ABSTRACT
Food preferences and dietary habits are heavily influenced by taste perception. There is growing interest in characterizing taste preferences based on genetic variation. Genetic differences in the ability to perceive key tastes may impact eating behavior and nutritional intake. Therefore, increased understanding of taste biology and genetics may lead to new personalized strategies, which may prevent or influence the trajectory of chronic disease risk. Recent advances show that single nucleotide polymorphisms (SNPs) in the CD36 fat taste receptor are linked to differences in fat perception, fat preference, and chronic-disease biomarkers. Genetic variation in the sweet taste receptor T1R2 has been shown to alter sweet taste preferences, eating behaviors, and risk of dental caries. Polymorphisms in the bitter taste receptor T2R38 have been shown to influence taste for brassica vegetables. Individuals that intensely taste the bitterness of brassica vegetables (“supertasters”) may avoid vegetable consumption and compensate by increasing their consumption of sweet and fatty foods, which may increase risk for chronic disease. Emerging evidence also suggests that the role of genetics in taste perception may be more impactful in children due to the lack of cultural influence compared to adults. This review examines the current knowledge of SNPs in taste receptors associated with fat, sweet, bitter, umami, and salt taste modalities and their contributions to food preferences, and chronic disease. Overall, these SNPs demonstrate the potential to influence food preferences and consequently health.
Abbreviations
| SNP | = | Single nucleotide polymorphism |
| CVD | = | Cardiovascular disease |
| T2D | = | Type II diabetes |
| MetS | = | Metabolic syndrome |
| CD36 | = | Cluster determinant 36 |
| T1R2 | = | Type 1 receptor member 2 |
| T2R38 | = | Type 2 receptor member 38 |
| ENaC | = | Epithelial sodium channel |
| TRPV1 | = | Transient receptor potential cation channel subfamily V member 1 |
| MAF | = | Minor allele frequency |
| TBC | = | Taste bud cell |
| GPCR | = | G-protein coupled receptors |
| ATP | = | Adenosine triphosphate |
| LCFA | = | Long-chain fatty acid |
| BMI | = | Body mass index |
| LD | = | Linkage disequilibrium |
| FFA | = | Free fatty acids |
| TG | = | Triglycerides |
| HDL-C | = | High-density lipoprotein cholesterol |
| LDL-C | = | Low-density lipoprotein cholesterol |
| PTC | = | Phenylthiocarbamide |
| PROP | = | 6-n-propylthiouracil |
| ST | = | Supertaster |
| SL | = | Sweet liker |
| MSG | = | Mono-sodium glutamate |
| GMP | = | 5′ribonucleotides guanosine monophosphate |
| IMP | = | Inosine monophosphate |
| AMP | = | Adenosine monophosphate |
| mGluR1 | = | Metabotropic glutamate receptor type 1 |
| PKDL | = | Polycystic kidney disease-like |
| AS | = | Amiloride-sensitive |
| AI | = | Amiloride-insensitive |
1. Introduction
Similar to many species, humans are often driven to select foods based on their hedonic characteristics (Rolls, Citation2012; Turner-McGrievy et al., Citation2013). As such, the taste of food is an important factor when parents create dietary habits for themselves and their children (Kral and Rauh, Citation2010). In modern society, it is easy to gain access to large amounts of highly palatable foods containing saturated fats and added sugars. Obesity, cardiovascular disease (CVD), type 2 diabetes (T2D), and metabolic syndrome (MetS—may include dyslipidemia, elevated blood pressure, insulin resistance, abdominal obesity, and pro-inflammatory and thrombotic states) are chronic pathologies that have been partially attributed to adverse eating behaviors in humans compelled by the rewarding experience of taste perception (Rolls, Citation2012). A growing body of knowledge demonstrates the importance of understanding individual responses to medicine and nutrition (Gibney et al., Citation2014). Emerging research suggests that genetic predisposition and non-genetic factors such as life stage, eating behavior, physical activity, and gut microbiota also determine taste perception differently for every individual (Fay and German, Citation2008; Grimm and Steinle, Citation2011). It is therefore possible that due to different perceptions of taste, individual food preferences are important determinants of chronic disease risk.
Taste receptors on the tongue can be characterized in terms of their genetic variation to determine their contributions to specific eating behaviors and potentially the development of chronic diseases. Variations in receptor function for sweet, fat, and bitter tastes form much of the basis for the known inter-individual differences in taste perception (Mennella et al., Citation2005). The putative fat taste receptor cluster determinant 36 (CD36), the sweet taste receptors type 1 member 2 (T1R2) and T1R3, the bitter taste receptor T2R38, the umami fat taste receptors T1R1 and T1R3, and the salt taste receptors epithelial sodium channel (ENaC), and transient receptor potential cation channel subfamily V member 1 (TRPV1) contain single nucleotide polymorphisms (SNPs), which may alter taste perception, food preference, and consequently metabolic and health outcomes. Polymorphisms in this review will be stated with their unique SNP identifier and, where available, the minor allele frequency (MAF) in the CEU population from the International HapMap Project is indicated (2003). The aim of this review is to summarize current knowledge of SNPs in taste receptors associated with fat, sweet, bitter, umami, and salt taste modalities and their contributions to food preferences and chronic disease.
2. Brief overview of human taste perception
Taste or gustation is one of the five traditional senses. Humans are able to taste food and other substances with the tongue papillae, which each consist of hundreds of taste buds (Daniel D. Chiras, Citation2005). Between 2000 and 5000 taste buds exist along the surface of the front and back of the tongue, and each taste bud is lined with 50–100 taste bud cells (TBCs) (Daniel Schacter, Citation2009). Nutrients dissolved in saliva interact with taste receptors located at the apical side of a TBC.
Taste bud cells can be separated into four morphological subtypes: Types I, II, III, and IV (Chaudhari and Roper, Citation2010) (). It is thought that Type I cells play a supporting role for other TBCs, similarly to glial cells of the central nervous system. As such, Type I cells express proteins capable of clearing or degrading neurotransmitters following a synaptic event (Bartel et al., Citation2006). Additionally, the Type I cells comprise membrane ion channels that allow for the perception of salty taste from sodium chloride (Vandenbeuch et al., Citation2008; Yoshida et al., Citation2009; Chandrashekar et al., Citation2010). Type II cells can be further categorized based on the expression of sweet, bitter, and umami taste receptors (Adler et al., Citation2000; Nelson et al., Citation2001). These receptors are seven-transmembrane G-protein coupled receptors (GPCR). Sweet taste is elicited by a heterodimer of T1R2 and T1R3, whereas umami taste is elicited by a heterodimer of T1R1 and T1R3. Bitter taste can be initiated by a number of T2R proteins, depending on the particular bitter substance consumed. All Type II cells are thought to express the signaling enzymes phospholipase C β2 and transient receptor potential channel M5 downstream of the GPCRs, while only a subset express the G protein subunit α-gustducin (Yarmolinsky et al., Citation2009). Type II taste cells do not form traditional synapses with afferent nerve fibers. However, Type III cells do display presynaptic features including synaptic vesicles and vesicle fusion machinery such as synaptosomal-associated protein 25 (e.g., Clapp et al., Citation2006; DeFazio et al., Citation2006). Some Type III cells are sensors for sour taste (Huang et al., Citation2006a). Type IV cells are basal cells, which have been suggested to contain precursor populations that can differentiate into other TBC types (Sullivan et al., Citation2010). It is not known which TBC types contain the putative fat taste receptor CD36, but it has been hypothesized that Type II and/or Type III cells contain this receptor (Simons et al., Citation2011). One study used the cell culture method to determine that CD36 is expressed in a taste bud cell line (HTC-8) similar to Type II cells (Hochheimer et al., Citation2014).
Table 1. Three types of taste bud cells and their known functions.
Small molecule neurotransmitters signal between taste cells and/or between taste cells and intragemmal nerve fibers. These molecules and their functions in taste perception have been reviewed in detail by Chaudhari and Roper (Citation2010). As mentioned, Type II cells do not exhibit typical synapses. Instead, Type II cells respond to stimuli by releasing adenosine triphosphate (ATP) and acetylcholine through hemichannels (Finger et al., Citation2005; Huang et al., Citation2007; Romanov et al., Citation2007; Dando and Roper, Citation2009), which, in turn, activate purinergic receptors on cranial nerve fibers (Bo et al., Citation1999; Ogura, Citation2002; Dando and Roper, Citation2012; Yang et al., Citation2012). Type III cells, however, respond to stimulation by releasing serotonin, norepinephrine, and L-aminobutyric acid (Obata et al., Citation1997; Huang et al., Citation2008a, Citation2008b, Citation2009, Citation2011; Cao et al., Citation2009; Dvoryanchikov et al., Citation2011). The primary neurotransmitter that allows for taste cells to associate with afferent nerves appears to be ATP (Finger et al., Citation2005), while other neurotransmitters may mediate important taste cell functions through autocrine and paracrine signaling. Thus, other neurotransmitters may also contribute to the output of the taste bud (Chaudhari and Roper, Citation2010). Fatty acids, sugars/sweeteners, bitter compounds, sodium chloride, monosodium glutamate, and protons are examples of stimuli, which have the potential to initiate synaptic events leading to the sensation of taste.
3. Eating behavior may differ in children and adults
The maturation of taste and its consequences on eating behavior appear to be influenced early in life by genetics, as well as by environmental and cultural experiences. In adulthood, environmental and/or cultural experiences rather than genetic predisposition appear to be more correlated to taste perception and eating behavior (Drewnowski et al., Citation1999; Connors et al., Citation2001; Drewnowski and Specter, Citation2004; Scheibehenne et al., Citation2007; Monge-Rojas et al., Citation2014). While not consistently reported, several studies on the genetics of taste suggest that learned behaviors can override genetic predisposition. This may explain, for example, why adults consume bitter vegetables, which confer health benefits despite the bitter taste that instinctively promotes avoidance (Glanville and Kaplan, Citation1965; Rodin et al., Citation1976; Forrai and Bankovi, Citation1984; Mattes, Citation1985; Lucas and Bellisle, Citation1987; Niewind et al., Citation1988; Mattes and Labov, Citation1989; Jerzsa-Latta et al., Citation1990; Zeinstra et al., Citation2007; Kulkarni et al., Citation2013; Sharma and Kaur, Citation2014).
Twin studies provide evidence that eating behavior in adults is contributed in part by environment and genetics. In a longitudinal adult study investigating eating styles in 39 female and 45 male monozygotic Finnish twin pairs discordant for obesity or overweight, normal-weight twins were observed to have different eating styles than their obese siblings. Specifically, the normal-weight twins were less likely to report restrictive eating, overeating, and unhealthful food choices (Keski-Rahkonen et al., Citation2007). In contrast, a Swedish adult male twin study demonstrated that the heritability of eating behavior such as cognitive restraint, emotional eating, and uncontrolled eating were 59%, 60%, and 45%, respectively (Tholin et al., Citation2005). Similarly, another adult male twin study from the United Kingdom and Finland showed that the heritability of cognitive restraint, emotional eating, and uncontrolled eating were 26–63%, 9–45%, and 45–69%, respectively (Keskitalo et al., Citation2008). It is important to note that the heritability of obesity in these discordant twin studies does not refer to the heritability of specific genes, such as taste receptor genes (Tholin et al., Citation2005; Keski-Rahkonen et al., Citation2007; Keskitalo et al., Citation2008). None of the heritable factors investigated in the twin studies have been attributed to specific polymorphic variations in taste receptors. Therefore, future studies in twins discordant for obesity should explore taste receptor genetic variation as a heritable factor, which influences eating behavior. Furthermore, no genetic analyses have been done in elderly participants in association with taste perception, taste preferences, nutritional status, or health outcomes. The reason for the lack of genetics studies could be that the loss of taste sensitivity, as a result of reduced TBC regeneration, in the elderly would strongly confound genetic components of taste as causes of change in taste outcomes and health outcomes (Fukunaga, Citation2005).
In children, studies examining the association between genetics of taste and eating behavior are limited and may not be generalized from adult studies. Compared to studies with adults, research with children shows stronger correlation between genetic predisposition for certain eating behaviors and their actual consumption patterns (Sharma and Kaur, Citation2014). This relationship is especially pronounced in the tasting of bitter compounds by young children. It is thought that aversion to bitter taste by young children is an evolutionary adaptation to prevent the ingestion of toxic plant compounds or other harmful bitter substances, which occur in nature (Zeinstra et al., Citation2007). These few observations present a potential new line of investigation to explore ways to develop positive eating habits in children through a better understanding of the genetics of taste on eating behavior (Zeinstra et al., Citation2007; Sharma and Kaur, Citation2014).
4. Fat taste
Oleogustus is the most recently identified taste modality (Running et al., Citation2015). The taste perception of long-chain fatty acids (LCFAs) was previously thought to only be dependent on the texture of the lipid, and the olfactory stimulus to a lesser extent (Ramirez, Citation1993; Greenberg and Smith, Citation1996; Tepper and Nurse, Citation1997). There is now an abundance of evidence supporting taste as a method of oral detection of dietary fat, including saturated fat () (Fukuwatari et al., Citation2003; Mattes, Citation2009; Running et al., Citation2015). LCFAs provide this stimulus in both rodents (Tsuruta et al., Citation1999; Takeda et al., Citation2000) and in humans (Chale-Rush et al., Citation2007). In the mouse, knockout experiments for LCFA receptor genes such as CD36 indicated a decrease in the spontaneous attraction for fat-enriched foods (Laugerette et al., Citation2005; Sclafani et al., Citation2007).
Table 2. Taste as a method of oral fat detection and the role of CD36.
The fatty acid translocase CD36 has been the subject of much of the research related to LCFA taste perception. CD36 is expressed in human TBCs (Simons et al., Citation2011), and has a strong affinity for LCFA (Baillie et al., Citation1996). CD36 knockout mice have been shown to lack the ability to detect LCFA in behavioral tests (Laugerette et al., Citation2005; Sclafani et al., Citation2007). The oral perception of linoleic acid (an n-6 polyunsaturated fatty acid), α-linolenic acid (an n-3 polyunsaturated fatty acid), and oleic acid (an n-9 monounsaturated fatty acid) has been attributed to the CD36 TBC receptor (Keller et al., Citation2012; Hochheimer et al., Citation2014). Additionally, a GPCR has been found to bind LCFA and influences the perception of lipids in the oral cavity. GPCR120-null mice show decreased preference for LCFA-enriched solutions in eating behavioral tests compared to wild type (Cartoni et al., Citation2010). Thus, LCFAs as gustatory stimuli are more complex than initially understood.
The consumption of energy-dense foods may be linked to enhanced palatability of dietary fats, which predispose individuals to metabolic complications (Stewart et al., Citation2011; Liang et al., Citation2012). The relationship between sensory fat perception and predisposition to weight gain has been shown in recent adult human studies (Stewart et al., Citation2010, Citation2011; Liang et al., Citation2012). This relationship outlines the remarkable difference in obese and lean subjects with respect to fat intake and taste sensitivity. In an obese rat model, an inverse correlation between gustatory sensitivity to LCFAs and dietary preference for fat was observed (Gilbertson et al., Citation1998). Correspondingly, a higher sensitivity to tasting LCFA has been correlated to reduced fat intake, total caloric intake, and body mass index (BMI; Stewart et al., Citation2010). Similarly, a high-fat diet decreased the taste sensitivity to oleic acid in lean, but not obese patients (Stewart and Keast, Citation2012). These observations suggest that a lack of sensitivity to fat taste increases fat intake, and may partly explain why obese adult individuals seem to consume fatty foods more frequently than lean patients (Stewart et al., Citation2011; Liang et al., Citation2012). Therefore, the implications of these findings on eating behavior and health have potential significance.
A study by Ma et al. investigated associations between common SNPs in the CD36 gene, lipid and glucose metabolism, and risk for cardiovascular disease (Ma et al., Citation2004). This group genotyped 21 polymorphisms, which evenly spanned the CD36 gene, revealing two linkage disequilibrium (LD) blocks represented by a haplotype. The haplotype comprised the following five SNPs: (1) −33137A/G (rs1984112; MAF = 0.3468), (2) −31118G/A (rs1761667; MAF = 0.3904), (3) 25444G/A (rs1527483; MAF = 0.1018), (4) 27645 deletion /insertion (del /in) (rs3840546; MAF not reported), and (5) 30294G/C (rs1049673; MAF = 0.3832). In a population of adults of European descent, the 30294C polymorphism was significantly associated with higher plasma free fatty acids (FFA), with a stronger association among men compared to women. A similar increase in plasma FFA levels was seen in men carrying the −33137A and −31118G polymorphisms. In the order of SNPs listed above, the haplotype of an individual can be represented by sequentially naming the nucleotide bases. Men with the AGGIG haplotype had 31% higher FFA and 20% higher plasma triglycerides (TG) than non-carriers. The AGGIG haplotype was also correlated to an increased risk of coronary artery disease in T2D American participants (n = 197) and Italian participants (n = 321). In the scope of this study by Ma et al., CD36 modulated lipid metabolism and CVD risk in adults of European descent. However, it is unclear whether this modulation occurred at the level of taste perception or metabolism. Further research is required to discern the effects of taste perception and lipid transport on the levels of FFA and TG in the plasma, or if this modulation would also be seen in other ethnicities.
CD36 plays an important role in lipid metabolism and polymorphisms within the CD36 gene have been linked to CVD risk factors (Noel et al., Citation2010). The relationship between genotypes and haplotypes of five SNPs in the CD36 gene (−33137G, −31118A, −22674C (rs2151916; MAF = 0.3339), 27645 del/ins and 30294C with lipid levels have been examined in young normal-weight subjects (Ramos-Arellano et al., Citation2013). High-density lipoprotein cholesterol (HDL-C) levels were lower in −22674C carriers than −22674T carriers, and TT homozygotes had lower oxidized low-density lipoprotein cholesterol (LDL-C) levels compared to −22674C allele carriers. LDL-C levels were higher in carriers of the CC genotype for the 30294C polymorphism compared to non-carriers. Subjects carrying the AATDC haplotype had 3.2 times higher risk of LDL-C > 100 mg/dL than those carrying the AGTIG haplotype, whereas subjects carrying the AATIC haplotype had 2.0 times higher risk of total cholesterol > 200 mg/dL than the AGTIC haplotype. Therefore, genetic variation in CD36 may modulate CVD risk by affecting lipid metabolism.
Single nucleotide polymorphisms in the CD36 gene have also been associated with metabolic disorders related to excess fat depots in adult populations (Lepretreet al., Citation2004a, Citation2004b; Corpeleijn et al., Citation2006; Love-Gregory et al., Citation2008; Ramos-Arellano et al., Citation2013), and a (TG)-repeat in intron 3 has been linked to elevated BMI in Korean patients with coronary heart disease (Yun et al., Citation2007). Bokor et al. assessed the link between CD36 SNPs and the risk of obesity in a case–control study comprising 307 obese (age = 15.0 ± 1.1 years) and 339 normal-weight (age = 14.6 ± 1.1 years) adolescents (Bokor et al., Citation2010). Four SNPs (rs3211867, rs3211883, rs3211908, and rs1527483) were associated with increased obesity risk, higher BMI, and higher percent body fat. A haplotype consisting of the minor alleles of these SNPs was also linked to obesity and excess adiposity (i.e., higher BMI and percent body fat).
Studies examining correlative relationships between genetic variations in CD36 and chronic disease have focused on predispositions to specific eating behaviors (). Three CD36 gene polymorphisms (−31118A and 25444A and 27645ins), which correspond to part of the haplotype studied by Ma et al., were investigated for their associations with changes in fat taste perception (Keller et al., Citation2012). Homozygotes for the A allele at locus −31118 reported greater perceived creaminess, independently of fat concentration of salad dressings and higher mean acceptance of added fats and oils compared to other CD36 genotypes. Participants with the CT or TT genotypes at 25444 also perceived greater fat content in the salad dressings, regardless of actual fat concentration. 27645 deletion homozygotes had higher waist circumference and BMI than ins/del or ins/ins individuals (p < 0.001, n = 2), however this particular observation requires further replication due to small sample size. Furthermore, Pepino et al. reported findings on the −31118A SNP and fat preference (Pepino et al., Citation2012). Twenty-one obese participants with the locus variations AA (n = 6), AG (n = 7), and GG (n = 8) were studied with respect to oleic acid and triolein orosensory detection thresholds. The major and minor alleles of this SNP vary depending on the population. In populations with African ancestry, the G allele is minor, whereas the opposite is true in individuals of European descent. G allele homozygotes had eight-fold lower oral detection thresholds (i.e., higher sensitivity) for oleic acid and triolein than the A allele, which was associated with a decrease in CD36 expression. Intermediate thresholds were observed in heterozygotes. Higher fat taste sensitivity is associated with a decrease in fat preference (Stewart et al., Citation2010). However, these findings conflict with other studies on the effect of the -31118 SNP on CD36 expression and function. For instance, Ma et al. hypothesized that the G homozygosity for the −31118 SNP is associated with decreased CD36 function, along with the other associated SNPs in LD (Ma et al., Citation2004). This was hypothesized due to higher plasma FFA and TG in the presence of normal glucose and insulin levels in G allele homozygotes. Higher plasma FFAs are indicative of defective CD36 due to the role of this receptor to translocate FFA into the muscle and other tissues from the plasma (Febbraio et al., Citation1999). The increased FFA in the plasma is redirected to the liver, which takes up FFAs independently of CD36 (Goudriaan et al., Citation2003). The liver can then synthesize TGs and release them into the plasma, which may also explain why TGs are high in G allele homozygotes. If the G allele of −31118 is associated with decreased function in the skeletal muscle, and the SNP is located in the promoter of the gene, one would hypothesize that the decreased function might be the result of a decreased expression of the receptor in skeletal muscle. Decreased expression of CD36 on the tongue should lead to decreased taste sensitivity to fat, hence an increase in fat intake and higher plasma FFAs and TGs. However, the oral fat perception studies that have examined the −31118 SNP have associated the A allele, not the G allele, with decreased fat taste sensitivity and greater preference for fat. This apparent discrepancy could be due to differences in CD36 promoter activity between skeletal muscle and tongue. This difference can be investigated in the future by assessing the expression of CD36 in these tissues in a group of individuals genotyped for the −31118 SNP as well as other associated SNPs in LD. This information would be useful in consolidating current studies on CD36 genetic variation and its effects on fat taste perception and metabolic profile. It would also serve to inform individuals on how they can optimize their diets to improve their metabolic profile based on their CD36 genetic profile (Madden et al., Citation2008).
Table 3. Associations between CD36 gene SNPs fat taste, eating behavior, and health outcomes.
Lean mice and humans can be more sensitive to LCFA-rich foods such as butter, oil, and fatty meat while obese individuals have decreased taste sensitivity to fatty acids due to particular CD36 variants. Therefore, CD36 genetic variation may have the potential to influence fat intake and weight gain by decreasing the oral taste sensitivity of LCFAs in individuals. CVD and dyslipidemia may result from this eating behavior associated with fat overconsumption, especially when the dietary fat predominantly consists of saturated fats (Colin-Ramirez et al., Citation2014). Although studies to date are limited, they provide promising data on the link between genetics, fat perception, and fat preference. It is important to characterize genetic variations in putative fat taste receptors in order to more clearly understand the role of fat taste perception in the development of chronic metabolic disease.
5. Sweet taste
The only receptors known to be involved in the perception of sweet taste are T1R2 and T1R3, which form a heterodimer (Li et al., Citation2002). These transmembrane proteins allow humans to taste a variety of sweet substances such as naturally occurring sugars (glucose, sucrose, fructose, and sugar alcohols), D-amino acids (D-tryptophan and D-phenylalanine), and glycosides (stevioside and glycyrrhizin), as well as alternative sweeteners such as sucralose, aspartame, neotame, saccharin sodium, acesulfame potassium, and cyclamate (Chandrashekar et al., Citation2006). Moreover, naturally occurring sweet proteins, such as brazzein, thaumatin, and monellin, and naturally occurring taste-modifying proteins, such as neoculin and miraculin, also bind to T1R2 and T1R3 (Temussi, Citation2002; Jiang et al., Citation2004; Cui et al., Citation2006; Nakajima et al., Citation2006; Walters and Hellekant, Citation2006; Ssadi-Porter et al., Citation2010). Although both receptors are needed to elicit sweet taste, T1R2 is the subunit specific to sweet taste perception because T1R3 is also involved in the detection of the umami taste when it dimerizes with T1R1 (Nelson et al., Citation2002).
Genetic variations in T1R2 and T1R3 are important to characterize because they pertain to changes in taste sensitivity to sugar (Reed et al., Citation1997; McDaniel and Reed, Citation2003; Reed et al., Citation2003,Citation2006; Kim et al., Citation2004; Drayna, Citation2005; Reed and McDaniel, Citation2006; Keskitalo et al., Citation2007a, Citation2007b, Citation2008), summarized in . In African, Asian, European, and Native American populations, all three T1R genes have a multitude of polymorphisms. These SNPs include non-synonymous SNPs that are located in the N-terminal extracellular domain, the part of the receptor where ligands for taste are thought to bind. Being particularly polymorphic in comparison with other human genes, T1R2 is within the top 5–10% of all human genes with regards to the reported number of polymorphisms. This increased polymorphic rate was hypothesized to be associated with variations in sweet taste perception (Kim et al., Citation2006). One study sought to determine whether Ile191Val variations in T1R2 were linked to differences in the consumption of sugars in 1037 diabetes-free young adults, and 100 individuals with T2D (Eny et al., Citation2010). They demonstrated an association between the Ile191Val SNP (rs35874116; MAF = 0.2670) in T1R2 and BMI in individuals with a BMI ≥ 25. In the diabetes-free individuals, Val carriers (homozygous or heterozygous) consumed significantly fewer sugars compared to homozygous Ile carriers. This finding was consistent in T2D individuals where Val carriers also consumed significantly less sugar compared to Ile homozygotes. Genetic variation in the other half of the heterodimer, T1R3, was explored in a separate study by Fushan et al. They showed that intronic SNPs in the T1R3 promoter were linked to sucrose taste sensitivity in humans by altering T1R3 transcription levels (Fushan et al., Citation2009). These SNPs accounted for 16% of the population's variability in sweet perception (Fushan et al., Citation2009). The associations between genetic variations in sweet taste receptors and consumption of sweet foods are important findings for individuals who are at risk for metabolic complications due to their eating behavior.
Table 4. Associations between T1R2/T1R3 gene SNPs and sweet taste, eating behavior, and health outcomes.
In children, the risk for developing dental caries is associated with the consumption of sweet foods (Weisberger, Citation1950). One study has investigated an association between the prevalence of dental caries and the Ile191Val polymorphism (T1R2) in 80 young adults of European descent (Kulkarni et al., Citation2013). Val carriers of the T1R2 SNP, that is, individuals who have been shown to have lower consumption of sweet foods, had lower risk of developing dental caries. A similar finding was observed in a study conducted in schoolchildren. A significant association between total caries was observed with the Ile191Val (T1R2) and the rs307355 SNPs (T1R3). Moderate number of caries (4–7 caries) was observed in rs307355 (MAF = 0.2364) heterozygotes, while high-risk caries (>8 caries) was observed in Val homozygotes of the T1R2 SNP (Haznedaroglu et al., Citation2015). Therefore, individuals who are genetically predisposed to prefer sweet foods should also consider that their eating behavior influences not only metabolic risk factors, but also dental health. The profundity of those implications must be verified with more studies that consider this T1R2 SNP and other polymorphisms in sweet-sensing genes. Nevertheless, these findings suggest a highly relevant outcome in children as well as young adults.
6. Bitter taste
Bitter and sweet taste profiles interact together to affect eating behavior, and this phenomenon is seen especially in children (Mennella et al., Citation2005). High concentrations of bitter substances generally result in food rejection, which corresponds to an evolutionary adaptation to avoid toxic substances such as rancid fat, hydrolyzed protein, and plant alkaloids (Glendenning, Citation1994; Hladik and Simmen, Citation1996). Higher sensitivity to bitter taste may cause individuals to avoid consuming vegetables rich in anti-tumor and anti-oxidant compounds and may, consequently, lead to a higher consumption of sweet and fatty foods as substitutes. This eating behavior has the potential to increase the risk of CVD, obesity, and cancer (Goldstein et al., Citation2005).
Although there are 25 different types of T2Rs for bitter taste perception on Type II TBCs (Dong et al., Citation2009), only T2R38 has been associated with a genetically predetermined bitter taste status (Kim et al., Citation2003). Kim et al. identified the bitter receptor T2R38 as being responsible for tasting phenylthiocarbamide (PTC) and 6-n-propylthiouracil (PROP) (Kim et al., Citation2003). Three common SNPs were found in this gene, which results in three amino acid substitutions at residues P49A (rs713598; MAF = 0.4952), A262V (rs1726866; MAF = 0.4255), and V296I (rs10246939; MAF = 0.4794). The bitter-tasting phenotype is attributed to a combination of these three SNPs resulting in a PAV amino acid haplotype, while non-tasters carry the AVI amino acid haplotype (Bufe et al., Citation2005). Homozygosity for the PAV haplotype is considered to be a marker of “supertaster” status while those who are homozygous for the AVI haplotype are considered “non-tasters,” and heterozygotes have an intermediate phenotype. This phenotype is also observable by simply assessing the intensity (or lack thereof) of the bitter taste impregnated on a strip of filter paper covered in PTC.
A supertaster is a person who experiences the sense of bitter taste with far greater intensity than average (Bartoshuk et al., Citation1994). This amplified oral perception appears to have a biological basis rather than being the result of response bias or a scaling artefact (Turner-McGrievy et al., Citation2013). The underlying cause of this relatively heightened oral response is not known, although it is thought to be due to both an increased number of fungiform papillae and the T2R38 phenotype (Duffy et al., Citation2004; Bufe et al., Citation2005); however, those factors do not completely explain the supertasting phenomenon as it may be attributed to other orosensory phenotypes (Hayes et al., Citation2008). Among adults, the T2R38 genotype has been linked to avoidance of alcoholic beverages (Duffy et al., Citation2004), increased prevalence of colon cancer through inadequate vegetable consumption (Glendenning, Citation1994), avoidance of cigarette smoking (Cannon et al., Citation2005), and a preference for sweetness (discussed in detail below).
A variety of studies, summarized in , have linked PTC taste status with eating behavior. Two studies involving young, healthy women observed an inverse relationship between bitter sensitivity and an acceptance of brassica vegetables, spinach, coffee, and tart citrus flavors (Drewnowski et al., Citation1998, Citation1999). Furthermore, supertaster women showed a decreased acceptance of sweet and fatty foods (Duffy and Bartoshuk, Citation2000). T2R38 taster status has also been associated with a preference for sweet-tasting foods in children, but not adults (Mennella et al., Citation2005; Timpson et al., Citation2005; Ooiet al., Citation2010); however, this result was not replicated in another study examining children (O'Brien et al., Citation2013). Bitter tasters ate fewer soy products and drank less green tea, and rated these foods to be more bitter than non-tasters (Gayathri et al., Citation1997). These eating behaviors, which are influenced by the T2R38 phenotype, can lead to adverse health effects (Goldstein et al., Citation2005). However, a study by Choi et al. did not observe any differences in food preference with varying bitter taste status based on a questionnaire (Choi, Citation2014).
Table 5. Associations between T2R38 gene SNPs and bitter/sweet taste, eating behavior, and health outcomes.
Turner McGrievy et al. investigated the relationship of supertasting (bitter) and sweet preference with MetS and dietary intake (Turner-McGrievy et al., Citation2013). In this study, 38% of the participants met criteria for MetS. After classifying adult study participants with respect to sweet liking (SL) or supertasting (ST), the study found a correlation between the combination of SL and ST statuses (SL+ST) and higher incidence of MetS. Participants who were either SL or ST had a significantly reduced risk of MetS compared to those who were SL and ST. In addition, those who were SL and ST consumed less fiber than SL + non-ST individuals. This study highlights an interaction between SL and ST statuses that may be linked with MetS. This is not the only study to indicate a possible relationship between taster status and weight. Indeed, Tepper et al. showed that those who are STs have a lower BMI than non-tasters (Tepper, Citation2008) while another study observed that mean stature of adult PTC non-tasters was higher than that of tasters, and tasters had higher body fat % than non-tasters (Sharma and Kaur, Citation2014).
Taken together, studies on bitter tasting phenotype suggest a complex relationship with other taste modalities. Despite the evidence presented thus far, the association between taster profile and health outcomes, weight, and dietary intake has been mixed, with studies showing variable results (Tepper and Nurse, Citation1998; Drewnowski et al., Citation2007; Tepper et al., Citation2008). Given that certain adverse eating behaviors may result from having a supertaster phenotype, further characterizing the effect of bitter taste receptor variation on food preferences has implications for metabolic and health outcomes.
7. Umami taste
The name “umami” originates from the Japanese language, which means “delicious savory taste” (Ikeda, Citation2002). The GPCR heterodimer complex of T1R1 and T1R3 elicits the umami taste when interacting with amino acids, typically mono-sodium glutamate (MSG), and this interaction occurs synergistically with 5′ribonucleotides guanosine monophosphate (GMP), inosine monophosphate (IMP), and adenosine monophosphate (AMP) (Nelson et al., Citation2002). The TBC receptors metabotropic glutamate receptor 1 (mGluR1) and mGluR4 have also been implicated in this taste modality (Toyono et al., Citation2002, Citation2003).
Perception of umami serves as an indicator of purine-rich foods, in particular, the purine-derived metabolite uric acid. Elevated serum urate (a salt derived from uric acid) has previously been considered benign except for carrying an increased risk for gout (varez-Lario and arron-Vicente, Citation2010). However, serum urate has recently been linked to a multitude of biologic effects including high blood pressure, increased hepatic lipogenesis, and insulin resistance (Baldwin et al., Citation2011; Lanaspa et al., Citation2012). These adverse health effects suggest that the consumption of certain umami tasting foods, similar to sweet and fatty foods, may increase metabolic disease risk.
There is evidence that differences in the perception of umami are common among individuals with taste threshold concentrations of umami stimuli varying up to five-fold (Lugaz et al., Citation2002). Similar to bitter taste perception, a fraction of the population may carry a “non-taster” phenotype for umami (Lugaz et al., Citation2002), but more research is needed to characterize this phenotype. Despite the lack of research linking differences in umami taste sensitivity to health outcomes, some studies have correlated certain SNPs in umami taste receptors to differences in taste perception (). One such study was conducted where 17 T1R1 and T1R3 SNPs were examined in Japanese adults (Shigemura et al., Citation2009). The purpose of the study was to demonstrate a link between SNPs and detection thresholds for IMP and MSG. Taste thresholds for three common SNPs: Gln12His (rs75881102; MAF = 0.0114) and Ala372Thr (rs34160967; MAF = 0.1354) in T1R1 and Arg757Cys (rs307377; MAF = 0.0481) in T1R3 were investigated. The T1R1 SNP Ala372Thr (rs34160967) appeared to be non-synonymous as the Thr allele was associated with increased sensitivity to umami. This association was significant for MSG as well as a mixture of MSG and IMP, but not for IMP alone. The T1R3 SNP Arg757Cys also appeared to be non-synonymous for all umami tasters, as the Cys757 allele yielded a reduced sensitivity for umami taste. Therefore, it is possible that the T1R1 and T1R3 heterodimer have separate binding sites for IMP and MSG, and the T1R3 757 and T1R1 372 SNPs are located in the active sites for IMP and MSG binding, respectively. Shigemura et al. have also shown that that these two SNPs are not in LD, but that their functions may overlap to a certain degree. Thus, having the T1R1 Thr372 SNP and the T1R3 Arg757 SNP causes an overall decrease in umami taste threshold, and having the T1R1 Ala372 SNP and the T1R3 Cys757 SNP causes an overall increase. This was confirmed in another study where the Cys757 variant reduced stimulation of the heterodimer by MSG (Raliou et al., Citation2011). More research is needed to better understand the influence of genetics on the preference or intake of MSG-rich foods. Further characterization of the genetic variation implicated in this taste modality would allow individuals to determine their predisposition to preferring umami foods. This knowledge may prove to be important given that moderating the consumption of umami foods may reduce the risk for metabolic complications.
Table 6. Associations between T1R1 and T1R3 gene SNPs and umami taste perception.
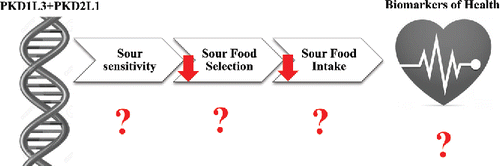
8. Sour taste
Taste receptor function for sour tasting, which involves GPCR-mediated signaling, is not well-characterized. Studies on the variation at the genetic loci for taste receptor genes involved in sour taste have yet to be undertaken—however, some important advances have been made with regards to this taste modality in a thorough and recent review by Garcia-Bailo et al. (Citation2009). Major findings were related to the biology of sour taste perception and the palatability of sour foods (Richter et al., Citation2003; Kim et al., Citation2004; Ishimaru et al., Citation2006; Lopez Jimenez et al., Citation2006; Huang et al., Citation2006b). In general, sour taste perception is thought to be elicited upon the stimulation of taste buds by acidic substances, leading to depolarization of TBCs. Many animals find mildly acidic foods to be palatable, but most mammals reject strong sour stimuli as a means to avoid the consumption of spoiled foods. Putative sour taste receptors PKD2L1 and PKD1L3 belong to the polycystic kidney disease-like (PKDL) subfamily of transient receptor potential ion channels and function as a heteromer to elicit sour taste. The PKD2L1 and PKD1L3 genes carry coding SNPs, which may influence sour taste perception. Exploring the effect of these SNPs on sour taste perception and subsequent food choices are important future directions for this taste modality.
9. Salt taste
Salt taste perception influences sodium intake, the excess of which corresponds to a major public health concern due to the risk of developing hypertension. Excessive sodium consumption may be attributed to greater individual preferences for salty foods, which may be linked to genetic factors such as salt taste receptor function. However, the source of inter-individual differences in salt preferences is controversial as environmental influences, such as dietary salt intake, may be more important than genetic predisposition to salt preference (Bertino et al., Citation1982; Wise et al., Citation2007). Thus, salt preference may be due to sensory habituation to high sodium foods and a subsequent increase in the preference of these foods. Similar to sour taste, salt taste receptors have yet to be characterized in sufficient detail to draw definitive conclusions about differences in salt taste receptor function and possible implications for differences in eating behavior and health outcomes. Due to the fundamental importance of electrolyte consumption for mammalian physiology, the desire to consume sodium-rich foods could be attributed to physiological factors rather than to the search for the sensory pleasure of salt taste (Wald and Leshem, Citation2003). The physiological basis for seeking electrolytes is supported by periodic fluctuations that occur in women more than men, likely due to hormonal cycling (Hayes et al., Citation2010).
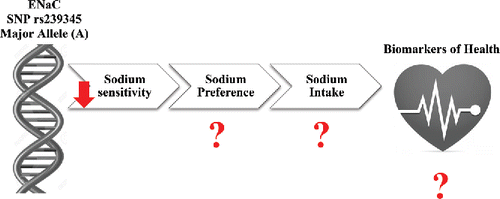
ENaC and TRPV1 are the two putative salt taste receptors (Dias et al., Citation2013). Studies have shown that nerve fibers responding to sodium can be classified according to their sensitivities to the ENaC blocker, amiloride, which distinguishes amiloride-sensitive (AS) and amiloride-insensitive (AI) groups (Yoshida et al., Citation2009). The AS fibers respond specifically to sodium chloride (NaCl) whereas AI fibers respond broadly to various electrolytes including NaCl. It has been proposed that while the AS mechanism of salt detection is dependent on ENaC, the AI mechanism is mediated by the function of TRPV1. Thus, it has been suggested that the appealing taste of sodium, representative of a lower taste concentration threshold, is mediated by the AS fibers and hence the ENaC protein while aversive reactions to tasting high concentrations of sodium are mediated by the AI fibers using TRPV1 (Yoshida et al., Citation2009). These data suggest that at least two different systems exist to facilitate salt taste transduction in taste cells via AS and AI fibers.
In a study by Dias et al., SNPs in ENaC and TRPV1 receptors were associated with differences in salt taste perception among adults (Dias et al., Citation2013). In the ENaC gene, two intronic SNPs modified salt taste sensitivity. Homozygotes for the A allele of rs239345 (A/T) (MAF = 0.2642) and the T allele of rs3785368 (C/T) (MAF = 0.1899) perceived salt solutions less intensely than carriers of the other respective alleles (p = 0.02; p = 0.03, respectively). In the TRPV1 gene, the Val585Ile polymorphism rs8065080 (C/T) (MAF = 0.3177) modified taste sensitivity where carriers of the T allele were significantly more sensitive to salt solutions than the CC genotype (p = 0.008). These findings suggest that genetic variation in the ENaC and TRPV1 genes may contribute to inter-individual differences in salt taste perception.
Altogether, environmental influences and genetic factors play a role in defining a salt-preferring phenotype, but environmental influences may have a more dominant role. Nevertheless, new genetic findings associating TRPV1 and ENaC to salt preference suggests that there is more to learn about genetic influences (Dias et al., Citation2013) (). Due to the novelty of this type of research, the extent to which such variation influences preference or intake of salty foods is unknown. Future studies are needed to investigate the role of salt taste receptor SNPs in sodium consumption and across the lifespan.
Table 7. Associations between ENaC and TRPV1 gene SNPs and salt taste perception.
10. Summary and conclusion
The relatively new and active field of taste research shows great promise to enhance our fundamental understanding of how taste receptors SNPs contribute to eating behavior, health, and disease. For illustrative purposes, a single candidate SNP is used to demonstrate the effect of genetic variation on taste perception, dietary intake, and health outcomes (). In particular, genetic variation in the CD36 LCFA receptor (), the T1R2 sweet taste receptor (), and the T2R38 bitter taste receptor () has important health implications because they may predispose individuals to the over-consumption of unhealthy foods and the under-consumption of healthy foods. Investigation into the roles of SNPs in taste receptor genes for umami, sour, and salt tastes () is also warranted due to their potential to influence taste preference, dietary intake, and health. An important consideration in future research requires attention to taste-driven food preferences in children and adults, which may be genetic in origin in children but uncoupled in adults due to cultural influences. Additional studies on taste preferences in children are needed, which may help establish healthy eating habits unique to every child (Nielsen and El-Sohemy, Citation2014; Nielsen et al., Citation2014). As food palatability often involves a combination of different types of taste, such as salt and fat, studies examining combinations of SNPs across different taste modalities are warranted. Sex-specific differences in various ethnicities are also important factors to be considered in future studies. Overall, new research in taste may aid in tailoring food choices and reducing the risk for obesity, T2D, CVD, and other chronic diseases.
Figure 1. Schematic representation of the effect of the rs1761667 SNP in the CD36 fat taste receptor gene on fat taste perception, fatty food selection, fat intake, and biomarkers of health such as blood TG levels, cholesterol levels, and anthropometrics. The effect of the G allele on CD36 function in the tongue as a fat taste receptor and in the muscle as a fatty acid transporter is controversial. While taste perception studies have been able to consistently show that G allele homozygotes have higher taste sensitivities to fatty acids due to the increased expression of the receptor, others have suggested that the G allele is associated with decreased function in the muscle as a fatty acid transporter. Therefore, there is a discrepancy between the expected decrease in plasma TGs due to taste effects and the observed increase in plasma TGs due to the lack of FA transport into the muscle.
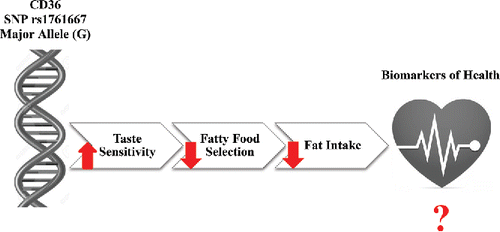
Figure 2. Schematic representation of the effect of the T allele in the rs35874116 SNP in the T1R2 sweet taste receptor gene on sweet taste perception, sweet food selection, sugar intake, and biomarkers of health such as dental caries and BMI. While this SNP has been related to increased preference as well as intake of sweet foods, the effect of the SNP on taste sensitivity has not been elucidated. This SNP has been linked to increased dental caries, and individuals with increase sweet preference and intake were also more likely to have higher BMIs. Importantly, a different SNP in the T1R2 sweet taste receptor (rs12033832) was found to be associated with altered sweet taste sensitivity.
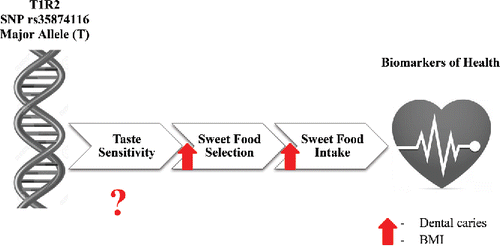
Figure 3. Schematic representation of the effect of the G allele in the rs713598 SNP in the T2R38 bitter taste receptor gene on bitter taste perception, bitter and sweet food selection, sugar, fruit, and vegetable intake, and biomarkers of health such as body fat % and plasma glucose. The G allele of this SNP is well established to be part of a haplotype, which causes the PTC non-taster phenotype. This phenotype, unlike the ‘‘supertaster” phenotype, is not prone to decreased vegetable preference and consumption or a compensatory increase in the preference and consumption of sweet food.
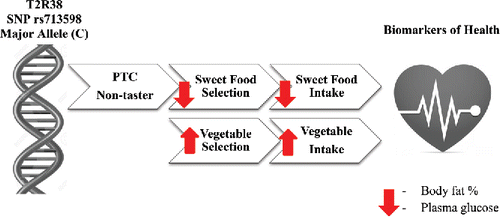
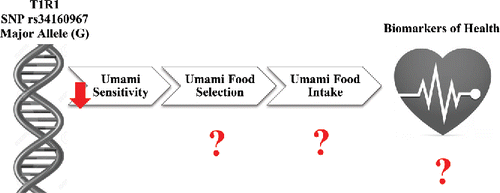
Figure 4. Schematic representation of the effect of the G allele in the rs34160967 SNP in the T1R1 umami taste receptor on umami taste perception, umami food selection, umami food intake, and biomarkers of health.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
E.C.: conception of review, design, analysis and interpretation of data, drafting of manuscript and critical revision. D.M.M.; E.A.V.; A.C.B.; A.M.D.; L.L.S.; J.H.: critical revision, and D.W.L.M.: conception of review, design, and critical revision.
Funding
Funding was received from the Health for Life Initiative at the University of Guelph.
References
- Adler, E., Hoon, M. A., Mueller, K. L., Chandrashekar, J., Ryba, N. J. and Zuker, C. S. (2000). A novel family of mammalian taste receptors. Cell 100:693–702.
- Baillie, A. G., Coburn, C. T. and Abumrad, N. A. (1996). Reversible binding of long-chain fatty acids to purified FAT, the adipose CD36 homolog. J. Membr. Biol. 153:75–81.
- Baldwin, W., McRae, S., Marek, G., Wymer, D., Pannu, V., Baylis, C., Johnson, R. J. and Sautin, Y. Y. (2011). Hyperuricemia as a mediator of the proinflammatory endocrine imbalance in the adipose tissue in a murine model of the metabolic syndrome. Diabetes 60:1258–1269.
- Bartel, D. L., Sullivan, S. L., Lavoie, E. G., Sevigny, J. and Finger, T. E. (2006). Nucleoside triphosphate diphosphohydrolase-2 is the ecto-ATPase of type I cells in taste buds. J. Comp. Neurol. 497:1–12.
- Bartoshuk, L. M., Duffy, V. B. and Miller, I. J. (1994). PTC/PROP tasting: Anatomy, psychophysics, and sex effects. Physiol. Behav. 56:1165–1171.
- Bertino, M., Beauchamp, G. K. and Engelman, K. (1982). Long-term reduction in dietary sodium alters the taste of salt. Am. J. Clin. Nutr. 36:1134–1144.
- Bo, X., Alavi, A., Xiang, Z., Oglesby, I., Ford, A. and Burnstock, G. (1999). Localization of ATP-gated P2×2 and P2×3 receptor immunoreactive nerves in rat taste buds. Neuroreport 10:1107–1111.
- Bokor, S., Legry, V., Meirhaeghe, A., Ruiz, J. R., Mauro, B., Widhalm, K., Manios, Y., Amouyel, P., Moreno, L. A., Molnàr, D., Dallongeville, J. and HELENA Study group. (2010). Single-nucleotide polymorphism of CD36 locus and obesity in European adolescents. Obesity (Silver Spring) 18:1398–1403.
- Bufe, B., Breslin, P. A., Kuhn, C., Reed, D. R., Tharp, C. D., Slack, J. P., Kim, U. K., Drayna, D. and Meyerhof, W. (2005). The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 15:322–327.
- Cannon, D. S., Baker, T. B., Piper, M. E., Scholand, M. B., Lawrence, D. L., Drayna, D. T., McMahon, W. M., Villegas, G. M., Caton, T. C., Coon, H. and Leppert, M. F. (2005). Associations between phenylthiocarbamide gene polymorphisms and cigarette smoking. Nicotine Tob. Res. 7:853–858.
- Cao, Y., Zhao, F. L., Kolli, T., Hivley, R. and Herness, S. (2009). GABA expression in the mammalian taste bud functions as a route of inhibitory cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 106:4006–4011.
- Cartoni, C., Yasumatsu, K., Ohkuri, T., Shigemura, N., Yoshida, R., Godinot, N., le Coutre, J., Ninomiya, Y. and Damak, S. (2010). Taste preference for fatty acids is mediated by GPR40 and GPR120. J. Neurosci. 30:8376–8382.
- Chale-Rush, A., Burgess, J. R. and Mattes, R. D. (2007). Evidence for human orosensory (taste?) sensitivity to free fatty acids. Chem. Senses 32:423–431.
- Chandrashekar, J., Hoon, M. A., Ryba, N. J. and Zuker, C. S. (2006). The receptors and cells for mammalian taste. Nature 444:288–294.
- Chandrashekar, J., Kuhn, C., Oka, Y., Yarmolinsky, D. A., Hummler, E., Ryba, N. J. and Zuker, C. S. (2010). The cells and peripheral representation of sodium taste in mice. Nature 464:297–301.
- Chaudhari, N. and Roper, S. D. (2010). The cell biology of taste. J. Cell Biol. 190:285–296.
- Choi, S. E. (2014). Racial differences between African Americans and Asian Americans in the effect of 6-n-propylthiouracil taste intensity and food liking on body mass index. J. Acad. Nutr. Diet. 114:938–944.
- Clapp, T. R., Medler, K. F., Damak, S., Margolskee, R. F. and Kinnamon, S. C. (2006). Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol. 4:7.
- Colin-Ramirez, E., Castillo-Martinez, L., Orea-Tejeda, A., Zheng, Y., Westerhout, C. M. and Ezekowitz, J. A. (2014). Dietary fatty acids intake and mortality in patients with heart failure. Nutrition 30:1366–1371.
- Connors, M., Bisogni, C. A., Sobal, J. and Devine, C. M. (2001). Managing values in personal food systems. Appetite 36:189–200.
- Corpeleijn, E., van der Kallen, C. J., Kruijshoop, M., Magagnin, M. G., de Bruin, T. W., Feskens, E. J., Saris, W. H. and Blaak, E. E. (2006). Direct association of a promoter polymorphism in the CD36/FAT fatty acid transporter gene with Type 2 diabetes mellitus and insulin resistance. Diabet. Med. 23:907–911.
- Cui, M., Jiang, P., Maillet, E., Max, M., Margolskee, R. F. and Osman, R. (2006). The heterodimeric sweet taste receptor has multiple potential ligand binding sites. Curr. Pharm. Des. 12:4591–4600.
- Dando, R. and Roper, S. D. (2009). Cell-to-cell communication in intact taste buds through ATP signalling from pannexin 1 gap junction hemichannels. J. Physiol. 587:5899–5906.
- Dando, R. and Roper, S. D. (2012). Acetylcholine is released from taste cells, enhancing taste signalling. J. Physiol. 590:3009–3017.
- Daniel, D. Chiras (2005). Human Biology. Jones and Bartlett Learning.Evergreen, Colorado.
- Daniel, Schacter (2009). Psychology, 2 ed., Worth Publishers. New York, New York.
- DeFazio, R. A., Dvoryanchikov, G., Maruyama, Y., Kim, J. W., Pereira, E., Roper, S. D. and Chaudhari, N. (2006). Separate populations of receptor cells and presynaptic cells in mouse taste buds. J. Neurosci. 26:3971–3980.
- Dias, A. G., Rousseau, D., Duizer, L., Cockburn, M., Chiu, W., Nielsen, D. and El-Sohemy, A. (2013). Genetic variation in putative salt taste receptors and salt taste perception in humans. Chem. Senses 38:137–145.
- Dong, D., Jones, G. and Zhang, S. (2009). Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol. Biol. 9:12.
- Drayna, D. (2005). Human taste genetics. Annu. Rev. Genom. Hum. Genet. 6:217–235.
- Drewnowski, A., Henderson, S. A. and Cockroft, J. E. (2007). Genetic sensitivity to 6-n-propylthiouracil has no influence on dietary patterns, body mass indexes, or plasma lipid profiles of women. J. Am. Diet. Assoc. 107:1340–1348.
- Drewnowski, A., Henderson, S. A., Levine, A. and Hann, C. (1999). Taste and food preferences as predictors of dietary practices in young women. Public Health Nutr. 2:513–519.
- Drewnowski, A., Henderson, S. A., Shore, A. B. and Barratt-Fornell, A. (1998). Sensory responses to 6-n-propylthiouracil (PROP) or sucrose solutions and food preferences in young women. Ann. N.Y. Acad. Sci. 855:797–801.
- Drewnowski, A. and Specter, S. E. (2004). Poverty and obesity: The role of energy density and energy costs. Am. J. Clin. Nutr. 79:6–16.
- Duffy, V. B. and Bartoshuk, L. M. (2000). Food acceptance and genetic variation in taste. J. Am. Diet. Assoc. 100:647–655.
- Duffy, V. B., Davidson, A. C., Kidd, J. R., Kidd, K. K., Speed, W. C., Pakstis, A. J., Reed, D. R., Snyder, D. J. and Bartoshuk, L. M. (2004). Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin. Exp. Res. 28:1629–1637.
- Dvoryanchikov, G., Huang, Y. A., Barro-Soria, R., Chaudhari, N. and Roper, S. D. (2011). GABA, its receptors, and GABAergic inhibition in mouse taste buds. J. Neurosci. 31:5782–5791.
- Eny, K. M., Wolever, T. M., Corey, P. N. and El-Sohemy, A. (2010). Genetic variation in TAS1R2 (Ile191Val) is associated with consumption of sugars in overweight and obese individuals in 2 distinct populations. Am. J. Clin. Nutr. 92:1501–1510.
- Fay, L. B. and German, J. B. (2008). Personalizing foods: Is genotype necessary? Curr. Opin. Biotechnol. 19:121–128.
- Febbraio, M., Abumrad, N. A., Hajjar, D. P., Sharma, K., Cheng, W., Pearce, S. F. and Silverstein, R. L. (1999). A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 274:19055–19062.
- Finger, T. E., Danilova, V., Barrows, J., Bartel, D. L., Vigers, A. J., Stone, L., Hellekant, G. and Kinnamon, S. C. (2005). ATP signaling is crucial for communication from taste buds to gustatory nerves. Science 310:1495–1499.
- Forrai, G. and Bankovi, G. (1984). Taste perception for phenylthiocarbamide and food choice–a Hungarian twin study. Acta Physiol. Hung. 64:33–40.
- Fukunaga, A. (2005). Age-related changes in renewal of taste bud cells and expression of taste cell-specific proteins in mice. Kokubyo Gakkai Zasshi 72:84–89.
- Fukuwatari, T., Shibata, K., Iguchi, K., Saeki, T., Iwata, A., Tani, K., Sugimoto, E. and Fushiki, T. (2003). Role of gustation in the recognition of oleate and triolein in anosmic rats. Physiol. Behav. 78:579–583.
- Fushan, A. A., Simons, C. T., Slack, J. P., Manichaikul, A. and Drayna, D. (2009). Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr. Biol. 19:1288–1293.
- Garcia-Bailo, B., Toguri, C., Eny, K. M. and El-Sohemy, A. (2009). Genetic variation in taste and its influence on food selection. OMICS. 13:69–80.
- Gayathri, D. A., Henderson, S. A. and Drewnowski, A. (1997). Sensory acceptance of Japanese green tea and soy products is linked to genetic sensitivity to 6-n-propylthiouracil. Nutr. Cancer 29:146–151.
- Gibney, M. J., McNulty, B. A., Ryan, M. F. and Walsh, M. C. (2014). Nutritional phenotype databases and integrated nutrition: from molecules to populations. Adv. Nutr. 5:352S–357S.
- Gilbertson, T. A., Liu, L., York, D. A. and Bray, G. A. (1998). Dietary fat preferences are inversely correlated with peripheral gustatory fatty acid sensitivity. Ann. N.Y. Acad. Sci. 855:165–168.
- Glanville, E. V. and Kaplan, A. R. (1965). Food preference and sensitivity of taste for bitter compounds. Nature 205:851–853.
- Glendenning, J. I. (1994). Is the bitter ejective response always adaptive? Physiol. Behav. 56:1217–1227.
- Goldstein, G. L., Daun, H. and Tepper, B. J. (2005). Adiposity in middle-aged women is associated with genetic taste blindness to 6-n-propylthiouracil. Obes. Res. 13:1017–1023.
- Goudriaan, J. R., Dahlmans, V. E., Teusink, B., Ouwens, D. M., Febbraio, M., Maassen, J. A., Romijn, J. A., Havekes, L. M. and Voshol, P. J. (2003). CD36 deficiency increases insulin sensitivity in muscle, but induces insulin resistance in the liver in mice. J. Lipid Res. 44:2270–2277.
- Greenberg, D. and Smith, G. P. (1996). The controls of fat intake. Psychosom. Med. 58:559–569.
- Grimm, E. R. and Steinle, N. I. (2011). Genetics of eating behavior: Established and emerging concepts. Nutr. Rev. 69:52–60.
- Hayes, J. E., Bartoshuk, L. M., Kidd, J. R. and Duffy, V. B. (2008). Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem. Senses 33:255–265.
- Hayes, J. E., Sullivan, B. S. and Duffy, V. B. (2010). Explaining variability in sodium intake through oral sensory phenotype, salt sensation and liking. Physiol. Behav. 100:369–380.
- Haznedaroglu, E., Koldemir-Gunduz, M., Bakir-Coskun, N., Bozkus, H. M., Cagatay, P., Susleyici-Duman, B. and Menteş, A. (2015). Association of Sweet Taste Receptor Gene Polymorphisms with Dental Caries Experience in School Children. Caries Res. 49:275–281.
- Hladik, C. M. and Simmen, B. (1996). Taste percpetion and feeding behavior in nonhuman primates and human populations. Evol. Anthropol. 5:58–71.
- Hochheimer, A., Krohn, M., Rudert, K., Riedel, K., Becker, S., Thirion, C. and Zinke, H. (2014). Endogenous gustatory responses and gene expression profile of stably proliferating human taste cells isolated from fungiform papillae. Chem. Senses 39:359–377.
- Huang, A. L., Chen, X., Hoon, M. A., Chandrashekar, J., Guo, W., Trankner, D., Ryba, N. J. and Zuker, C. S. (2006a). The cells and logic for mammalian sour taste detection. Nature 442:934–938.
- Huang, A. L., Chen, X., Hoon, M. A., Chandrashekar, J., Guo, W., Trankner, D., Ryba, N. J. and Zuker, C. S. (2006b). The cells and logic for mammalian sour taste detection. Nature 442:934–938.
- Huang, Y. A., Dando, R. and Roper, S. D. (2009). Autocrine and paracrine roles for ATP and serotonin in mouse taste buds. J. Neurosci. 29:13909–13918.
- Huang, Y. A., Maruyama, Y. and Roper, S. D. (2008a). Norepinephrine is coreleased with serotonin in mouse taste buds. J. Neurosci. 28:13088–13093.
- Huang, Y. A., Maruyama, Y., Stimac, R. and Roper, S. D. (2008b). Presynaptic (Type III) cells in mouse taste buds sense sour (acid) taste. J. Physiol. 586:2903–2912.
- Huang, Y. A., Pereira, E. and Roper, S. D. (2011). Acid stimulation (sour taste) elicits GABA and serotonin release from mouse taste cells. PLoS One. 6:e25471.
- Huang, Y. J., Maruyama, Y., Dvoryanchikov, G., Pereira, E., Chaudhari, N. and Roper, S. D. (2007). The role of pannexin 1 hemichannels in ATP release and cell-cell communication in mouse taste buds. Proc. Natl. Acad. Sci. U.S.A. 104:6436–6441.
- Ikeda, K. (2002). New seasonings. Chem. Senses 27:847–849.
- Ishimaru, Y., Inada, H., Kubota, M., Zhuang, H., Tominaga, M. and Matsunami, H. (2006). Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc. Natl. Acad. Sci. U.S.A. 103:12569–12574.
- Jerzsa-Latta, M., Krondl, M. and Coleman, P. (1990). Use and perceived attributes of cruciferous vegetables in terms of genetically-mediated taste sensitivity. Appetite 15:127–134.
- Jiang, P., Ji, Q., Liu, Z., Snyder, L. A., Benard, L. M., Margolskee, R. F. and Max, M. (2004). The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J. Biol. Chem. 279:45068–45075.
- Keller, K. L., Liang, L. C., Sakimura, J., May, D., van, B. C., Breen, C., Driggin, E., Tepper, B. J., Lanzano, P. C., Deng, L. and Chung, W. K. (2012). Common variants in the CD36 gene are associated with oral fat perception, fat preferences, and obesity in African Americans. Obesity (Silver Spring) 20:1066–1073.
- Keski-Rahkonen, A., Bulik, C. M., Pietilainen, K. H., Rose, R. J., Kaprio, J. and Rissanen, A. (2007). Eating styles, overweight and obesity in young adult twins. Eur. J. Clin. Nutr. 61:822–829.
- Keskitalo, K., Knaapila, A., Kallela, M., Palotie, A., Wessman, M., Sammalisto, S., Peltonen, L., Tuorila, H. and Perola, M. (2007a). Sweet taste preferences are partly genetically determined: Identification of a trait locus on chromosome 16. Am. J. Clin. Nutr. 86:55–63.
- Keskitalo, K., Tuorila, H., Spector, T. D., Cherkas, L. F., Knaapila, A., Kaprio, J., Silventoinen, K. and Perola, M. (2008). The Three-Factor Eating Questionnaire, body mass index, and responses to sweet and salty fatty foods: A twin study of genetic and environmental associations. Am. J. Clin. Nutr. 88:263–271.
- Keskitalo, K., Tuorila, H., Spector, T. D., Cherkas, L. F., Knaapila, A., Silventoinen, K. and Perola, M. (2007b). Same genetic components underlie different measures of sweet taste preference. Am. J. Clin. Nutr. 86:1663–1669.
- Kim, U. K., Breslin, P. A., Reed, D. and Drayna, D. (2004). Genetics of human taste perception. J. Dent. Res. 83:448–453.
- Kim, U. K., Jorgenson, E., Coon, H., Leppert, M., Risch, N. and Drayna, D. (2003). Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299:1221–1225.
- Kim, U. K., Wooding, S., Riaz, N., Jorde, L. B. and Drayna, D. (2006). Variation in the human TAS1R taste receptor genes. Chem. Senses 31:599–611.
- Kral, T. V. and Rauh, E. M. (2010). Eating behaviors of children in the context of their family environment. Physiol. Behav. 100:567–573.
- Kulkarni, G. V., Chng, T., Eny, K. M., Nielsen, D., Wessman, C. and El-Sohemy, A. (2013). Association of GLUT2 and TAS1R2 genotypes with risk for dental caries. Caries Res. 47:219–225.
- Lanaspa, M. A., Sanchez-Lozada, L. G., Choi, Y. J., Cicerchi, C., Kanbay, M., Roncal-Jimenez, C. A., Ishimoto, T., Li, N., Marek, G., Duranay, M., Schreiner, G., Rodriguez-Iturbe, B., Nakagawa, T., Kang, D. H., Sautin, Y. Y. and Johnson, R. J. (2012). Uric acid induces hepatic steatosis by generation of mitochondrial oxidative stress: Potential role in fructose-dependent and -independent fatty liver. J. Biol. Chem. 287:40732–40744.
- Laugerette, F., Passilly-Degrace, P., Patris, B., Niot, I., Febbraio, M., Montmayeur, J. P. and Besnard, P. (2005). CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J. Clin. Invest. 115:3177–3184.
- Lepretre, F., Cheyssac, C., Amouyel, P., Froguel, P. and Helbecque, N. (2004a). A promoter polymorphism in CD36 is associated with an atherogenic lipid profile in a French general population. Atherosclerosis 173:375–377.
- Lepretre, F., Linton, K. J., Lacquemant, C., Vatin, V., Samson, C., Dina, C., Chikri, M., Ali, S., Scherer, P., Séron, K., Vasseur, F., Aitman, T. and Froguel, P. (2004b). Genetic study of the CD36 gene in a French diabetic population. Diabetes Metab. 30:459–463.
- Li, X., Staszewski, L., Xu, H., Durick, K., Zoller, M. and Adler, E. (2002). Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. U.S.A. 99:4692–4696.
- Liang, L. C., Sakimura, J., May, D., Breen, C., Driggin, E., Tepper, B. J., Chung, W. K. and Keller, K. L. (2012). Fat discrimination: A phenotype with potential implications for studying fat intake behaviors and obesity. Physiol. Behav. 105:470–475.
- LopezJimenez, N. D., Cavenagh, M. M., Sainz, E., Cruz-Ithier, M. A., Battey, J. F. and Sullivan, S. L. (2006). Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J. Neurochem. 98:68–77.
- Love-Gregory, L., Sherva, R., Sun, L., Wasson, J., Schappe, T., Doria, A., Rao, D. C., Hunt, S. C., Klein, S., Neuman, R. J., Permutt, M. A. and Abumrad, N. A. (2008). Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum. Mol. Genet. 17:1695–1704.
- Lucas, F. and Bellisle, F. (1987). The measurement of food preferences in humans: Do taste-and-spit tests predict consumption? Physiol. Behav. 39:739–743.
- Lugaz, O., Pillias, A. M. and Faurion, A. (2002). A new specific ageusia: Some humans cannot taste L-glutamate. Chem. Senses 27:105–115.
- Ma, X., Bacci, S., Mlynarski, W., Gottardo, L., Soccio, T., Menzaghi, C., Iori, E., Lager, R. A., Shroff, A. R., Gervino, E. V., Nesto, R. W., Johnstone, M. T., Abumrad, N. A., Avogaro, A., Trischitta, V. and Doria, A. (2004). A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum. Mol. Genet. 13:2197–2205.
- Madden, J., Carrero, J. J., Brunner, A., Dastur, N., Shearman, C. P., Calder, P. C. and Grimble, R. F. (2008). Polymorphisms in the CD36 gene modulate the ability of fish oil supplements to lower fasting plasma triacyl glycerol and raise HDL cholesterol concentrations in healthy middle-aged men. Prostaglandins Leukot. Essent. Fatty Acids 78:327–335.
- Mattes, R. and Labov, J. (1989). Bitter taste responses to phenylthiocarbamide are not related to dietary goitrogen intake in human beings. J. Am. Diet. Assoc. 89:692–694.
- Mattes, R. D. (1985). Gustation as a determinant of ingestion: methodological issues. Am. J. Clin. Nutr. 41:672–683.
- Mattes, R. D. (2009). Is there a fatty acid taste? Annu. Rev. Nutr. 29:305–327.
- McDaniel, A. H. and Reed, D. R. (2003). Genomics and Proteomics in Nutrition. Marcel Dekker, New York.
- Mennella, J. A., Pepino, M. Y. and Reed, D. R. (2005). Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics 115:e216–e222.
- Monge-Rojas, R., Mattei, J., Fuster, T., Willett, W. and Campos, H. (2014). Influence of sensory and cultural perceptions of white rice, brown rice and beans by Costa Rican adults in their dietary choices. Appetite 81:200–208.
- Nakajima, K., Asakura, T., Oike, H., Morita, Y., Shimizu-Ibuka, A., Misaka, T., Sorimachi, H., Arai, S. and Abe, K. (2006). Neoculin, a taste-modifying protein, is recognized by human sweet taste receptor. Neuroreport 17:1241–1244.
- Nelson, G., Chandrashekar, J., Hoon, M. A., Feng, L., Zhao, G., Ryba, N. J. and Zuker, C. S. (2002). An amino-acid taste receptor. Nature 416:199–202.
- Nelson, G., Hoon, M. A., Chandrashekar, J., Zhang, Y., Ryba, N. J. and Zuker, C. S. (2001). Mammalian sweet taste receptors. Cell 106:381–390.
- Nielsen, D. E. and El-Sohemy, A. (2014). Disclosure of genetic information and change in dietary intake: A randomized controlled trial. PLoS One. 9:e112665.
- Nielsen, D. E., Shih, S. and El-Sohemy, A. (2014). Perceptions of genetic testing for personalized nutrition: A randomized trial of DNA-based dietary advice. J. Nutrigenet. Nutrigen. 7:94–104.
- Niewind, A., Krondl, M. and Shrott, M. (1988). Genetic influences on the selection of Brassica vegetables by elderly individuals. Nutr. Res. 8:13–20.
- Noel, S. E., Lai, C. Q., Mattei, J., Parnell, L. D., Ordovas, J. M. and Tucker, K. L. (2010). Variants of the CD36 gene and metabolic syndrome in Boston Puerto Rican adults. Atherosclerosis 211:210–215.
- Obata, H., Shimada, K., Sakai, N. and Saito, N. (1997). GABAergic neurotransmission in rat taste buds: Immunocytochemical study for GABA and GABA transporter subtypes. Brain Res. Mol. Brain Res. 49:29–36.
- O'Brien, S. A., Feeney, E. L., Scannell, A. G., Markey, A. and Gibney, E. R. (2013). Bitter taste perception and dietary intake patterns in Irish children. J. Nutrigenet. Nutrigen. 6:43–58.
- Ogura, T. (2002). Acetylcholine increases intracellular Ca2+ in taste cells via activation of muscarinic receptors. J. Neurophysiol. 87:2643–2649.
- Ooi, S. X., Lee, P. L., Law, H. Y. and Say, Y. H. (2010). Bitter receptor gene (TAS2R38) P49A genotypes and their associations with aversion to vegetables and sweet/fat foods in Malaysian subjects. Asia Pac. J. Clin. Nutr. 19:491–498.
- Pepino, M. Y., Love-Gregory, L., Klein, S. and Abumrad, N. A. (2012). The fatty acid translocase gene CD36 and lingual lipase influence oral sensitivity to fat in obese subjects. J. Lipid Res. 53:561–566.
- Raliou, M., Grauso, M., Hoffmann, B., Schlegel-Le-Poupon, C., Nespoulous, C., Debat, H., Belloir, C., Wiencis, A., Sigoillot, M., Bano, S. P., Trotier, D., Pernollet, J. C., Montmayeur, J. P., Faurion, A. and Briand, L. (2011). Human genetic polymorphisms in T1R1 and T1R3 taste receptor subunits affect their function. Chem. Senses 36:527–537.
- Ramirez, I. (1993). Role of olfaction in starch and oil preference. Am. J. Physiol. 265:R1404–R1409.
- Ramos-Arellano, L. E., Salgado-Bernabe, A. B., Guzman-Guzman, I. P., Salgado-Goytia, L., Munoz-Valle, J. F. and Parra-Rojas, I. (2013). CD36 haplotypes are associated with lipid profile in normal-weight subjects. Lipids Health Dis. 12:167.
- Reed, D. R., Bachmanov, A. A., Beauchamp, G. K., Tordoff, M. G. and Price, R. A. (1997). Heritable variation in food preferences and their contribution to obesity. Behav. Genet. 27:373–387.
- Reed, D. R., Li, X., Bachmanov, A. A., Mascioli, K. and Beauchamp, G. K. (2003). Progress in Obesity Research, Vol. 9, John Libbey Eurotext Ltd., London.
- Reed, D. R. and McDaniel, A. H. (2006). The human sweet tooth. BMC Oral Health 6(Suppl 1):S17.
- Reed, D. R., Tanaka, T. and McDaniel, A. H. (2006). Diverse tastes: Genetics of sweet and bitter perception. Physiol. Behav. 88:215–226.
- Richter, T. A., Caicedo, A. and Roper, S. D. (2003). Sour taste stimuli evoke Ca2+ and pH responses in mouse taste cells. J. Physiol. 547:475–483.
- Rodin, J., Moskowitz, H. R. and Bray, G. A. (1976). Relationship between obesity, weight loss, and taste responsiveness. Physiol. Behav. 17:591–597.
- Rolls, E. T. (2012). Taste, olfactory and food texture reward processing in the brain and the control of appetite. Proc. Nutr. Soc. 71:488–501.
- Romanov, R. A., Rogachevskaja, O. A., Bystrova, M. F., Jiang, P., Margolskee, R. F. and Kolesnikov, S. S. (2007). Afferent neurotransmission mediated by hemichannels in mammalian taste cells. EMBO J. 26:657–667.
- Running, C. A., Craig, B. A. and Mattes, R. D. (2015). Oleogustus: The unique taste of fat. Chem. Senses 40:507–516.
- Scheibehenne, B., Miesler, L. and Todd, P. M. (2007). Fast and frugal food choices: Uncovering individual decision heuristics. Appetite 49:578–589.
- Sclafani, A., Ackroff, K. and Abumrad, N. A. (2007). CD36 gene deletion reduces fat preference and intake but not post-oral fat conditioning in mice. Am. J. Physiol. Regul. Integr. Comp Physiol. 293:R1823–R1832.
- Sharma, K. and Kaur, G. K. (2014). PTC bitter taste genetic polymorphism, food choices, physical growth in body height and body fat related traits among adolescent girls from Kangra Valley, Himachal Pradesh (India). Ann. Hum. Biol. 41:29–39.
- Shigemura, N., Shirosaki, S., Sanematsu, K., Yoshida, R. and Ninomiya, Y. (2009). Genetic and molecular basis of individual differences in human umami taste perception. PLoS One. 4:e6717.
- Simons, P. J., Kummer, J. A., Luiken, J. J. and Boon, L. (2011). Apical CD36 immunolocalization in human and porcine taste buds from circumvallate and foliate papillae. Acta Histochem. 113:839–843.
- Ssadi-Porter, F. M., Maillet, E. L., Radek, J. T., Quijada, J., Markley, J. L. and Max, M. (2010). Key amino acid residues involved in multi-point binding interactions between brazzein, a sweet protein, and the T1R2-T1R3 human sweet receptor. J. Mol. Biol. 398:584–599.
- Stewart, J. E., Feinle-Bisset, C., Golding, M., Delahunty, C., Clifton, P. M. and Keast, R. S. (2010). Oral sensitivity to fatty acids, food consumption and BMI in human subjects. Br. J. Nutr. 104:145–152.
- Stewart, J. E., Feinle-Bisset, C. and Keast, R. S. (2011). Fatty acid detection during food consumption and digestion: Associations with ingestive behavior and obesity. Prog. Lipid Res. 50:225–233.
- Stewart, J. E. and Keast, R. S. (2012). Recent fat intake modulates fat taste sensitivity in lean and overweight subjects. Int. J. Obes. (Lond.) 36:834–842.
- Sullivan, J. M., Borecki, A. A. and Oleskevich, S. (2010). Stem and progenitor cell compartments within adult mouse taste buds. Eur. J. Neurosci. 31:1549–1560.
- Takeda, M., Imaizumi, M. and Fushiki, T. (2000). Preference for vegetable oils in the two-bottle choice test in mice. Life Sci. 67:197–204.
- Temussi, P. A. (2002). Why are sweet proteins sweet? Interaction of brazzein, monellin and thaumatin with the T1R2-T1R3 receptor. FEBS Lett. 526:1–4.
- Tepper, B. J. (2008). Nutritional implications of genetic taste variation: The role of PROP sensitivity and other taste phenotypes. Annu. Rev. Nutr. 28:367–388.
- Tepper, B. J., Koelliker, Y., Zhao, L., Ullrich, N. V., Lanzara, C., d'Adamo, P., Ferrara, A., Ulivi, S., Esposito, L. and Gasparini, P. (2008). Variation in the bitter-taste receptor gene TAS2R38, and adiposity in a genetically isolated population in Southern Italy. Obesity (Silver Spring) 16:2289–2295.
- Tepper, B. J. and Nurse, R. J. (1997). Fat perception is related to PROP taster status. Physiol. Behav. 61:949–954.
- Tepper, B. J. and Nurse, R. J. (1998). PROP taster status is related to fat perception and preference. Ann. N.Y. Acad. Sci. 855:802–804.
- The International HapMap Project (2003). Nature 426:789–796.
- Tholin, S., Rasmussen, F., Tynelius, P. and Karlsson, J. (2005). Genetic and environmental influences on eating behavior: The Swedish Young Male Twins Study. Am. J. Clin. Nutr. 81:564–569.
- Timpson, N. J., Christensen, M., Lawlor, D. A., Gaunt, T. R., Day, I. N., Ebrahim, S. and Davey Smith, G. (2005). TAS2R38 (phenylthiocarbamide) haplotypes, coronary heart disease traits, and eating behavior in the British Women's Heart and Health Study. Am. J. Clin. Nutr. 81:1005–1011.
- Toyono, T., Seta, Y., Kataoka, S., Harada, H., Morotomi, T., Kawano, S., Shigemoto, R. and Toyoshima, K. (2002). Expression of the metabotropic glutamate receptor, mGluR4a, in the taste hairs of taste buds in rat gustatory papillae. Arch. Histol. Cytol. 65:91–96.
- Toyono, T., Seta, Y., Kataoka, S., Kawano, S., Shigemoto, R. and Toyoshima, K. (2003). Expression of metabotropic glutamate receptor group I in rat gustatory papillae. Cell Tissue Res. 313:29–35.
- Tsuruta, M., Kawada, T., Fukuwatari, T. and Fushiki, T. (1999). The orosensory recognition of long-chain fatty acids in rats. Physiol. Behav. 66:285–288.
- Turner-McGrievy, G., Tate, D. F., Moore, D. and Popkin, B. (2013). Taking the bitter with the sweet: Relationship of supertasting and sweet preference with metabolic syndrome and dietary intake. J. Food Sci. 78:S336–S342.
- Vandenbeuch, A., Clapp, T. R. and Kinnamon, S. C. (2008). Amiloride-sensitive channels in type I fungiform taste cells in mouse. BMC Neurosci. 9:1.
- Varez-Lario, B. and Arron-Vicente, J. (2010). Uric acid and evolution. Rheumatology (Oxford) 49:2010–2015.
- Wald, N. and Leshem, M. (2003). Salt conditions a flavor preference or aversion after exercise depending on NaCl dose and sweat loss. Appetite 40:277–284.
- Walters, D. E. and Hellekant, G. (2006). Interactions of the sweet protein brazzein with the sweet taste receptor. J. Agric. Food Chem. 54:10129–10133.
- Weisberger, D. (1950). A role of glucose in the production of artificial caries. J. Dent. Res. 29:14–22.
- Wise, P. M., Hansen, J. L., Reed, D. R. and Breslin, P. A. (2007). Twin study of the heritability of recognition thresholds for sour and salty taste. Chem. Senses 32:749–754.
- Yang, R., Montoya, A., Bond, A., Walton, J. and Kinnamon, J. C. (2012). Immunocytochemical analysis of P2×2 in rat circumvallate taste buds. BMC Neurosci. 13:51.
- Yarmolinsky, D. A., Zuker, C. S. and Ryba, N. J. (2009). Common sense about taste: From mammals to insects. Cell 139:234–244.
- Yoshida, R., Horio, N., Murata, Y., Yasumatsu, K., Shigemura, N. and Ninomiya, Y. (2009). NaCl responsive taste cells in the mouse fungiform taste buds. Neuroscience 159:795–803.
- Yun, Y. M., Song, E. Y., Song, S. H., Song, J. and Kim, J. Q. (2007). CD36 polymorphism and its relationship with body mass index and coronary artery disease in a Korean population. Clin. Chem. Lab Med. 45:1277–1282.
- Zeinstra, G. G., Koelen, M. A., Kok, F. J. and de, G. C. (2007). Cognitive development and children's perceptions of fruit and vegetables; a qualitative study. Int. J. Behav. Nutr. Phys. Act. 4:30.