinterpretations can occur when using apo(a) measurements based on mass, depending on whether the patient possesses the bigger less atherogenic apo(a) isoforms or the smaller more atherogenic isoforms. Some commercially available assays require a conversion from mass units to molar units. Unfortunately, one cannot use conversion factors without knowing what isoform(s) is (are) present in patients. Accordingly, a single conversion factor between assays is inadequate. It was previously advised to use a conversion factor of 2.85 for small Lp(a) isoforms and 1.85 for large Lp(a) isoforms, with a mean of 2.4 [Citation6]. It is still noted that the immunoassay measurements do not take into account the isoform size and that this differentiated conversion factor is therefore impractical, incorrect and manufacturer-dependent. Marcovina and coworkers confirmed that the use of a conversion factor, regardless of how it is computed, ought to be abandoned [Citation70]. Lp(a) concentrations should not be converted from nmol/L to mg/dL or vice versa. The selectivity of current Lp(a) immunoassay-based tests is imperfect as KIV2-dependent polyclonal antibodies are used [Citation95]. Also, the product calibrators cannot be a perfect mimic of the apo(a) isoform composition in the native specimens to be analyzed.
4. Analytical challenges for accurate lp(a) measurements with regard to a measurand, antibody, calibration type, and calibration number
Here, a structured overview of parameters that potentially explain Lp(a) result variability among Lp(a) field methods will be discussed.
4.1. Analytical challenges in Lp(a) field methods
Because of its clinical relevance as an independent risk factor for CVD serum/plasma Lp(a) is increasingly measured in medical laboratories using polyclonal anti-apo(a) based immunoassays on high throughput platforms. The specific epitopes that have been used by IVD manufacturers for antibody generation should ideally enable selective identification and accurate quantification of Lp(a) by targeting a single apo(a) molecule per Lp(a) particle, independent of its number of KIV2 repeats. As experienced in the Physician Health Studies and the Framingham Study () [Citation28,Citation71,Citation72], kringle IV2-sensitive tests, especially in combination with serially diluted single calibrators, typically result in underestimation of high Lp(a) levels (with low molecular weight apo(a) isoforms) and overestimation of low Lp(a) concentrations (with high molecular weight apo(a) isoforms). [Citation28,Citation71,Citation72] Monoclonal antibodies against a unique apo(a) epitope would theoretically be ideally suited, but their application lacks analytical sensitivity on routine clinical chemistry analyzers, as only a single antibody binds per Lp(a) particle [Citation72]. As the polyclonal antibodies used in almost all Lp(a) field methods detect the apo(a)’s repeating KIV2 structure, these tests are all apo(a) isoform dependent [Citation73]. Yet, in the majority of current Lp(a) tests, KIV2 dependence has been elegantly diminished within allowable limits of MU by using multiple (5 at least), independent and well-selected product calibrators with typical isoform composition across the measuring range. [Citation95] Accuracy-related characteristics of current Lp(a) field tests are shown in . Calibration of Lp(a) field tests is typically performed using a multi-point calibration with at least five independent calibrators or using serial dilutions from a single high calibrator. Calibrators are nowadays selected to mimic the relation between KIV2 repeats and Lp(a) concentration in patient specimens, in order not to underestimate the association between Lp(a) and CVD [Citation6]. So far it has been documented that the Denka Seiken test is the least apo(a) isoform-sensitive test (Denka Seiken Co. Ltd., Japan). Note that several commercial Lp(a) tests are Denka Seiken-based [Citation71]. If the Lp(a) field tests generate molar test results that are traceable to SRM2B and the gold standard RMP within allowable MU, valid and accurate Lp(a)/apo(a) tests results are generally assured within the measuring range in undiluted specimens.
Alternative calibration strategies for immunoassays could be based on either recombinant protein or native serum samples. When deciding upon a calibration strategy using native serum samples, the samples must be selected carefully to truthfully reflect the KIV2 size polymorphism to Lp(a) concentration. A pool of serum samples could aid here, as the size polymorphism is averaged out, making the isoform composition more similar to that of the test sample, thus improving isoform insensitivity [Citation31]. When using recombinant protein, multiple constructs with variable KIV2 repeats are required [Citation74], and the effects of lipid-free apo(a) on the assay had to be diminishable.
4.2. The search for specific measurands and accurate apo(a) results
The large heterogeneity in apo(a) size within and between individuals due to the inheritance of two different apo(a) alleles has been a huge challenge for the accurate measurement of Lp(a). Specifically, variation is introduced by two alleles and the variable number of the KIV2 repeats that may result in apo(a) with a molecular weight spanning from 250 to 800 kDa [Citation75]. Moreover, fragmented and lipid-free apo(a) may further add to the variation in the measurand since these species will likely interfere with initial antibody capture. Furthermore, the principle of the read-out method used has an effect on the recovery of the measurands. For example, there is always the possibility of interference in methods based on antibody capture combined with light absorption/scatter, where the signal and accompanying read-out would be underestimated as a result. The possibility of interference could be due to LDL-unbound apo(a) particles or free apo(a) [Citation47]. Notably, when looking into the instructions for use (IFU) of commonly used field methods, most fail to disclose their antibodies’ ability to bind lipid-free apo(a) or truncated apo(a) (see ). Size differences could generate different results when providing results in mass instead of in particle concentration. Additionally, not all immunoturbidimetric assays (ITAs) are identical. We can subdivide into direct immunoturbidimetry, PETIA, and latex-enhanced immunoturbidimetry. In direct immunoturbidimetry, antibodies directly connect to the targeting antigen to produce an immunological complex. In PETIA, target antibodies coated particles form complexes with the antigen in the sample. The formed complexes amplify the signal and as such are very helpful when the antigen is present in low quantity. The principle of latex-enhanced immunoturbidimetry is similar as it also results in amplified signals by using latex particles instead of coated particles. Different modes of action imply the possibility of variability among result read-outs, and one must thus carefully specify the measurand.
4.3. Alternative Lp(a) quantification using protein mass spectrometry
As an alternative to immunoassay-based methods, Lp(a) can be quantified using protein MS which will be detailed in Section 7. In this approach, apo(a) is directly quantified via unique proteotypic peptide fragments which have the inherent advantage of being antibody and kringle size independent. Protein MS requires expertise and high-end equipment and is therefore considered a costly procedure compared to conventional field methods. Moreover, sample throughput is lower when using quantitative proteomics compared to the field methods. However, once the inherent multiplexing capabilities of this approach are utilized (i.e. quantify multiple proteins), this procedure may turn financially viable for large clinical trials and/or reference services. Specific challenges need to be addressed when using a peptide-centric (or bottom-up) procedure, such as completeness of digestion, matrix effects and optimal chromatographic separation (further discussed in Section 7). Earlier, we compared apo(a) test results between MS-based and immunoassay-based tests in CVD patients. Although the results from the MS-based and ITAs were equivalent and correlated well, the method comparison data also demonstrated scatter, which could perhaps be related to different recoveries of free apo(a) [Citation76].
The earlier mentioned RMS developed by Marcovina and coworkers includes a traceability chain up to a secondary RM () [Citation28,Citation29]. This material was assembled from 17 pooled blood donors and had an assigned value of 0.1071 nanomoles per vial. The Lp(a) test systems for value assignment included ten ITAs, eight INAs, two fluorescence immunoassays (FIAs), one electro-immunodiffusion assay, and one ELISA, and the median within-assay CV was 5.1% [Citation29]. This RM was accompanied by a “golden standard” RMP consisting of a KIV2-independent ELISA-based approach [Citation51,Citation95]. The apo(a) quantity that inherently correlates to Lp(a) concentrations allows for quantification that is traceable to the WHO-IFCC secondary RM WHO/IFCC SRM®-2B. The monoclonal antibody used in the ELISA was generated using purified native Lp(a) and recognizes a unique KIV9 epitope (i.e. LETPTVV), using 35 measurements in each of five quality control samples, a within-assay CV ranging from 2.3% to 4.0% was determined [Citation28]. Currently, the WHO/IFCC SRM®-2B material has run out of stock and the calibration lab has been closed. For reasons of continuity, an IFCC working group was established to set up a next-generation RMS for Lp(a), ultimately aiming for the traceability of Lp(a) test results to SI units [Citation26].
5. Moving toward the next generation Lp(a) reference measurement system based on quantitative protein mass spectrometry and molar units
As Lp(a) recently arose as a Phoenix from its ashes, the availability of an internationally endorsed RMS for guaranteeing Lp(a) test result equivalence among commercial Lp(a) tests in time and space is key [Citation77]. As the current “gold standard”, ELISA-based RMP at Northwest Lipid Research Laboratory, University of Washington, Seattle, USA, is no longer operational and the secondary RM, WHO/IFCC SRM-2B, has run out of stock, the IFCC Scientific Division had decided (after IFCC Executive Board approval) to establish an international working group [https://www.ifcc.org/ifcc-scientific-division/sd-working-groups/wg-apo-ms/]. This group is entrusted to develop a next-generation SI-traceable RMS for Lp(a)/apo(a), as displayed in (right). So far, the conceptual approach for establishing the apo(a) RMS, the developed apo(a) candidate RMP, and the commutability study data that gives insight into the selection of suitable secondary matrix-based RMs have been published [Citation26].
Meanwhile, several non-IFCC-related initiatives are also ongoing. As such, the former RMP has served as a starting point for the creation of a candidate RMP referred to as the LPA4/LPA-KIV9 ELISA [Citation78]. This second ELISA designed by Marcovina, Tsimikas, and coworkers exhibits a larger bias as compared to the first RMP, which was explained by the development process that was based on a truncated recombinant apo(a) instead of native purified Lp(a) [Citation78]. It should be noted that the use of an immunoassay for reference standardization of an Lp(a) test is not a prerequisite. On the contrary, in our opinion, the quantification of Lp(a) levels can much better be based on methods that consider the multiple sequence variations, isoforms, and PTMs on apo(a). Applications derived from MS-based bottom-up proteomics are ideally suited for this purpose [Citation30]. The advantages of applying MS instead of immunoassays for protein quantification were identified in the early days of proteomics [Citation79]. In a bottom-up proteomics strategy, a protein is enzymatically converted (i.e. digested) into multiple peptides, followed by MS-based quantification of specific proteotypic peptides. For this purpose, trypsin is commonly used to yield peptides with either arginine or lysine residues at the C-terminus [Citation80]. The simultaneously identified and quantified proteotypic peptides correlate to the quantities of endogenous corresponding proteins. MS has proven to be feasible for quantifying protein biomarkers and evaluating proteins from complex biological matrices in a clinical chemistry setting [Citation8,Citation81]. To this end, the first quantitative protein MS test for the quantitation of apo(a) has been reported in 2014.
Marcovina and coworkers have also created a targeted liquid chromatography-mass spectrometry (LC-MS) candidate RMP based on the bottom-up proteomics strategy that applies proteolytic peptides as surrogates for protein identification and quantification [Citation59]. Here apo(a) levels are determined via peptides that are selected and relatively quantified (i.e. normalized) using stable isotope-labeled peptide analogues. Next, the calibration is based on in-house-produced recombinant human apo(a) containing 14 KIV repeats, with valueassignment through SI-traceable amino acid analysis [Citation59]. The proteotypic peptides that are used in this procedure are TPAYYPNAGLIK, TPENYPNAGLTR, and GISSTTVTGR, located on KIV4, KIV5, and KIV9, respectively. The mean response of these three peptides is used to determine the apo(a) level and to derive the Lp(a) concentration. Importantly, the method is validated for EDTA-plasma and thereby suitable as an end users’ procedure for the quantitation of apo(a); however, this method may not hold for serum or lithiumheparin-plasma matrices.
This principle of using MS for global standardization of apo(a)/Lp(a) tests at the molar level has great potential, provided the method can be established for serum and/or lithiumheparin-plasma as these are the common matrices collected for lipid profiling in medical laboratories [Citation59,Citation60,Citation76,Citation82]. Clearly, to circumvent apo(a) polymorphisms, proteotypic peptides need to be selected that are not present in KIV2 repeats. Moreover, protein quantification through its surrogate measurands is only accurate when the peptides are fully released from the protein backbone during proteolysis, in other words, when the conversion is equimolar. Within the IFCC WG APO-MS, a candidate RMP has been developed and analytically validated. The peptide-based calibration strategy is being developed within a collaboration of three IFCC calibration laboratories and quantifies the peptides LFLEPQADIALLK, GISSTTVTGR. and TPENYPNAGLTR, from the protease, KIV9, and KIV5 domains, respectively, relative to their stable isotope labelled internal standards. As the peptide-based primary reference materials are under development, the method was validated using a native serum-based calibrator provisionally value assigned with Roche ITA (with an apo(a) concentration of 94.6 nmol/L) and hence indirectly traceable to SRM2B [Citation76]. The linear measuring range is between 3.8 nmol/L and 450 nmol/L, with a limit of quantification (LoQ) of 3.8 nmol/L and peptides could be measured with an average imprecision of 8.9% which is well within the set APS of 12% [Citation76].
A prerequisite for peptide-based calibration of a candidate RMP aimed toward SI traceability of the measurands is an equimolar conversion of the protein of interest into its quantifying peptides. To that end, digestion should be fully understood and carefully controlled. Earlier studies on the digestion kinetics of a protease such as trypsin have focused on the specificity of the enzyme and how this specificity is influenced by other residues in close proximity to the cleavage site [Citation80]. Moreover, protein digestion can be influenced by local conformation, tertiary structure, and experimental conditions and to that end identifications of peptides with unexpected cleavages are of interest. In a similar way, for quantification purposes, it is informative to focus on peptides that contain so-called missed cleavages. The presence of missedcleavages in the proximity of the proteotypic peptides is an indicator that digestion has not been “completed” at that position in the protein [Citation80]. Earlier, we reported three proteotypic peptides of apo(a), namely LFLEPTQADIALLK (LFLEP in short), TPENYNAGLTR (TPENY in short), and GISSTVTGR (GISST in short), that all reached a plateau in a digestion curve [Citation76]. Although informative, a digestion plateau does not warrant the full conversion of the protein into peptides. Therefore, the presence or absence of missed cleavages in apo(a) digest is currently under investigation.
6. Conclusions and anticipated impact of the next-generation apo(a) RMS on clinical practice
Lp(a) has made a major comeback as a clinically useful risk factor for CVD. The availability of the first gold standard RMS was of pivotal importance in re-assessing Lp(a) data from the Physicians Health Study and the Framingham Study. A consistent relation with CVD was revealed and it was clarified that apo(a) isoform dependency, polyclonal antibodies, and inadequate selection of calibrators and/or calibration procedures had been responsible for weakening or even masking the Lp(a)-CVD relationship. Multiple studies have demonstrated that elevated levels of Lp(a) are associated with a significantly increased risk of CVD, independent of other traditional risk factors like LDL. It has become apparent that serum lipids do not tell the full story and underestimate CVD risk, leading to underdiagnosis and undertreatment of patients. The latest EAS consensus statement recommends quantification of Lp(a) at least once in all individuals to identify those at enhanced cardiovascular risk. The here-described pilgrimage toward the development of an SI-traceable next-generation apo(a) RMS based on quantitative protein MS and molar units includes various analytical challenges that IVD manufacturers must overcome to develop safe and clinically effective Lp(a) tests. The envisioned comprehensive RMS is developed timely as it should guide manufacturers to design Lp(a) tests that are fit-for-purpose now that its clinical relevance has been unequivocally demonstrated and with new promising Lp(a) lowering drugs under development. The conceptual approach and sequential steps taken by the different stakeholders involved in the setup of the apo(a) RMS have recently been described [Citation26,Citation76,Citation83]. It is anticipated that the Lp(a)/apo(a) RMS will soon be operational because the candidate reference measurement procedure (cRMP) for apo(a) is endorsed by the IFCC. Moreover, selections for suitable matrix-based secondary RMs have been completed based on the previous commutability study.
Lp(a) particles are widely measured using immunoassay-based tests with the apo(a) protein as the antigen. Apo(a) is present in highly variable forms due to a size polymorphism in KIV2 and PTMs such as glycosylation, as well as lipid-free truncated and fragmented species. The insufficiently defined and heterogeneous character of the apo(a) measurands were and are major determinants of inaccurate and flawed Lp(a) test results, notwithstanding the availability of a well-functioning (i.e. the gold standard) ELISA-based RMS [Citation28,Citation84]. Other confounders that may invalidate Lp(a) tests results are apo(a) isoform-sensitive reagents, cross-reacting polyclonal anti-apo(a) antibodies, insufficiently wide measuring ranges of immunoassay-based tests that necessitate automatic dilution to the wrong calibrator isoforms, serial dilutions of a single high calibrator, prozone effects, and matrix effects caused by anticoagulants and other preanalytical conditions such as freeze/thaw cycles. All these insights were clarified thanks to the availability of the former RMS and the recent head-to-head comparisons between representative indirect immunoassays and direct protein measurements via quantitative protein MS. The latter enables an unequivocal molecular definition of the measurand(s), namely apo(a). A recent IFCC initiative, which aims to reestablish standardization of Lp(a) tests, is focused on the development of a cRMP applying a quantitative protein MS strategy [Citation76]. To specifically measure apo(a), peptides can be selected that are not present in KIV2 and that are not modified post-translationally. Lipid-free apo(a) is included in the measurements as well as truncated apo(a) and apo(a) fragments. For global standardization of IVDs and in order not to confuse manufacturers, it is strongly recommended that RMs, RMPs, and reference measurement services are endorsed by official international organizations such as the IFCC and listed in the Joint Committee on Traceability in Laboratory Medicine (JCTLM) database [Citation85], after passing peer review by an expert panel.
The availability of listed reference measurement services in itself does not yet accomplish test standardization. Commitment from IVD manufacturers to adopt and implement JCTLM-listed RMS is the key to success. Therefore, the anticipated transition from the former ELISA-based RMS to the next-generation SI-traceable apo(a) RMS requires guidance and education of IVD manufacturers, the pharma industry, clinicians, and laboratory specialists through this metrological labyrinth and scientific change process. It is foreseen that the change to molar units and the transition from former to new RMS will have an impact on the results generated by the IFCC-based cRMP compared to SRM2B-traceable test results.
The apo(a) isoform-sensitivity of previous Lp(a) tests has been overcome to a large extent by most IVD manufacturers of Lp(a) tests by means of multipoint calibration with multiple independent native human calibrators. In order to evaluate new Lp(a) lowering drugs in the near future, it is essential to have a sustainable Lp(a) RMS in place, implemented in a network of calibration labs using harmonized procedures. To that end, the IFCC WG APO-MS is preparing apo(a) cRMP, primary peptide-based calibrators, and secondary matrix-based RMs as previously reported in a multistep approach.
The IVD manufacturers will be invited to reestablish the metrological traceability of their Lp(a) test results and to verify whether their Lp(a) tests are still fit-for-clinical-purpose and compliant with desirable APS deduced from biological variation data. In the first step, the transition to molar units according to the old RMS is anticipated. In a second step, traceability to the new SI-traceable apo(a) RMS based on quantitative protein MS will be prepared. Meanwhile, IVD manufacturers are invited to improve persisting inadequacies in their Lp(a) immunoassay-based tests (e.g. implementation of multiple independent native calibrators, an extension of the Lp(a) measuring range, replacing erroneous Lp(a) mass reporting by molar units, etc.) to guarantee accurate Lp(a) test results within allowable MU for adequate CVD risk assessment or evaluation of therapeutic effects of Lp(a) lowering drugs. Importantly, IVD manufacturers should be committed to implementing the future apo(a) RMS when available, for the purpose of universal clinical decision limits and exchangeable treatment goals in clinical trials and guidelines.
Lp(a) tests have the reputation of being the most misunderstood metric in clinical chemistry labs. Unfamiliarity with the heterogeneity of the apo(a) measurand(s), the selection of the ISO 17511 calibration model and/or the implementation of a higher order metrological traceability chain, as well as cross-reactivity of the polyclonal antibodies, combined with flawed designs of immunoassay-based tests (such as reporting Lp(a) mass instead of Lp(a) particle number or apo(a) concentration) have all hampered the validity of the produced Lp(a) data in the past. Thanks to the evolution in science and metrology, and the establishment of valuable RMS by Marcovina in the 1990s and by the IFCC WG APO-MS nowadays, key determinants of Lp(a) test inaccuracy were identified. In the past, IVD manufacturers solved several analytical challenges, especially apo(a) isoform sensitivity and serial dilutions of single high calibrators. The problem-solving measures such as the introduction of multipoint calibration procedures with independent native calibrators and representative apo(a) isoforms enabled the reporting of accurate results traceable to SRM2B (in nmol/L) and the ELISA-based RMP. Desirable analytical performance and adequate product calibration strategies assured that most Lp(a) field tests are fit-for-clinical-purpose in undiluted specimens within the measuring range. Both the former ELISA-based RMS and the foreseen SI-traceable RMS based on quantitative protein MS from the IFCC WG APO-MS are instrumental for ongoing Lp(a) test standardization and are considered the holy grail for standardizing Lp(a) field methods.
Finally, to enable the proper implementation of EAS consensus statements and clinical guidelines, laboratory specialists, clinicians, and IVD industry should be aware of the strict inter-dependence between the analytical and clinical performance of medical tests in general, but especially for the polymorphic apo(a) in Lp(a). It is a joint responsibility toward our patients to manufacture and implement safe and clinically effective Lp(a) tests, and by doing so, to enable a more personalized approach for predicting, diagnosing, treating, and monitoring patients at risk for CVD.
| Abbreviations | ||
| Apo(a) | = | apolipoprotein(a) |
| ApoB | = | apolipoprotein B |
| ApoB100 | = | apolipoprotein B100 |
| APS | = | analytical performance specifications |
| BAPS | = | bias |
| BIVAC | = | Biological Variation Data Critical Appraisal Checklist |
| cRMP | = | candidate reference measurement procedure |
| CV | = | Coefficient of variation |
| CVa | = | analytical variation |
| CVi | = | intra-individual variation |
| CVG | = | within-group variation |
| CPS | = | clinical performance specifications |
| CVD | = | cardiovascular disease |
| DEIFA | = | dissociation-enhanced lanthanide fluorescence immunoassays |
| EAS | = | European Atherosclerosis Society |
| ESC | = | European Society of Cardiology |
| EFLM | = | European Federation of Clinical Chemistry and Laboratory Medicine |
| ELISA | = | enzyme-linked immunosorbent assay |
| FIA | = | fluorescence immunoassay |
| GUM | = | Guide to the Expression of Uncertainty in Measurement |
| IFCC | = | International Federation of Clinical Chemistry and Laboratory Medicine |
| INA | = | immunonephelometric assay |
| ITA | = | immunoturbidimetric assay |
| ISO | = | International Organization for Standardization |
| IVD | = | in vitro diagnostics |
| JCTLM | = | Joint Committee on Traceability in Laboratory Medicine |
| JRC | = | Joint Research Center |
| KIV1-10 | = | kringle-IV subunits 1–10 |
| LC-MS | = | liquid chromatography-mass spectrometry |
| LDL | = | low-density lipoprotein |
| LDL-c | = | LDL cholesterol |
| LNE | = | Laboratoire National de Métrologie et d‘Essais |
| Lp(a) | = | Lipoprotein(a) |
| Lp(a)-c | = | Lp(a) cholesterol |
| LoQ | = | limit of quantification |
| MI | = | myocardial infarction |
| MU | = | measurement uncertainty |
| MS | = | mass spectrometry |
| NIBSC | = | National Institute for Biological Standards and Control |
| NIST | = | National Institute of Standards and Technology |
| PETIA | = | particle-enhanced immunoturbidimetric assay |
| PTM | = | post-translational modification |
| RCT | = | randomized control trial |
| RIA | = | radioimmunoassay |
| RID | = | radial immunodiffusion |
| RM | = | reference material |
| RMP | = | reference measurement procedure |
| RMS | = | reference measurement system |
| TEa | = | total allowable error |
| VIM | = | International Vocabulary of Metrology |
| WHO | = | World Health Organization |
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Berg K. A new serum type system in man–the lp system. Acta Pathol Microbiol Scand. 1963;59:369–382.
- Ridker PM, Hennekens CH, Stampfer MJ. A prospective study of lipoprotein(a) and the risk of myocardial infarction. JAMA. 1993; 270(18):2195–2199.
- Ridker PM, Stampfer MJ, Hennekens CH. Plasma concentration of lipoprotein(a) and the risk of future stroke. JAMA. 1995; 273(16):1269–1273.
- Ridker PM, Stampfer MJ, Rifai N. Novel risk factors for systemic atherosclerosis: a comparison of C-reactive protein, fibrinogen, homocysteine, lipoprotein(a), and standard cholesterol screening as predictors of peripheral arterial disease. JAMA. 2001; 285(19):2481–2485.
- Rifai N, Ma J, Sacks FM, et al. Apolipoprotein(a) size and lipoprotein(a) concentration and future risk of angina pectoris with evidence of severe coronary atherosclerosis in men: the physicians’ health study. Clin Chem. 2004;50(8):1364–1371.
- Marcovina SM, Albers JJ. Lipoprotein (a) measurements for clinical application. J Lipid Res. 2016; 57(4):526–537.
- Pearson GJ, Thanassoulis G, Anderson TJ, et al. 2021 Canadian cardiovascular society guidelines for the management of dyslipidemia for the prevention of cardiovascular disease in adults. Can J Cardiol. 2021;37(8):1129–1150.
- Grundy SM, Stone NJ, Bailey AL, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: executive summary: a report of the American college of cardiology/American heart association task force on clinical practice guidelines. Circulation. 2019;139(25):e1046–e1081.
- Arnett DK, Blumenthal RS, Albert MA, et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: executive summary: a report of the American college of cardiology/American heart association task force on clinical practice guidelines. J Am Coll Cardiol. 2019;74(10):1376–1414.
- Kronenberg F, Mora S, Stroes ESG, et al. Lipoprotein(a) in atherosclerotic cardiovascular disease and aortic stenosis: a European atherosclerosis society consensus statement. Eur Heart J. 2022;43(39):3925–3946.
- Authors/Task Force M, Guidelines ESCCfP. Societies ESCNC. 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Atherosclerosis. 2019; 290:140–205.
- Puri R, Mehta V, Duell PB, et al. Evidence for intensive LDL-C lowering for acute coronary syndrome: recommendations from the lipid association of India. J Clin Lipidol. 2022;16(3):261–271.
- Chinese Society of Cardiology of Chinese Medical A, Cardiovascular Disease P, Rehabilitation Committee of Chinese Association of Rehabilitation M, et al. [Chinese guideline on the primary prevention of cardiovascular diseases]. Zhonghua Xin Xue Guan Bing Za Zhi. 2020; 48(12):1000–1038.
- Clarke R, Peden JF, Hopewell JC, et al. Genetic variants associated with lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361(26):2518–2528.
- Burgess S, Ference BA, Staley JR, et al. Association of LPA variants With risk of coronary disease and the implications for lipoprotein(a)-lowering therapies: a Mendelian randomization analysis. JAMA Cardiol. 2018;3(7):619–627.
- Patel AP, Wang M, Pirruccello JP, et al. Lp(a) (lipoprotein[a]) concentrations and incident atherosclerotic cardiovascular disease: new insights From a large national biobank. Arterioscler Thromb Vasc Biol. 2021;41(1):465–474.
- Kamstrup PR, Benn M, Tybjaerg-Hansen A, et al. Extreme lipoprotein(a) levels and risk of myocardial infarction in the general population: the copenhagen city heart study. Circulation. 2008;117(2):176–184.
- Nordestgaard BG, Langsted A. Lipoprotein (a) as a cause of cardiovascular disease: insights from epidemiology, genetics, and biology. J Lipid Res. 2016; 57(11):1953–1975.
- Rhainds D, Brodeur MR, Tardif JC. Lipoprotein (a): when to measure and how to treat? Curr Atheroscler Rep. 2021; 23(9):51.
- Cobbaert C, Arentsen JC, Mulder P, et al. Significance of various parameters derived from biological variability of lipoprotein(a), homocysteine, cysteine, and total antioxidant status. Clin Chem. 1997;43(10):1958–1964.
- Sandberg S, Fraser CG, Horvath AR, et al. Defining analytical performance specifications: consensus statement from the 1st strategic conference of the European federation of clinical chemistry and laboratory medicine. Clin Chem Lab Med. 2015; 53(6):833–835.
- Langlois MR, Nordestgaard BG, Langsted A, et al. Quantifying atherogenic lipoproteins for lipid-lowering strategies: consensus-based recommendations from EAS and EFLM. Clin Chem Lab Med. 2020;58(4):496–517.
- Aarsand AKF-CP, Webster C, Coskun A, et al. The EFLM biological variation database. https://biologicalvariation.eu/. EFLM; 2019.
- Ricos CA, Cava F, Garcia-Lario JV, et al. Current databases on biologic variation: pros, cons and progress. https://www.westgard.com/biodatabase1.htm. : Scand J Clin Lab Invest 1999; 2014.
- Ruhaak LR, Cobbaert CM. Quantifying apolipoprotein(a) in the era of proteoforms and precision medicine. Clin Chim Acta. 2020;511:260–268.
- Cobbaert CM, Althaus H, Begcevic Brkovic I, et al. Towards an SI-traceable reference measurement system for seven serum apolipoproteins using bottom-up quantitative proteomics: conceptual approach enabled by cross-disciplinary/cross-sector collaboration. Clin Chem. 2021;67(3):478–489.
- Tsimikas S, Fazio S, Ferdinand KC, et al. NHLBI working group recommendations to reduce lipoprotein(a)-mediated risk of cardiovascular disease and aortic stenosis. J Am Coll Cardiol. 2018;71(2):177–192.
- Marcovina SM, Albers JJ, Gabel B, et al. Effect of the number of apolipoprotein(a) kringle 4 domains on immunochemical measurements of lipoprotein(a). Clin Chem. 1995;41(2):246–255.
- Dati F, Tate JR, Marcovina SM, et al. First WHO/IFCC international reference reagent for lipoprotein(a) for immunoassay–lp(a) SRM 2B. Clin Chem Lab Med. 2004;42(6):670–676.
- Cobbaert C, Deprez L, Ruhaak LR. On the way to a next generation lp(a) reference measurement system based on quantitative protein mass spectrometry and molar units. In Kostner K, Kostner G, Toth P, editors. Lipoprotein(a). Contemporary cardiology. Cham, Switzerland: Humana; 2023.
- Clouet-Foraison N, Vaisar T, Marcovina SM. Standardization of analytical methods for the measurement of lipoprotein(a): bridging Past and future initiatives. In: Kostner K, Kostner GM, Toth PP, editors. Lipoprotein(a). Cham: Springer International Publishing; 2023. p. 297–323.
- Vesper HW, Thienpont LM. Traceability in laboratory medicine. Clin Chem. 2009; 55(6):1067–1075.
- ISO. (International Organization for Standardization) 17511. 2020. Requirements for establishing metrological traceability of values assigned to calibrators, trueness control materials and human samples.
- Joint Committee for Guides in Metrology, International Vocabulary of Metrology [accessed 15 Jan 2023.
- SI. Brochure: The International System of Units [accessed on 12 December 2012]; 2023.
- ISO. (International, Organization, for, Standardization) 15195; 2018. Laboratory medicine—Requirements for the competence of calibration laboratories using reference measurement procedures. https://www.iso.org/standard/69824.html.
- ISO. (International, Organization, for, Standardization) 15193; 2009. in vitro diagnostic medical devices—measurement of quantities in samples of biological origin—Requirements for content and presentation of reference measurement procedures.
- Tsimikas S, Fazio S, Viney NJ, et al. Relationship of lipoprotein(a) molar concentrations and mass according to lipoprotein(a) thresholds and apolipoprotein(a) isoform size. J Clin Lipidol. 2018;12(5):1313–1323.
- McConnell JP, Guadagno PA, Dayspring TD, et al. Lipoprotein(a) mass: a massively misunderstood metric. J Clin Lipidol. 2014;8(6):550–553.
- Koschinsky ML, Marcovina SM. Structure-function relationships in apolipoprotein(a): insights into lipoprotein(a) assembly and pathogenicity. Curr Opin Lipidol. 2004; 15(2):167–174.
- Koschinsky ML, Marcovina SM. Lipoprotein(a): structural implications for pathophysiology. Int J Clin Lab Res. 1997;27(1):14–23.
- Enas EA, Varkey B, Dharmarajan TS, et al. Lipoprotein(a): an independent, genetic, and causal factor for cardiovascular disease and acute myocardial infarction. Indian Heart J. 2019;71(2):99–112.
- Kronenberg F. Human genetics and the causal role of lipoprotein(a) for various diseases. Cardiovasc Drugs Ther. 2016; 30(1):87–100.
- Tsimikas S, Brilakis ES, Miller ER, et al. Oxidized phospholipids, lp(a) lipoprotein, and coronary artery disease. N Engl J Med. 2005;353(1):46–57.
- Subramanian SP, Gundry RL. The known unknowns of apolipoprotein glycosylation in health and disease. iScience. 2022; 25(9):105031.
- Garner B, Merry AH, Royle L, et al. Structural elucidation of the N- and O-glycans of human apolipoprotein(a): role of o-glycans in conferring protease resistance. J Biol Chem. 2001;276(25):22200–22208.
- Gries A, Nimpf J, Nimpf M, et al. Free and apo B-associated lpa-specific protein in human serum. Clin Chim Acta. 1987;164(1):93–100.
- Cauza E, Kletzmaier J, Bodlaj G, et al. Relationship of non-LDL-bound apo(a), urinary apo(a) fragments and plasma lp(a) in patients with impaired renal function. Nephrol Dial Transplant. 2003;18(8):1568–1572.
- McLean JW, Tomlinson JE, Kuang WJ, et al. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 1987; 330(6144):132–137.
- Lawn RM, Schwartz K, Patthy L. Convergent evolution of apolipoprotein(a) in primates and hedgehog. Proc Natl Acad Sci U S A. 1997; 94(22):11992–11997.
- Parson W, Kraft HG, Niederstatter H, et al. A common nonsense mutation in the repetitive kringle IV-2 domain of human apolipoprotein(a) results in a truncated protein and low plasma lp(a). Hum Mutat. 2004;24(6):474–480.
- Hoff HF, O'Neil J, Smejkal GB, et al. Immunochemically detectable lipid-free apo(a) in plasma and in human atherosclerotic lesions. Chem Phys Lipids. 1994;67-68:271–280.
- Mooser V, Marcovina SM, White AL, et al. Kringle-containing fragments of apolipoprotein(a) circulate in human plasma and are excreted into the urine. J Clin Invest. 1996;98(10):2414–2424.
- Fortunato JE, Bassiouny HS, Song RH, et al. Apolipoprotein (a) fragments in relation to human carotid plaque instability. J Vasc Surg. 2000;32(3):555–563.
- ISO. (International, Organization, for, Standardization), 20914. 2019. Medical laboratories – practical guidance for the estimation of measurement uncertainty.
- Lord SJ, St John A, Bossuyt PM, et al. Setting clinical performance specifications to develop and evaluate biomarkers for clinical use. Ann Clin Biochem. 2019;56(5):527–535.
- White GH. Basics of estimating measurement uncertainty. Clin Biochem Rev. 2008; 29 Suppl 1(Suppl 1):S53–S60.
- Jones GRD, Albarede S, Kesseler D, et al. Analytical performance specifications for external quality assessment - definitions and descriptions. Clin Chem Lab Med. 2017;55(7):949–955.
- Marcovina SM, Clouet-Foraison N, Koschinsky ML, et al. Development of an LC-MS/MS proposed candidate reference method for the standardization of analytical methods to measure lipoprotein(a). Clin Chem. 2021;67(3):490–499.
- Renee Ruhaak L, van der Laarse A, Cobbaert CM. Apolipoprotein profiling as a personalized approach to the diagnosis and treatment of dyslipidaemia. Ann Clin Biochem. 2019; 56(3):338–356.
- Harris EK. Proposed goals for analytical precision and accuracy in single-point diagnostic testing. Theoretical basis and comparison with data from college of American pathologists proficiency surveys. Arch Pathol Lab Med. 1988; 112(4):416–420.
- Clouet-Foraison N, Marcovina SM, Guerra E, et al. Analytical performance specifications for lipoprotein(a), apolipoprotein B-100, and apolipoprotein A-I using the biological variation model in the EuBIVAS population. Clin Chem. 2020;66(5):727–736.
- Greif M, Arnoldt T, von Ziegler F, et al. Lipoprotein (a) is independently correlated with coronary artery calcification. Eur J Intern Med. 2013; 24(1):75–79.
- Jurgens G, Hermann A, Aktuna D, et al. Dissociation-enhanced lanthanide fluorescence immunoassay of lipoprotein(a) in serum. Clin Chem. 1992;38(6):853–859.
- Lequin RM. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin Chem. 2005; 51(12):2415–2418.
- Albers JJ, Hazzard WR. Immunochemical quantification of human plasma lp(a) lipoprotein. Lipids. 1974; 9(1):15–26.
- Albers JJ, Adolphson JL, Hazzard WR. Radioimmunoassay of human plasma lp(a) lipoprotein. J Lipid Res. 1977; 18(3):331–338.
- Yeang C, Witztum JL, Tsimikas S. Novel method for quantification of lipoprotein(a)-cholesterol: implications for improving accuracy of LDL-C measurements. J Lipid Res. 2021; 62:100053.
- Yeang C, Witztum JL, Tsimikas S. LDL-C’ = LDL-C + lp(a)-C: implications of achieved ultra-low LDL-C levels in the proprotein convertase subtilisin/kexin type 9 era of potent LDL-C lowering. Curr Opin Lipidol. 2015; 26(3):169–178.
- Brown WV, Ballantyne CM, Jones PH, et al. Management of lp(a). J Clin Lipidol. 2010;4(4):240–247.
- Cegla J, France M, Marcovina SM, et al. Lp(a): when and how to measure it. Ann Clin Biochem. 2021;58(1):16–21.
- Kronenberg F, Tsimikas S. The challenges of measuring lp(a): a fight against hydra? Atherosclerosis. 2019; 289:181–183.
- Kronenberg F. Lipoprotein(a) measurement issues: Are we making a Mountain out of a molehill? Atherosclerosis. 2022; 349:123–135.
- Kang C, Dominguez M, Loyau S, et al. Lp(a) particles mold fibrin-binding properties of apo(a) in size-dependent manner: a study with different-length recombinant apo(a), native lp(a), and monoclonal antibody. Arterioscler Thromb Vasc Biol. 2002;22(7):1232–1238.
- Utermann G, Menzel HJ, Kraft HG, et al. Lp(a) glycoprotein phenotypes. Inheritance and relation to lp(a)-lipoprotein concentrations in plasma. J Clin Invest. 1987;80(2):458–465.
- Ruhaak L, Renee RF, Brkovic BZ, Dittrich JUHL, et al. Development of an LC-MRM-MS based candidate reference measurement procedure for standardization of serum apolipoprotein (a) tests. Clin Chem. 2023; 69(3):251–261.
- Kostner KM, Kostner GM, Wierzbicki AS. Is lp(a) ready for prime time use in the clinic? A pros-and-cons debate. Atherosclerosis. 2018; 274:16–22.
- Marcovina SM, Navabi N, Allen S, et al. Development and validation of an isoform-independent monoclonal antibody-based ELISA for measurement of lipoprotein(a). J Lipid Res. 2022;63(8):100239.
- Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods. 2009; 347(1-2):3–11.
- Siepen JA, Keevil EJ, Knight D, et al. Prediction of missed cleavage sites in tryptic peptides aids protein identification in proteomics. J Proteome Res. 2007;6(1):399–408.
- Smit NPM, Ruhaak LR, Romijn F, et al. The time has come for quantitative protein mass spectrometry tests that target unmet clinical needs. J Am Soc Mass Spectrom. 2021;32(3):636–647.
- Smit NPM, Romijn F, van Ham VJJ, et al. Quantitative protein mass-spectrometry requires a standardized pre-analytical phase. Clin Chem Lab Med. 2023;61(1):55–66.
- Dikaios IA, Angles-Cano E, Ceglarek U, et al. Commutability assessment of candidate reference materials for lipoprotein(a) by comparison of a MS-based candidate reference measurement procedure with immunoassays. Clin Chem Forthcoming. Clin Chem. 2023; 69(3):262–272.
- Marcovina SM, Albers JJ, Scanu AM, et al. Use of a reference material proposed by the international federation of clinical chemistry and laboratory medicine to evaluate analytical methods for the determination of plasma lipoprotein(a). Clin Chem. 2000;46(12):1956–1967.
- JCTLM. JCTLM Database: higher-order reference materials, methods and services. [cited 2023 Jan14] Available from: https://www.jctlmdb.org/#/app/home2022.
- Nordestgaard BG, Chapman MJ, Ray K, et al. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur Heart J. 2010;31(23):2844–2853.
- Piepoli MF, Hoes AW, Agewall S, et al. 2016 European guidelines on cardiovascular disease prevention in clinical practice: the sixth joint task force of the European society of cardiology and other societies on cardiovascular disease prevention in clinical practice (constituted by representatives of 10 societies and by invited experts)developed with the special contribution of the European association for cardiovascular prevention & rehabilitation (EACPR). Eur Heart J. 2016;37(29):2315–2381.
- Cardiovascular disease: risk assessment and reduction, including lipid modification. London: National Institute for Health and Care Excellence (NICE); 2023.
- Cegla J, Neely RDG, France M, et al. HEART UK consensus statement on lipoprotein(a): A call to action. Atherosclerosis. 2019;291:62–70.
- Correction to: 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease Executive summary: a report of the American college of cardiology/American heart association task force on clinical practice guidelines. Circulation. 2020; 141(16):e773.
- Ponte-Negretti CI, Isea-Perez JE, Lorenzatti AJ, et al. Atherogenic dyslipidemia in Latin America: prevalence, causes and treatment: expert’s position paper made by The Latin American academy for the study of lipids (ALALIP) endorsed by the Inter-American society of cardiology (IASC), the South American society of cardiology (SSC), the Pan-American college of endothelium (PACE), and the international atherosclerosis society (IAS). Int J Cardiol. 2017;243:516–522.
- Rhee EJ, Kim HC, Kim JH, et al. 2018 Guidelines for the management of dyslipidemia in Korea. J Lipid Atheroscler. 2019;8(2):78–131.
- Kinoshita M, Yokote K, Arai H, et al. Japan atherosclerosis society (JAS) guidelines for prevention of atherosclerotic cardiovascular diseases 2017. J Atheroscler Thromb. 2018;25(9):846–984.
- Klug E, et al. South African dyslipidaemia guideline consensus statement. S Afr Med J. 2012; 102(3 Pt 2):178–187.
- O'Callaghan CJ, Rong P, Goh MY. National guidelines for the management of absolute cardiovascular disease risk. Med J Aust. 2014; 200(8):454, 456.
- Scharnagl H, Stojakovic T, Dieplinger B, et al. Comparison of lipoprotein (a) serum concentrations measured by six commercially available immunoassays. Atherosclerosis. 2019;289:206–213.
- Wyness SP, Genzen JR. Performance evaluation of five lipoprotein(a) immunoassays on the roche cobas c501 chemistry analyzer. Pract Lab Med. 2021; 25:e00218.
- Lapic I, Segulja D, Dukic K, et al. Analytical validation of 39 clinical chemistry tests and 17 immunoassays on the alinity analytical system. Scand J Clin Lab Invest. 2022; 82(3):199–209.
