Abstract
The human genome contains a large number of retroviral elements acquired over the process of evolution, some of which are specific to primates. However, as many of these are defective or silenced through epigenetic changes, they were historically considered “junk DNA” and their potential role in human physiology or pathological circumstances have been poorly studied. The most recently acquired, human endogenous retrovirus-K (HERV-K), has multiple copies in the human genome and some of them have complete open reading frames that are transcribed and translated, especially in early embryogenesis. Phylogenetically, HERV-K is considered a supergroup of viruses. One of the subtypes, termed HML-2, seems to be the most active and hence, it is the best studied. Aberrant expression of HML-2 in adult tissues has been associated with certain types of cancer and with neurodegenerative diseases. This review discusses the discovery of these viruses, their classification, structure, regulation and potential for replication, physiological roles, and their involvement in disease pathogenesis. Finally, it presents different therapeutic approaches being considered to target these viruses.
1. Introduction
Retroviruses and retroviral elements have been associated with a wide variety of diseases; most often with cancer, but also with immune disorders and neurodegenerative diseases. This presents an interesting paradox: the same virus can cause both a proliferative disorder in non-nervous systems and a degenerative disorder in the nervous system. For example, human T-lymphotropic virus I (HTLV-I) can cause T-cell leukaemia/lymphoma and myelopathy/tropical spastic paraparesis, a progressive neurodegenerative disorder (Barmak et al. Citation2003). Similarly, murine and feline retroviruses have been associated with leukaemia, breast cancer and neurodegenerative diseases (Li XJ et al. Citation2009).
An interesting feature of some retroviruses, e.g. Mouse Mammary Tumour Virus (MMTV) and Murine Leukaemia Virus (MLV), is that they exist in both exogenous and endogenous forms (Holt et al. Citation2013). The human genome also harbours a large number of retroviruses that have been acquired throughout the process of evolution; however, to date there is no evidence for horizontal transmission of these viruses. Of all the human endogenous retroviruses (HERVs), one group termed HERV-K is the most recently acquired and the most transcriptionally active. It is the only group of endogenous retroviruses (ERVs) known to have human-specific members (Medstrand and Mager Citation1998; Buzdin et al. Citation2003). In recent years, it has become clear that this group of ERVs play a critical role in embryogenesis, but their expression is silenced in most cell types in healthy adults (Grow et al. Citation2015). However, its reactivation has been associated with several types of cancer (Agoni et al. Citation2013; Cegolon et al. Citation2013; Bhardwaj et al. Citation2015; Argaw-Denboba et al. Citation2017) and with the neurodegenerative disorder amyotrophic lateral sclerosis (ALS) (Douville et al. Citation2011; Li W et al. Citation2015). HERV-K is present in hundreds of copies in the human genome (Wildschutte et al. Citation2016). The scientific interest in this retrovirus has increased in the last few years, as evidenced by the number of publications. Hence, a comprehensive review to address all the aspects of this complex retrovirus is much needed. In the present review, we provide a historical perspective on the discovery of these viruses and discuss their classification, genomic organization, regulation and their capability to form viral proteins. Finally, we critically examine the evidence for a role in human physiology and disease pathogenesis and identify areas of knowledge where there are still important gaps.
2. Historical perspective
Transposable elements were first described by McClintock (Citation1950) in 1950 as DNA sequences that are capable of transposition. Fifty years after that discovery, completion of the human genome project provided unexpected results: 45% of our genome was composed of transposable elements, or more accurately, “transposed elements” (TE): mutated or truncated copies of transposable elements that have been rendered immobile (Faulkner and Carninci Citation2009). ERVs are long terminal repeat (LTR) retrotransposons (class I of TEs), which originated from exogenous retroviruses that infected the germ line throughout evolution.
ERVs were discovered in the late 1960s and early 1970s (Weiss Citation2006), when three types of ERVs were reported by independent groups within a few years of each other; avian leukosis virus (ALV) (Weiss Citation1969a, Citation1969b), MLV (Aaronson et al. Citation1971), and MMTV (Bentvelzen et al. Citation1970).
The first HERV was reported in 1981 (Martin et al. Citation1981), using probes against MLV in human DNA. Following that, numerous groups of HERVs were discovered in the human genome. The HERV-K family, and especially its subgroup HML-2, is the youngest and most active group. Although all HML-2 proviruses are defective in at least one gene (Subramanian et al. Citation2011), many of them possess complete open reading frames (ORFs) with coding capability and HML-2 transcripts and proteins have been detected in healthy tissues (Ehlhardt et al. Citation2006), especially in embryonic cells (Fuchs et al. Citation2013), and in malignancies (Buscher et al. Citation2006). It is thought that mutations accumulate in ERV sequences over time through subsequent generations, which would eventually lead to non-coding DNA. Whether the maintenance of coding ORFs in the case of HML-2 results from evolutionary conservation or they simply exist because there has not been enough time for all of them to accumulate ORF-destroying mutations remains unclear. However, the protein expression in healthy tissues might point to the conservation of such genes for specific physiological roles in human cells, as it has been demonstrated for other HERV proteins such as syncytin-1 of the HERV-W family (Blond et al. Citation2000). Interestingly, HML-2 can also form viral-like particles, which have been detected in various cells, particularly teratocarcinomas and melanomas (Lower et al. Citation1984; Buscher et al. Citation2005; Schmitt et al. Citation2013). Although these viral-like particles are considered non-infectious, two independent research groups have demonstrated that the consensus sequence of HML-2 provirus can produce infectious particles (Dewannieux et al. Citation2006; Lee and Bieniasz Citation2007). Because of its evolutionary and clinically interesting features, this review is mostly dedicated to the HML-2 group.
3. Classification and nomenclature
The nomenclature and classification of ERVs is complex and confusing. There is no consistency between different sources and very often similar sequences receive different names. Repbase (https://www.girinst.org/repbase) probably provides the most complete classification: (1) ERV1, which consists of gammaretroviruses, similar to MLV; (2) ERV2, which clusters as betaretroviruses, as they are similar to MMTV; (3) ERV3, or Spumaretrovirus-like elements, which are similar to Simian foamy virus (Wilkinson et al. Citation1994; Blomberg et al. Citation2009); (4) Lentivirus; and (5) ERV4, which do not have any described exogenous counterparts. However, the databases Repeatmasker (http://www.repeatmasker.org) and DFAM (http://www.dfam.org) call the betaretrovirus class “ERVK”, instead of “ERV2” and use the term “ERVL” for the Spumaretrovirus class.
HERV-K may be the best example of confusing nomenclature among ERVs as it has received multiple names since its discovery. It was first described by Callahan et al. (Citation1985), who isolated a human DNA sequence (clone HLM-2) with similarities to MMTV. Later, Ono et al. (Citation1986) reported the complete nucleotide sequence of an almost full-length provirus that was found to be relatively uninterrupted by stop codons. They named it HERV-K10 and acknowledged its similarities with clone HLM-2. A few years later, Medstrand and Blomberg (Citation1993) introduced for the first time the term HML when they amplified sequences similar to MMTV and HERV-K10 and classified them in 6 groups (HML-1 to HML-6), based on alignments of the rt region. HML stands for “human endogenous MMTV-like” and by chance the acronym resembles Callahan’s clone name HLM-2, which adds even more confusion to the terminology. Subsequent analysis of HERV-K sequences by different researchers resulted in the following designations for the same HERV group: HLM-2, HML-2, HERV-K10, HTDV/HERV-K, HERV-K (HML-2), HERV-K, HERVK or ERVK (Manghera et al. Citation2015).
Phylogenetically, the HERV-K group belongs to the ERV2 or Class II or Betaretrovirus-like supergroup. Currently, the HERV-K clade contains 10 groups (from HML-1 to HML-10) (Medstrand and Blomberg Citation1993; Andersson et al. Citation1996, Citation1999; Medstrand et al. Citation1997; Yin et al. Citation1999), which are classified based on the reverse transcriptase gene (rt) sequence. It is worth noting that one study from Coffin’s group determined that two proviruses (located in chromosome bands 17p13.1 and 8p22), previously assigned to the HML-2 clade, were very dissimilar to this group when aligning the sequences of other viral genes. Thus, they proposed these loci be moved to a new group named HML-11 (Subramanian et al. Citation2011). Traditionally, ERVs have been named per the tRNA that binds their RT enzyme and the primer binding site (PBS) (Cohen and Larsson Citation1988). Thus, HERV-K is named after lysine-tRNA. However, this nomenclature is imprecise because analysis of larger sequences show that some members have PBSs for tRNAs other than lysine, while phylogenetically they belong to the same supergroup (Reus, Mayer, Sauter, Zischler et al. Citation2001; Blikstad et al. Citation2008).
3.1. HML-2 subtype classification
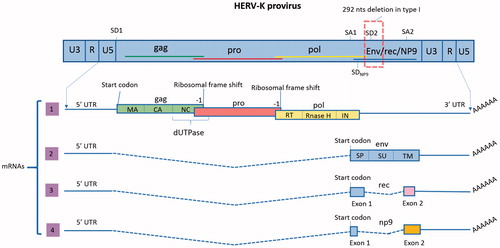
HML-2 proviruses have been classified as type 1 or type 2 (Lower et al. Citation1993), depending on the presence or absence of a 292-bp deletion in the pol–env junction (). Type 2 proviruses, which do not have the deletion, encode the protein Rec, which is an accessory protein that binds to viral transcripts to facilitate their nucleocytoplasmic transport (Wodrich and Krausslich Citation2001), and the envelope (Env) (Magin et al. Citation1999; Magin-Lachmann et al. Citation2001; Mayer et al. Citation2004). Due to the deletion, type 1 proviruses have lost a splice donor (SD) site and are incapable of encoding Rec or Env. An alternative SD site located just upstream of the 292-bp deletion is instead used to splice a mRNA that encodes a ∼9-kDa protein named Np9 (Armbruester et al. Citation2002; Buscher et al. Citation2006) (). Some HML-2 proviruses have larger deletions and they have not been classified (Subramanian et al. Citation2011).
Figure 1. Proviral organization of HML-2 and transcripts. In the proviral form of HML-2, the sequences of the four ORFs overlap (shown in the scheme by the colored lines). Splice donor (SD) and splice acceptor (SA) sites are shown. LTRs are composed of the U3 and U5 regions separated by the R segment. As opposed to canonical retroviruses, which include the R segment in their transcripts, HML-2 transcription starts after the R. Transcript 1 has three ORFs to encode proteins GAG, PRO and POL. In this transcript, only gag has a start codon (AUG); pro and pol translation is mediated by two ribosomal frame shifts (–1). As a result, and despite having overlapping DNA sequences, the three proteins do not have any amino acid sequence in common. The figure also shows the ORFs organization to encode the different final proteins and domains (matrix (MA), capsid (CA) and nucleocapsid (NC) in gag, dUTPase in Gag-Pro junction and reverse transcriptase (RT), Rnase H and integrase (IN) in pol). The functional proteins will be formed by proteolysis of the polyproteins Gag, Gag-Pro and Gag-Pro-Pol. Transcript 2 encodes the protein Env, which has three different domains: the signal peptide (SP), surface (SU) and transmembrane (TM). Transcript 3 is the product of alternative splicing of the env ORF and encodes Rec. This transcript is only produced by type 2 HML-2 proviruses, which do not have the 292 nts deletion. Rec has 87 amino acids in common with Env, corresponding to its first exon. The second exon starts in a different frame and therefore, the amino acid sequence is not shared with Env. Transcript 4 is only produced by type 1 proviruses, which contain a deletion of 292 nts in the pol–env junction. As a result, the SD2 site is lost and an alternative SD (SDNP9) is used for the splicing. Due to this change only the first 14 amino-acids of NP9 are shared with Env and Rec.
Based on phylogenetic analysis of the LTR sequences, HML-2 can be also classified into three subgroups: LTR5Hs, LTR5A, and LTR5B (Buzdin et al. Citation2003; Macfarlane and Simmonds Citation2004). LTR5Hs viruses are the most recently acquired, whereas the LTR5A or LTR5B subgroups come from older integrations (Buzdin et al. Citation2003). Type 1 proviruses have been exclusively found in the LTR5Hs subgroup and are scattered across all the subclades suggesting frequent recombination (Subramanian et al. Citation2011). For an unambiguous identification of HML-2 proviruses, Hughes and Coffin (Citation2001) have proposed to name them based upon chromosome band location. In the case of multiple proviruses in the same chromosome band the Coffin laboratory has suggested labelling each provirus with an “a”, “b”, “c”, etc. depending upon their order within the band (Subramanian et al. Citation2011).
4. Genomic distribution, transcriptional activity and unfixed copies
ERV insertions in the genome can be found as proviral forms (full-length or almost complete proviruses) or as solitary (solo) LTRs, resulting from homologous recombination between the two LTRs flanking the provirus and subsequent deletion of the internal sequence (Hughes and Coffin Citation2004). Considerable differences exist in the total number of HML-2 proviruses and solo LTRs reported in publications. Such discrepancies are due to the different criteria used to identify the loci among the studies. Most recent estimates suggest that there are at least 89 HML-2 proviruses, of which 26% are type 1 and 74% are type 2, and there are 944 (Subramanian et al. Citation2011) to 1200 solo LTRs (Babaian and Mager Citation2016).
It seems that mutations in HERV sequences are more likely to affect their coding capacity due to nonsense mutations than to disrupt their promoter activity (Flockerzi et al. Citation2008). Studies investigating the expression of individual HML-2 proviruses (Flockerzi et al. Citation2008) found up to 23 transcriptionally active HML-2 proviruses in the human genome. In , we summarize the published data on the HML-2 proviruses with ORFs for the different viral genes. However, it is important to note that not all studies analysed whether the ORFs have coding capability or contain premature stop codons in their sequences. To determine the genomic origin of the HML-2 proteins involved in physiological studies or disease pathophysiology, such analysis is needed.
Table 1. HML-2 proviruses with ORFs for the main viral genes and some domains.
The involvement of HML-2 in pathological conditions, especially in malignancies, has been the focus of numerous publications. However, expression profiles of healthy tissues such as brain, heart, peripheral blood mononuclear cells (PBMCs), lung, liver and breast (Flockerzi et al. Citation2008; Schmitt et al. Citation2015) indicate that several HML-2 proviruses are transcriptionally active, although the magnitude of expression in most healthy tissues is low. This is also applicable for other groups of HERVs (Flockerzi et al. Citation2008), as several HERV families are expressed in all investigated human tissues in health and disease (Seifarth et al. Citation2005; Muradrasoli et al. Citation2006). Placenta and testicles seem to be privileged tissues for HERV expression, most likely due to decreased epigenetic regulation and specific transcription factors. Androgens can stimulate HML-2 expression (Hanke, Chudak, et al. Citation2013) and LTR hypomethylation has been described in these tissues (Perot et al. Citation2012).
Recently, several HML-2 copies not present in the reference genome were identified. These polymorphic loci can be found as almost complete full-length provirus, solo LTR and unoccupied sites. One study that analysed more than 2500 sequenced genomes identified 36 non-referenced insertions with frequencies ranging from <0.0005 to 0.75 (Wildschutte et al. Citation2016). Previous studies had identified 17 of those polymorphic HML-2 loci by different methods (Turner et al. Citation2001; Kidd et al. Citation2008; Lee E et al. Citation2012; Marchi et al. Citation2014).
5. HML-2 structure: genes, transcripts and proteins
5.1. Proviral organization
HML-2 proviral organization is described in . In the proviral form of HML-2, the sequences of the four ORFs overlap, flanked by a 5′ and a 3′ LTR. There are two SD and two splice acceptor (SA) sites. As explained previously, there are two types of proviruses characterized by the presence or absence of a 292 bp-deletion in the pol–env junction. Four mRNAs have been described as products of HML-2 transcription (Hohn et al. Citation2013) (). One transcript has three ORFs to encode polyproteins Gag, protease (Pro) and polymerase (Pol). In this transcript, only gag has a start codon (AUG); pro and pol translation is mediated by two ribosomal frame shifts (–1). As a result, and despite having overlapping DNA sequences, the three proteins do not have any amino acid sequence in common. Gag, Pro and Pol are translated as polyproteins. Gag contains the sequences to form the structural proteins matrix (MA), capsid (CA) and nucleocapsid (NC) and Pol forms the reverse trancriptase (RT), which has Rnase H and polymerase domains, and the integrase (IN). The pro ORF also encodes the enzyme dUTPase. The functional proteins are formed by proteolysis of the polyproteins Gag, Gag-Pro and Gag-Pro-Pol. Another mRNA encodes the protein Env, which has three different domains: the signal peptide (SP), surface unit (SU) and transmembrane domain (TM). The third transcript is the product of alternative splicing of the env ORF and encodes Rec. This transcript is only produced by type 2 HML-2 proviruses, which do not have the 292-bp deletion. Rec has 87 amino acids in common with Env, corresponding to its first exon. The second exon starts in a different frame and therefore the amino acid sequence is not shared with Env. The fourth transcript, np9, is only produced by type 1 proviruses. As a result of the deletion at the pol–env junction, the SD2 site is lost and an alternative SD is used for the splicing (Lower et al. Citation1995; Armbruester et al. Citation2002) (). Due to this change only the first 14 amino-acids of Np9 are shared with Env and Rec.
5.2. Long terminal repeats
The gene expression of HML-2 is under the direct control of the LTRs, the 5′ LTR promotes transcription of the viral genome. Each flanking viral LTR consists of a U3, R, and U5 regions in 5′ to 3′ direction (). HML-2 LTRs possess promoter and enhancer elements, multiple transcription factor binding sites, and polyadenylation signal (Kovalskaya et al. Citation2006). The Rec binding site or Rec Responsive Element (RcRE) is a highly structured RNA region that has been identified in the U3R segment of the 3′ LTR (Magin et al. Citation1999). We have included a list of the published transcription factor binding sites in the consensus sequence of the HML-2 LTR (). However, it is important to highlight that due to the multiple sequence variations among the genomic LTRs, not all these binding sites may be present in each particular locus. These variations probably underlie the differential gene expression in different cell types or after different stimuli, reflecting expression of different proviruses with different patterns of mutations in their LTRs.
Table 2. Transcription factors binding to different HML-2 LTRs.
Solo LTRs were created during evolution via homologous recombination between 5′ and 3′ LTRs of an integrated provirus and subsequent loss of the internal sequence. These solo LTRs retain some of their promoter/enhancer function and hence have gene regulatory capacity. At least 50% of the HML-2 LTRs possess promoter activity and are differentially expressed in normal and cancer tissues (Buzdin et al. Citation2006) and some elements are methylated in a tissue-specific manner (Khodosevich et al. Citation2004). Some LTRs have bi-directional promoter activity (Domansky et al. Citation2000) and can regulate the function and expression of proximal down- and upstream genes (see Section 7.5). The transcription in canonical retroviruses starts between U3 and R elements, so R is part of the transcripts. In a study using a consensus sequence of the HML-2 Hs family, authors determined that, surprisingly, the transcriptional starting point is located at the very 3′ end of the LTR R region. Therefore, the R region is excluded from transcripts (Kovalskaya et al. Citation2006). In another study using the promoter of the K108 provirus, it was found that the LTR acts as a TATA-less promoter, since mutation of the TATA motif did not decrease promoter function. Instead, significantly increased transcriptional activity was observed (Fuchs et al. Citation2011).
5.3. HERV-K proteins
5.3.1. Gag
As described in , two –1 ribosomal frameshifts are needed to translate the 160-kDa HML-2 Gag–Pro–Pol precursor protein. Analysis of the proteolysis of HML-2 Gag () showed that the 74 kDa polyprotein is processed by the viral protease to yield the matrix protein (MA) (15 kDa), SP1 (spacer peptide of 14 amino acids), a peptide of 15 kDa, capsid (CA) (27 kDa), nucleocapsid (NC) (10 kDa) and two C-terminally encoded glutamine- and proline-rich peptides, QP1 and QP2, spanning 23 and 19 amino acids, respectively (George et al. Citation2011).
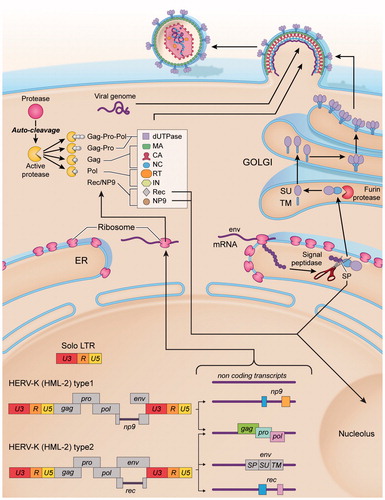
Figure 2. Molecular biosynthesis of HML-2. In the human genome HML-2 sequences can be found in the form of two types of proviruses (type 1 and type 2) as well as solo LTRs, resulting from homologous recombination between the two LTRs of a provirus and subsequent loss of the internal sequence. The two types of proviruses differ in the presence (type 1) or absence (type 2) of a 292 nts-deletion in the pol–env junction. Because of such deletion, type 1 proviruses are unable to encode the Env protein and the accessory protein Rec, encoding NP9 instead. In addition to transcripts with coding capability, non-coding RNAs are produced, resulting from the transcription of loci with premature stop codons. The consensus sequence of HML-2 provirus (type 2) presents a 5′LTR, a PBS and four main ORFs: (1) gag, encoding the structural proteins: matrix (MA), capsid (CA) and nucleocapsid (NC); (2) protease (pro), also encoding the enzyme dUTPase; (3) polymerase (pol), encoding the reverse transcriptase (RT), RNase H and integrase (IN) and (4) Envelope (env); a polypurine tract (PPT) and a 3′LTR. HML-2 gag, pro and pol transcripts are translated as polyproteins. The position in the reading frames indicates that ribosomal frameshifting is performed to synthesize Gag–Pro and Gag–Pro–Pol polyproteins. After autocleavage, the viral protease (Pro) will proteolyze the precursors, yielding the mature structural proteins of the matrix (MA), capsid (CA) and nucleocapsid (NC), and the active enzymes reverse transcriptase (RT) and integrase (IN). Env transcript can undergo alternative splicing, generating either env or rec/np9 mRNAs (depending on the type (1 or 2) of provirus). HML-2 Env is synthesized as a polyprotein that follows the secretory pathway. It has a signal peptide (SP) to direct the protein to the endoplasmic reticulum, where it is cleaved by the signal peptidase. Then Env is cleaved by furin host proteases into a surface unit (SU), and a transmembrane unit (TM). SU and TM are non-covalently associated and will possibly trimerize in the Golgi apparatus, resulting in a trimer of heterodimers. Env anchors into the cell membrane via the TM subunit, then traffics to the plasma membrane and studs the surface of the newly budding virus particles.
Without the ribosomal frameshifts, only a Gag protein of 76 kDa is translated (Bannert and Kurth Citation2004). The processing takes place after the viral particle buds from the producer cell, during the maturation process. It results in a condensed core morphology (Boller et al. Citation1993). Several HML-2 proviruses possess complete gag ORFs (). Gag-encoding transcripts have been detected in many cells and tissues from diseased and healthy individuals (Ishida et al. Citation2008). Gag proteins can be detected in teratocarcinoma cell lines and in testicular tumour cells by immunoperoxidase staining or immunogold labelling in electron micrographs (Boller et al. Citation1993; Citation1997).
5.3.2. Protease (Pro)
The protease of HML-2 is similar to retrovirus aspartic proteinases. In general, in mammalian retroviruses the proteinase is encoded as part of the Gag–Pol polyprotein. In contrast, HML-2 proteinase is encoded by its own reading frame located between gag and pol (Schommer et al. Citation1996). It can be translated as Gag–Pro or Gag–Pro–Pol polyproteins (Mueller-Lantzsch et al. Citation1993). The protease undergoes autocatalytic cleavage into an 18-kDa fragment that cleaves Gag (Mueller-Lantzsch et al. Citation1993; Schommer et al. Citation1996).
5.3.3. dUTPase
The dUTPase domain is found in almost all HML-2 loci (Mayer and Meese Citation2003). This enzyme has two important functions: it removes dUTP from the deoxynucleotide pool, reducing the probability of uracil incorporation into the DNA, and it produces the dTTP precursor dUMP (Vertessy and Toth Citation2009). In all betaretroviruses the dUTPase is a proteolytic product of the Gag–Pro polyprotein precursor. Proviruses belonging to the HERV-K(HML-2) family also contain dUTPase motifs (Mayer and Meese Citation2003). In MMTV, the betaretrovirus most closely related to HERV-K, the ribosomal frameshift occurring at the gag–pro junction is responsible for the expression of a 30-kDa transframe protein, which contains the nucleocapsid protein domain of Gag fused to 154-amino acid residues derived from the 5′ region of the pro ORF. This transframe product has been identified as the viral MMTV dUTPase (Koppe et al. Citation1994). In a study using primers against the HERV-K10 sequence, the authors amplified a 513-nts-amplicon from human DNA, corresponding to the 3′ region of gag and the 5′ region of pro, containing the five essential dUTPase motifs (Harris et al. Citation1997). They cloned and sequenced 22 copies of the HERV-K dUTPase gene and deducted a consensus sequence from them. Although enzymatic activity was reported for such consensus sequence of the dUTPase region of the HML-2 group (Harris et al. Citation1997), it is it is uncertain whether any of the real predicted dUTPse proteins of the HML-2 proviruses are enzymatically active (Ariza and Williams Citation2011).
5.3.4. Polymerase (Pol): reverse transcriptase (RT), integrase (IN) and RNase H
RT gives retroviruses their name, based on the conversion of RNA into DNA. Reverse transcription of retroviruses occurs by the concerted action of the RT with the RNase H, which degrades the template RNA of the RNA–DNA heteroduplex after it is copied by the polymerase. The polymerase domain of the RT enzyme then forms the second strand of DNA, which is integrated into the host genome by the integrase (IN). In HML-2, the pol gene encodes the RT enzyme with a RNase H domain (Ono et al. Citation1986; Berkhout et al. Citation1999), and the integrase (IN) (Kitamura et al. Citation1996). Several HML-2 loci appear to encode functional RT proteins (Berkhout et al. Citation1999; Contreras-Galindo et al. Citation2017) with RNase H activity (Berkhout et al. Citation1999) (). In one study, six full-length HML-2 RT genes were cloned and several of the HERV-K RT enzymes produced exhibited polymerase as well as RNase H activity (Berkhout et al. Citation1999). Those HML-2 RT enzymes were strictly dependent on Mg2+, and did not show polymerase activity with other divalent cations (Berkhout et al. Citation1999). The integrase of HML-2 (HERV-K10) has also been cloned; it showed both terminal cleavage and strand transfer in the presence of Mn2+ of the HML-2 LTR substrate (Kitamura et al. Citation1996; Bray et al. Citation2016). In the human genome (GRCh38) only eight HML-2 proviruses have intact integrase motifs (Bray et al. Citation2016).
5.3.5. Envelope (Env)
Retroviral Env proteins are generally synthesized as precursors that trimerize in the endoplasmic reticulum and are cleaved by cellular furin-proteases in the late Golgi apparatus to generate two subunits: SU, which interacts with the receptor on target cells, and TM, which mediates fusion and is anchored in the viral envelope near its C terminus. After cleavage, SU and TM remain associated either non-covalently or via an intersubunit disulfide bond, resulting in trimers of heterodimers that traffic to the cellular plasma membrane and become part of the envelope in budding viral particles (Henzy and Coffin Citation2013).
Several lines of evidence suggest that the exogenous ancestors of HML-2 used the Env protein to infect germ cells via a cellular receptor (Turner et al. Citation2001). In HML-2 genomes, the pol and env reading frames partially overlap, and the Env protein is translated from a singly spliced transcript (). Several studies have identified HML-2 proviruses with complete ORFs for the Env protein (). In a study, the authors cloned 6 HML-2 env sequences from genomic loci to test their coding capability and functionality. The sequence of the putative proteins encoded by the six complete HERV-K env genes were highly conserved, with more than 97% identity at the amino acid level (de Parseval et al. Citation2003). The structural organization of the HML-2 Env proteins encoded by the six genes cloned was determined to be canonical, with a SP (although longer than canonical retroviruses) at the N-terminus, a consensus cleavage site for the cellular furin protease that splits the SU and TM subunits (), a hydrophobic fusion domain at the TM subunit and a hydrophobic TM anchor domain (Dewannieux et al. Citation2005). Of all loci tested, the authors found that only one, locus K108L in chromosome 7p22.1a, encoded a functional Env protein (Dewannieux et al. Citation2005). The Env protein encoded by this locus underwent cleavage into SU and TM subunits, could be exported to the cell surface and finally generated infectious particles when pseudotyped with env-deficient constructs derived from simian immunodeficiency virus, HIV or MLV. The infection was restricted to certain types of cells, possibly indicating the necessity of a particular receptor (Dewannieux et al. Citation2005), which has not yet been identified. The processing of the Env protein encoded by the same locus (K108L; 7p22.1a) was confirmed in another study (Ruggieri et al. Citation2009). In the endoplasmic reticulum, the Env precursor undergoes cleavage by the signal peptidase releasing a 13-kDa SP and the 90-kDa Env precursor. The Env precursor follows the maturation pathway to the Golgi where it is further cleaved by a furin-like endoprotease into two N-glycosylated domains, a 55-kDa SU and a 39-kDa TM subunit (Ruggieri et al. Citation2009) (). Then, the SU and TM subunits of HML-2 Env associate non-covalently as predicted by the two-cysteine motif of the TM sequence (Henzy and Coffin Citation2013). Although studies on the formation of trimers by HML-2 Envs remain to be performed, since all the loci studied have a canonical sequence, it is reasonable to assume that HML-2 Env heterodimers will associate in trimers as canonical betaretroviruses (Aydin et al. Citation2014).
Despite only locus K108L has been found to encode a functional Env, in another study, the infectivity of the K113 element could be restored by reversing some mutations in its ORF (Hanke et al. Citation2009).
In a study by Ruggieri et al. (Citation2009) the authors attempted to characterize the long SP predicted by the sequence. SPs assist in transportation of the Env precursors to the endoplasmic reticulum to follow the secretory pathway. Then, SP is cleaved by the signal peptidase and is degraded. The SP of HML-2 Env has 96 amino acids organized into three domains. Amino acids 1–75 form the N extension, amino acids 76–90 form the hydrophobic domain and amino acids 91–96 form the polar domain. The arginine rich nuclear localization signal (amino acids 13–20) and the leucine rich nuclear export signal (amino acids 54–60) are located within the N extension region (Ruggieri et al. Citation2009). The authors determined that HML-2 SP (locus K108L; 7p22.1a) has a long half-life compared to conventional SPs and is translocated to the granular component of the nucleoli after being cleaved. The authors speculated that HML-2 SP exerts an unknown activity in the nucleolus that is yet to be characterized (Ruggieri et al. Citation2009).
5.3.6. Rec
Rec is encoded by two exons ( and ) and consists of 105 amino acids. It is an accessory protein that binds to viral transcripts to facilitate their nucleocytoplasmic transport and incorporation of the viral genome into particles in the cytoplasm (Wodrich and Krausslich Citation2001). It functions like the Rev protein of human immunodeficiency virus (HIV) and the Rex protein of human T cell leukemia virus (HTLV). Besides assisting in nuclear RNA export, these proteins also facilitate translation of the transported mRNAs by enhancing the association with polysomes and accelerating the encapsidation of viral transcripts (Blissenbach et al. Citation2010). However, the functions of HML-2 Rec in this regard have not yet been explored. Rec is a 14-kDa protein that accumulates primarily in the nucleoli but is also present in the cytoplasm (Hanke, Chudak et al. Citation2013). It contains an arginine-rich nuclear location signal (NLS), which interacts with importin-β, mediating entry into the nucleus. The NLS also recognizes the Rec-responsive element (RcRE) within the viral RNAs (vRNAs). Rec forms tetramers and, presumably, three tetramers bind to purine-rich stretches within the RcRE (Langner et al. Citation2012). To achieve its nucleocytoplasmic function Rec interacts with Staufen-1, which is also involved in utilization of the cargo RNA for efficient transport and particle encapsidation (Hanke, Hohn, et al. Citation2013).
5.3.7. Np9
In contrast to Rec, Np9 has no known function in HML-2. Np9 is expressed in various types of cancer (Armbruester et al. Citation2002; Chen et al. Citation2013), and in healthy tissues (Schmitt et al. Citation2015). Like Rec, Np9 translocates to the nucleoli (Armbruester et al. Citation2004) and can activate several oncogenic pathways (see Section 8.1.1.2).
6. HML-2 silencing, activation and potential for replication
6.1. Silencing
HML-2 expression is highly regulated, temporally and spatially, during human development. It is highly expressed at the 8-cell stage in the embryo and the expression continues throughout formation of the blastocyst (Grow et al. Citation2015). As development progresses, HML-2 is silenced by mechanisms such as CpG methylation, deamination (Manghera and Douville Citation2013) and alteration of histones (Grow et al. Citation2015). The reconstituted HML-2 consensus sequence (Lee and Bieniasz Citation2007) is sensitive to the retroviral restriction factor APOBEC3F, but not to APOBEC3G or TRIM5α (Lee and Bieniasz Citation2007).
It would be of interest to test whether the Krueppel-associated box (KRAB) zinc finger protein ZFP809 (Wolf et al. Citation2015), which has been reported to play a role in ERV silencing, might also repress HML-2. Previous studies using mouse embryonic stem cells (ESCs) have shown that the KRAB zinc finger proteins target TRIM28, which in turn recruits histone methyltransferase SETDB1 (ESET) that deposits histone 3 lysine 9 trimethylation at the proviral genes (Gautam et al. Citation2017). This histone modification is critical for gene repression. In mouse ESCs, ChIP-seq analysis demonstrated transcriptional repression of ERVS through direct recruitment of the histone chaperone CHAF1A and the sumoylation factor Sumo2 to those sequences. Sumo2 represses the provirus by sumoylation of TRIM28 and CHAF1A reinforces transcriptional repression via its interaction with members of the NuRD complex (KDM1A, HDAC1/2) and ESET (Yang et al. Citation2015). The KRAB domain is known for its repressor activity mediated by a tandem array of up to 40 zinc finger domains which bind to specific DNA sequences. The DNA-binding motifs for the KRAB zinc finger proteins C2H2 are listed in Barazandeh et al. (Citation2018).
6.2. Transcription
Both epigenetic modifications and transcription factor binding regulate HML-2 expression. It has been shown that a functional HML-2 promoter cannot be activated in a cell lacking the appropriate transcription factors (Fuchs et al. Citation2011). Due to the variations in the sequence of the HML-2 LTR among the different loci, it is important to point out that different proviruses vary considerably in their patterns of transcription factor binding sites, no doubt leading to the alterations in expression of different viral genes (from different proviruses) in various cell types. For example, during human preimplantation development, transcripts originating from LTR5HS, but not LTR5A or LTR5A are preferentially expressed (Grow et al. Citation2015). Sequence analysis showed that OCT4 has a binding site in diverse LTR5HS sequences that it is not present in LTR5A or LTR5B. ChIP–qPCR analysis showed OCT4 binding in the predicted group of LTR5HS, but not in LTR5A/LTR5B in human embryonic carcinoma cells (Grow et al. Citation2015).
Using a construct based on the sequence of the K108 LTR (chr 7) provirus, transcription factors Sp1 and Sp3 were found to regulate their transcription (Fuchs et al. Citation2011). It is worth noting that the K108 locus on chromosome 7 contains a tandem HML-2 repeat with three LTRs, sharing more than 99% of homology (Reus, Mayer, Sauter, Scherer, et al. Citation2001), and it is not clear from the published data which LTR was used in the study by Fuchs et al. (Citation2011).
A corticoid responsive site has been found in the LTRs of HERV-K18 and HERV-K10 (Ono Citation1986). Its expression is stimulated by progesterone and testosterone (Hanke, Chudak, et al. Citation2013). Furthermore, Rec upregulates androgen receptor (AR) activity and thus, enhances AR-mediated activation of the HML-2 LTR (Hanke, Chudak, et al. Citation2013). The consensus sequence of HML-2 LTR has binding sites for inflammatory transcription factors (Manghera and Douville Citation2013). Moreover, HML-2 may also indirectly enhance its own expression through cellular components such as TAR (trans-activation-responsive) DNA binding protein 43 (TDP-43), which binds to the consensus HML-2 LTR and upregulates its transcription (Li et al. Citation2015). HIV Tat has been shown to activate HML-2 expression at the level of the consensus transcriptional promoter (Gonzalez-Hernandez et al. Citation2012). Since the LTR has a Rec-responsive element (Magin et al. Citation1999), it would be interesting to study whether Rec affects the activation of HML-2 LTRs. A list of the transcription factors confirmed to bind to the consensus and certain genomic HML-2 LTRs sequences is included in .
6.3. Reverse transcription and particle formation
Until recently, all retroviruses, including HML-2, were considered to have a general life cycle that included packaging of a RNA genome that is reverse transcribed in the target cell and subsequently integrated in the host genome. However, recent discoveries have demonstrated alternative replication patterns among retroviruses. In fact, the retroviral genus of spumaviruses produce viral particles containing both RNA and DNA genomes (Yu et al. Citation1996, Citation1999; Linial Citation1999; Rethwilm Citation2003); the reverse transcription of the RNA occurs in the virus-producing cell (late reverse transcription), prior to budding and release (Yu et al. Citation1996, Citation1999) and the DNA-containing particles appear to be more infectious than the RNA ones (Yu et al. Citation1996, Citation1999; Delelis et al. Citation2003). HIV also has partial reverse transcribed DNA in some viral particles upon release from the cells, although the majority of DNA sequences found in the virions are short, with only a fraction of them extending beyond the minus-strand strong stop (Lori et al. Citation1992; Trono Citation1992).
Studies using uracil N-glycosylase (UNG) have suggested that HML-2 is a genomically versatile virus containing either RNA or DNA genomes within viral particles. UNG specifically hydrolyzes uracil–glycosidic bonds in DNA but not in RNA. While genomic DNA contains little or no uracil, reverse transcribed DNA can contain significant amounts (Longo et al. Citation1990). When HML-2 viral particles are treated with UNG it results in a significant decrease in viral load (Laderoute et al. Citation2007, Citation2015; Dube et al. Citation2014).
Using an HML-2 consensus construct and by measuring infectivity after treating virion-producing and/or target cells with antiretrovirals, it has been suggested that HML-2 reverse transcription can take place in three distinct times and locations (1) in the virus-producing cell, prior to viral release (late reverse transcription); (2) within the extracellular virus particle itself; and (3) in the target cell, after a RNA-containing viral particle enters the cell (early reverse transcription) (Dube et al. Citation2014). This would result in viral particles containing either RNA or DNA genomes. In the same study, authors showed that both RNA and DNA-containing viral particles could infect target cells (Dube et al. Citation2014). However, it is important to note that this concept is not widely accepted as the possibility of contamination with genomic DNA in the assay cannot be entirely ruled out. More studies from independent groups are needed to confirm these results.
6.4. Integration and episomal DNA
Reconstruction of HML-2 using a consensus sequence derived from proviral sequences in the human genome produced retrovirus-like particles that were stably integrated after undergoing reverse transcription (Lee and Bieniasz Citation2007). It is important to note that this property of infection has only been demonstrated in vitro with a reconstructed virus using a consensus sequence (HML-2Con) and not with any of the actual sequences of HML-2 present in the human genome. When cell lines were infected with HML-2Con, it preferentially integrated in gene-rich regions near active transcriptional units and regulatory regions. In contrast, genomic HML-2 insertions are located preferentially in intergenic regions. Among the HML-2 loci that are inserted in genes, the majority are in opposite orientation relative to the host gene. This orientation is thought to be minimally disruptive to the host mRNA synthesis. In contrast, the consensus sequence integrated randomly in vitro in antisense and sense orientation. These findings have been interpreted to suggest that initial integration of HML-2 into the human genome occurred in transcriptionally active units but through a process of selection there was a loss of proviruses that were disruptive to gene function (Brady et al. Citation2009).
In another study, an HML-2 probe derived from provirus K113 was created to study the packaging and transmission of HERV-K sequences. This resulted in abundant episomal proviral DNA after the viral particles entered the target cells and underwent reverse transcription, but integration of the probe into the chromosomal DNA was not detected (Contreras-Galindo et al. Citation2015). However, the HML-2 episomes were transcribed and translated into proteins that were packaged into new particles, as the probes were able to produce a protein that conferred G418 resistance to the target cells. If, as suggested by Contreras-Galindo et al. (Citation2015), HML-2 is not able to integrate in the genome due to a lack of essential viral factors or by repressor mechanisms of the host cell, insertional mutagenesis cannot be a mechanism for cancer pathogenesis caused by HML-2.
6.5. Viral entry
For all retroviruses, the Env protein is responsible for the initial events that lead to endogenization, from cell binding to membrane fusion. The first step for effective viral infection is the fusion between the viral Env and the cellular membrane. HML-2 enters the cells via an endocytic pathway which requires dynamin-mediated membrane scission and endosomal acidification, but does not require macropinocytosis or actin polymerization (Robinson and Whelan Citation2016). HML-2 Env imparts broad species tropism to the virus, by being able to enter numerous mammalian and non-mammalian cell lines. This suggests that the receptors used by HML-2 to enter the cells are conserved throughout amniotes or that it can use multiple pathways (Robinson and Whelan Citation2016).
7. Possible physiological functions of HML-2
The main focus of HML-2 research has been their involvement in diseases, leading to the erroneous idea that they are only reactivated under pathological conditions. However, as mentioned above, HERVs are constitutively expressed at low levels in every human tissue studied and may provide potential benefits to their hosts (Kurth and Bannert Citation2010). In the sections below, we review the possible physiological roles of those genes.
7.1. Maintenance of pluripotency
To understand their role in oncogenesis, possibly the most relevant function of HERV-K and other ERVs is their involvement in the maintenance of cell pluripotency. High levels of HML-2 transcripts and proteins are found in undifferentiated ESCs and induced pluripotent stem cells. Induction of differentiation rapidly silences this expression (Fuchs et al. Citation2013). The role of HERVs in maintenance of the undifferentiated phenotype and cell proliferation is supported by the observation that some HERV LTRs contain binding sites for p53 and OCT4 (Grow et al. Citation2015). In fact, HERV LTRs account for over 30% of all p53 binding sites genome-wide (Wang et al. Citation2007). The exact function of p53 in the regulation of HML-2 expression has not been studied, but possibly the tumour suppressor p53 could act as a transcriptional repressor of HERVs.
7.2. Neurotrophic effects
Human neural stem cell lines transfected with HML-2 Env exhibited increased NGF and BDNF expression, which promoted cellular viability and prevented neurotoxicity. In mice, HML-2 Env expressed by neural stem cells implanted in the brain suppressed TNF-α expression and microglial activation while also improving neurobehavioral deficits in vpr/RAG1−/− mice (Bhat et al. Citation2014). These findings in neural stem cells suggest that HML-2 may have opposite effects in undifferentiated and differentiated cells, as its expression has been shown to be pathogenic for neurons (Li W et al. Citation2015).
7.3. Antiviral defense
Viral infection of a cell prevents infection from other related viruses (Moelling and Broecker Citation2015). Thus, it has been hypothesized that ERVs have been co-opted by vertebrates to protect them against infections of related exogenous retroviruses. For example, expression of the Env protein of the Fv-4 ERV on the surface of mouse cells competes for the receptors of related viruses, preventing them from entering the cell (Ikeda and Odaka Citation1984). The expression of a truncated Gag from the Fv-1 ERV also protects against some strains of MLV (Best et al. Citation1997). Similarly, the Env protein of HERV-W induces cellular resistance to spleen necrosis virus (Ponferrada et al. Citation2003). This suggests that HML-2 activation in HIV-infected patients may also confer similar protective effects. However, this potential antiviral effect of HML-2 has not been fully studied. Interestingly, none of the three clades of HERVs (gamma, beta and spuma-like retroviruses) have exogenous counterparts that are infectious for humans, as spuma-like viruses only infect humans by zoonosis. Similarly, exogenous retroviruses that are able to infect human cells (lenti and deltaretroviruses) do not have endogenous representatives in humans (van der Kuyl Citation2012). This fact could support the hypothesis of HERVs co-opted as antiviral defense against related exogenous retroviruses.
7.4. Placenta formation
The most characteristic physiological function of HERVs is the involvement of the Env proteins in the formation of the placenta, where they are highly expressed. Syncytin-1 and 2, which are the Env proteins of HERV-W and HERV-FRD, respectively, have preserved fusogenic capacity that allows the formation of the syncytiotrophoblast layer and its junction with the cytotrophoblast (Blond et al. Citation2000; Blaise et al. Citation2003). HML-2 Env, however, does not form syncytia. It is expressed in villous and extravillous cytotrophoblast cells in the placenta, but not in the syncytiotrophoblast (Kammerer et al. Citation2011).
7.5. Effects of HML-2 on host genome function
ERVs can affect the expression of the genes in their proximity. LTRs can act as alternative promoters or enhancers not only of the genes downstream in sense orientation (Schulte et al. Citation1996), but also upstream, as they can have bidirectional activity (Feuchter and Mager Citation1990; Dunn et al. Citation2006). Such LTRs with promoter functions can influence genes at a distance of up to 100 kb (Whitelaw and Martin Citation2001). For example, an antisense-oriented HERV-H LTR serves as an alternative promoter for the gene GSDML (Huh et al. Citation2008). The LTRs of HERVs might influence the expression of neighbour genes due to the presence of regulatory elements in their sequences, such as enhancers, promoters, splice sites (Kapitonov and Jurka Citation1999), and polyadenylation signals. Interestingly, two HML-2 LTRs situated in the introns of genes SLC4A8 (sodium bicarbonate cotransporter) and IFT172 (intraflagellar transport protein 172) in the antisense orientation serve in vivo as promoters for generating RNAs complementary to the exons of those genes. The antisense transcripts formed from the LTRs decreased the mRNA level of the corresponding genes (Gogvadze et al. Citation2009). The LTR of an HML-2 provirus in chromosome 22 has been shown to regulate the expression of proline dehydrogenase gene (PRODH) in the hippocampus, creating a tissue-specific enhancer (Suntsova et al. Citation2013).
8. HML-2 in pathological conditions
8.1. HML-2 and cancer
The fact that some retroviruses can cause cancer has been known since the discovery that Rous sarcoma virus causes sarcoma in chickens. Two other well-known onco-retroviruses are HTLV-1, which causes adult T-cell lymphoma in 2–7% of infected individuals (Watanabe Citation2017), and MMTV, which causes mammary tumours in mice and is also a beta-retrovirus that can be carried endogenously like HML-2.
HML-2 has been implicated in cancer development, as its expression has been associated with many cancer types such as teratocarcinoma, germ cell tumours, melanoma, ovarian, and prostate cancer (Lower et al. Citation1984; Herbst et al. Citation1996; Muster et al. Citation2003; Buscher et al. Citation2005, Citation2006; Wang-Johanning et al. Citation2007; Kurth and Bannert Citation2010), and with various features of malignant cells (Oricchio et al. Citation2007; Reis et al. Citation2013; Schmitt et al. Citation2013; Wildschutte et al. Citation2014; Bhardwaj et al. Citation2015).
8.1.1. Potential Mechanisms of HML-2-Induced Carcinogenesis
8.1.1.1. Chromosomal rearrangements and LTR-induced upregulation of oncogenes
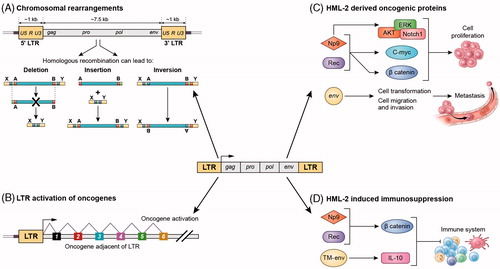
Cancer arises from abnormal expression of oncogenes, as well as inactivation of tumour suppressor genes. Aberrant expression of coding genes or long non-coding RNAs (lncRNAs) with oncogenic properties can be caused by epigenetic changes, translocations, point mutations and other less characterized mechanisms. Due to their sequence similarities, homologous recombination can occur between two different LTRs. Thus, many HERVs exist as solo LTRs and lack their internal proviral sequence due to homologous recombination between the 5′ and 3′ LTRs. In fact, solo LTRs outnumber full length proviruses by a factor of 10 (Subramanian et al. Citation2011). The recombination can also occur between distant LTRs, even on different chromosomes. Phylogenetic and sequence analyses have shown that some HML-2 loci have contributed to human genome evolution through large-scale chromosomal rearrangements (Hughes and Coffin Citation2001), although probably to a lesser extent than other transposable elements like Alus (Ade et al. Citation2013), simply due to their much lower copy number. Thus, it is possible for HERVs to contribute to carcinogenesis through homologous recombination resulting in chromosomal rearrangements (), although it is likely a rare event. Additionally, as explained in Section 5.2, HML-2 LTRs contain all the regulatory elements to act as promoter of the provirus. There is a selective transcriptional repression of LTRs residing in gene introns (Buzdin et al. Citation2006) and at least half of the human specific HML-2 LTRs seem to serve in vivo as active promoters in non-repetitive regions of the genome (Buzdin et al. Citation2006). If an HML-2 LTR is de-repressed, it could act as alternative promoter of adjacent host genes (Buzdin et al. Citation2006; Fuchs et al. Citation2011; Katoh et al. Citation2011) (). Although it has not yet been described for HML-2, LTRs in their natural positions in the genome have been shown to drive ectopic expression of genes in cancer, possibly due to epigenetic de-repression (reviewed in Babaian and Mager Citation2016). Regarding HERV-K, one study on prostate cancer found that chromosomal rearrangements caused the ETV1 gene to fuse with the LTR of HML-2 22q11.23, causing an aberrant overexpression of truncated ETV1. ETV1 encodes a transcription factor that controls cellular proliferation, differentiation, development, transformation, and apoptosis (Tomlins et al. Citation2007). The fusion transcript included the upstream 5′LTR, providing evidence that the HERV-K LTR controls the expression of ETV1. Another example of HML-2 participating in an oncogenic gene fusion after chromosomal translocation was found in the stem-cell myeloproliferative disorder linked to the 8p12 chromosomal region. This syndrome is caused by the aberrant expression of the FGFR1 gene, which encodes one of the tyrosine kinase receptors for fibroblast growth factors (Chaffanet et al. Citation1998). Several FGFR1 fusion genes formed by translocation have been described in this lymphoproliferative disorder (Guasch et al. Citation2003). By cloning one translocation breakpoint, a fusion of a HERV-K LTR on chromosome 19 to FGFR1 sequences was revealed. Sequence analysis showed nucleotide similarities with the sequence of HERV-K113 (Chr 19) (Guasch et al. Citation2003). However, while it was suggested by the authors that the LTR promoter may contribute to the expression of the fusion gene, in this case no supporting evidence was presented.
Figure 3. Potential mechanisms for HML-2-induced carcinogenesis. (A) Chromosomal rearrangements: HML-2 LTRs may contribute to carcinogenesis by facilitating homologous recombination, resulting in deletions, duplications, inversions or fusions of interspersed genomic sequence. (B) LTR-induced upregulation of adjacent oncogenes: If the LTR of HML-2 is de-repressed, it could recruit transcription factors and serve as an alternative promoter of adjacent host genes involved in cell proliferation. (C) HML-2-derived oncoproteins: NP9 and Rec contribute to cellular proliferation, by activation of ERK, AKT and Notch1, c-myc and beta-catenin. HML-2 Env has been shown to promote cell transformation and increase cellular migration and invasion, which could lead to metastasis. (D) HML-2 immunosuppression: NP9 and Rec can upregulate beta-catenin, which induce immune tolerance to tumours. HML-2 Env has an immunosuppressive domain in the TM subunit that triggers upregulation of IL-10.
8.1.1.2. HML-2-derived oncoproteins
HML-2 encodes several proteins that have been implicated in cancer such as Np9, Rec and Env, which could be contributing to cellular proliferation (). Np9 has been found to be expressed in various tumour tissues such as breast cancer, ovarian cancer, and leukaemia (Armbruester et al. Citation2002; Chen et al. Citation2013). Np9 promotes growth of myeloid and lymphoblastic leukaemia cells by activation of ERK, AKT, and Notch1 pathways and by upregulation of β-catenin (Chen et al. Citation2013). Moreover, like Rec, Np9 interacts with PLZF, which has been linked to prostate cancer (Robinson et al. Citation2015). Np9 interaction with PLZF leads to increased transcription of the gene c-myc, which in turn leads to enhanced cell growth and reduced apoptosis (Denne et al. Citation2007).
Another way Rec may contribute to prostate cancer is by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT), which is a known regulator of the androgen receptor (AR). When Rec binds to hSGT, AR activity is increased. Interestingly, AR has been shown to activate HML-2 LTR, and this activation may lead to a “vicious cycle” that could result in increased cell proliferation and tumorigenesis (Hanke, Chudak et al. Citation2013).
HML-2 Env has also been suggested as a potential oncogenic stimulus. Using a MCF10A cell line, HML-2 Env (K108L) expression from a lentiviral construct endowed the cells with the ability to transition from epithelial cells to mesenchymal cells; the cells displayed an increase in cellular migration and invasion (Lemaitre et al. Citation2017). Additionally, a recent study showed that downregulation of Env prompted a decrease in cell proliferation and a concomitant reduction of RAS, p-ERK, and p-AKT expression (Li M et al. Citation2017).
8.1.1.3. HML-2-mediated immunosuppression
Immunosuppressive properties are characteristic of many retroviruses and HML-2 has retained this feature (Denner Citation1998, Citation2000). HML-2 particles released from a human teratocarcinoma cell line, a recombinant Env transmembrane (TM) protein, and a peptide corresponding to a highly-conserved region of the TM were all able to inhibit human immune cell proliferation, change the expression of numerous cytokines, such as increasing IL-10, and affect gene expression (Morozov et al. Citation2013). Similarly, Rec has been shown to interact with β-catenin, which induces immune tolerance to tumours (Liang et al. Citation2014; Suryawanshi and Manicassamy Citation2015), contributing to carcinogenesis (Gonzalez-Cao et al. Citation2016). Currently, HML-2 proteins and transcripts are being considered as potential targets for treatment in several cancer types.
8.2. HML-2 polymorphism-associated diseases
Sequence variations and insertional polymorphisms of HERV-K have been associated with diabetes and other autoimmune diseases. A HERV-K18 haplotype located in the intron of CD48 on chromosome band 1q has been reported to present a weak association with type 1 diabetes (Marguerat et al. Citation2004). In addition, low copies of the insertional polymorphism HERV-K(C4), which belongs to the HML10 group, on chromosome 6 has been associated with an increased risk for diabetes. This copy of HERV-K is present in ∼70% of the population in intron 9 of the C4 complement gene, in opposite orientation to the host gene. However, this association was not confirmed by another study in Germany (Pani et al. Citation2002). HERV-K113 and HERV-K115 are other insertional polymorphisms found in 29 and 16% of individuals, respectively. Both are full length proviruses and have ORFs for all their genes except for a frame shift mutation in the gag gene of HERV-K115. The presence of HERV-K113 has been associated with an increased risk for certain autoimmune diseases (Krzysztalowska-Wawrzyniak et al. Citation2011). HERV-K113 is capable of forming a complete viral particles but the envelope is non-fusogenic (Moyes et al. Citation2007).
8.3. HML-2 and neurological diseases
The evidence for the role of HML-2 in the pathophysiology of sporadic ALS is strong. Several groups have identified the presence of RT activity in the blood and cerebrospinal fluid of patients with ALS (Viola et al. Citation1975; Steele et al. Citation2005; McCormick et al. Citation2008). HML-2 gene products, gag, pol and env can be detected in the brains of ALS patients, and RT and Env proteins are expressed in cortical neurons (Douville et al. Citation2011; Li W et al. Citation2015). This expression was specific for ALS, since it could not be found in patients with Parkinson’s or Alzheimer’s disease. Multiple active HML-2 loci have been identified, although loci in chromosome 7 seem to be differentially expressed in patients and controls (Douville et al. Citation2011). Forced expression of HML-2 in neurons, either by transfection with the complete consensus sequence or by activation of the endogenous proviruses with a CRISPR/dCAS9 targeting HML-2 LTRs, lead to neuronal injury and cell death. Transgenic mice in which HML-2 Env (consensus) was expressed under a neuronal promoter developed progressive motor dysfunction, with specific loss of neurons in the motor cortex and the anterior horn of the spinal cord (Li W et al. Citation2015). Although the mechanism of pathogenicity is not entirely clear, it may be partly mediated by TDP-43 which has several putative binding sites on the LTR of HML-2. Signs of nucleolar dysfunction were also observed, as evidenced by redistribution of the nucleolar marker nucleophosmin into the cytoplasm of Env-expressing neurons (Li W et al. Citation2015). Additional evidence comes from rare cases of HIV-infected patients who also develop ALS. HIV infection has been associated with increasing levels of HML-2. When HIV-ALS patients were treated with antiretroviral drugs early in the course of the neurological manifestations, ALS symptoms could be reversed or slowed in a subset of patients (Alfahad and Nath Citation2013). The activation of HML-2 found in the blood of some of these patients decreased following treatment with antiretroviral drugs (Bowen et al. Citation2016). Besides ALS, activation of HML-2 and polymorphisms of HERV-K18 and K115 have been associated with schizophrenia (Otowa et al. Citation2006; Dickerson et al. Citation2008). A nearly full length HML-2 has also been identified in the PRODH gene where its enhancer activity is regulated by methylation. PRODH is expressed in multiple regions of the brain, with highest levels in the hippocampus where the HML-2 is hypomethylated (Suntsova et al. Citation2013). PRODH encodes proline oxidase, a mitochondrial enzyme that regulates proline catabolism, which is vital for brain function. Polymorphisms in this gene have been associated with schizophrenia (Kempf et al. Citation2008). However, in schizophrenia the association does not seem to be specific for HERV-K, as HERV-W and ERV-9 had also elevated levels (Diem et al. Citation2012).
8.4. Interactions between HML-2 and HIV
It is well established that HIV infection can increase HML-2 mRNA levels in PBMCs (Contreras-Galindo, Almodovar-Camacho, et al. Citation2007; Ormsby et al. Citation2012; Bhardwaj et al. Citation2014) and the activation of certain HML-2 loci can be cell-type specific (Vincendeau et al. Citation2015). The possible mechanisms of induction of HML-2 by HIV are explained in . However, the level of HML-2 induction in HIV infection remains unclear. Several studies have found that HML-2 RNA was readily detectable in plasma of HIV-infected individuals (Contreras-Galindo et al. Citation2006, Citation2012; Contreras-Galindo, Almodovar-Camacho, et al. Citation2007; Contreras-Galindo, Lopez, et al. Citation2007; Esqueda et al. Citation2013); however, other investigators were unable to confirm these findings (Bhardwaj et al. Citation2014; Karamitros et al. Citation2016). It has been suggested that contamination with genomic DNA is largely responsible for detection of HML-2 in blood, since rigorous DNA digestion with DNase I eliminated HML-2 from almost all patient samples (Esqueda et al. Citation2013; Bhardwaj et al. Citation2014; Karamitros et al. Citation2016). However, as described above (Section 6.3), if it is true that some HML-2 virions do in fact contain reverse-transcribed DNA in addition to viral RNA (Dube et al. Citation2014), then DNAse treatment would degrade the viral DNA and, hence, prevent detection of extracellular virus. Nevertheless, the hypothesis that HML-2 viral particles might contain DNA genomes is still far from being confirmed. Furthermore, intracellular HML-2 transcripts may be elevated in PBMCs from HIV-positive individuals compared to controls even when HML-2 RNA is undetectable in plasma (Sugimoto et al. Citation2001; Bhardwaj et al. Citation2014; Brinzevich et al. Citation2014).
Figure 4. Interactions between HML-2 and HIV. Productive infection with HIV upregulates HML-2 expression at the mRNA level. Upregulation occurs at least partly as a result of the HIV regulatory protein Tat, which activates the NFkB and NF-AT pathways that enhance both HIV and HML-2 transcription. Similarly, HIV Rev binds to the Rev response element (RRE) in the env region of unspliced and partially spliced HIV transcripts to mediate nuclear export. HML-2 Env proteins may also be produced and expressed on the surface of HIV-infected lymphocytes. Increased levels of HML-2 Env-specific antibodies have been observed in HIV-positive individuals. HML-2-Gag and Env proteins may also be processed intracellularly for presentation in the context of MHC class I molecules, leading to activation of HML-2 specific cytotoxic T cell responses. Similarly, the HML-2 integrase can mediate integration of an integrase-deficient HIV provirus under in vitro conditions. HML-2 co-assembles with HIV Gag in vitro impairing HIV assembly and release. Expression of HML-2-Env from specific type 2 proviruses (K108, K109) also inhibits HIV release. In contrast, Env derived from a consensus HML-2 sequence can substitute for the HIV Vpu protein by downregulating the HIV restriction factor tetherin, leading to enhanced viral release.
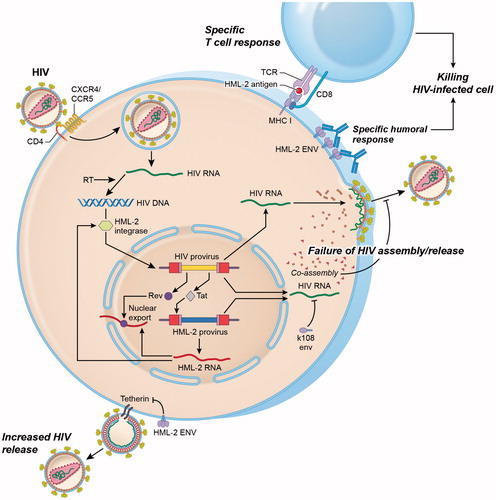
Regarding the influence of HML-2 on HIV pathogenesis, it is not clear whether expression of HML-2 helps control HIV or if it contributes to its pathogenicity (). HML-2 specific antibodies have been found in the blood of HIV-infected individuals at higher titers than uninfected controls (Michaud et al. Citation2014). “Elite controller” individuals, who control HIV replication in the absence of treatment had higher titers of antibodies to HML-2 compared to their antiretroviral-treated counterparts (Michaud et al. Citation2014), indicating that immune responses against HML-2 may correlate with HIV suppression. However, other groups could not find differences in HML-2 antibody levels between HIV patients and uninfected controls (Vogetseder et al. Citation1993). Similarly, several groups have reported T cell responses to HML-2 in HIV-infected patients (Garrison et al. Citation2007; SenGupta et al. Citation2011; Tandon et al. Citation2011; Jones et al. Citation2012) that are associated with better virologic control (Garrison et al. Citation2007; SenGupta et al. Citation2011) and higher T cell counts (SenGupta et al. Citation2011). Collectively, these findings suggest that HML-2 transcripts induced by HIV infection may be translated into viral proteins that can generate a humoral and/or cytotoxic T cell response.
The presence of HML-2 proteins produced in response to HIV infection also introduces the possibility that these proteins modulate HIV replication. It has been suggested that HIV and other primate lentiviruses, which do not encode dUTPase, can use the host enzyme encoded by HERVs (McIntosh and Haynes Citation1996; Harris et al. Citation1997). Similarly, some HML-2 loci encode a functional integrase protein that can act on the HIV-1 LTR (Kitamura et al. Citation1996), which suggests that HML-2 integrase may substitute for HIV integrase under specific conditions ().
HML-2 Gag can co-assemble with HIV Gag in vitro, which results in diminished HIV assembly and release (Monde et al. Citation2012, Citation2017). However, two studies have revealed functional differences between the consensus/reconstituted and native HML-2 Env sequences; the consensus sequence promotes HIV infection by antagonizing the restriction factor tetherin (Lemaitre et al. Citation2014; Terry et al. Citation2017), while Env produced from type 2 endogenous proviruses K108 and K109 significantly diminished HIV Gag protein levels and inhibited HIV release (Terry et al. Citation2017). In contrast, plasmids encoding functional HML-2 Env proteins derived from either the consensus sequence (type 2) or native HERV-K18 (type 1) “rescue” Env-deficient HIV in co-transfection experiments, allowing assembly and release of infectious viral particles (Brinzevich et al. Citation2014). Given that HML-2 Env appears to bind to the surface of many different cell types (Kramer et al. Citation2016), HIV virions “pseudotyped” with HML-2 Env could exhibit altered cellular tropism. These experiments highlight the need to better define which HML-2 loci are upregulated following HIV infection, and whether this induction results in translation of functional HML-2 proteins such as Gag and Env.
To date there is no evidence that the endogenous retroviral elements can rescue defective HIV virions in vivo. However, the possibility exists that recombination events might occur between the activated endogenous retroviral elements themselves or between the endogenous and exogenous retroviruses which may aid in their evolution.
9. Therapeutic approaches
Since increased activation of HML-2 has been associated with several pathologies, it raises the question of whether these genes can be silenced to affect the outcome of these diseases. Gene silencing techniques such as antisense, siRNA or shRNA could be used, although delivery to the target cells can be challenging, particularly within the brain. However, new viral vectors such as the adeno-associated viruses and nanoparticle delivery mechanisms hold promise. Even if its participation in carcinogenesis is not entirely clear, HML-2 is consistently over-expressed in certain cancer types. Therefore, cell-mediated immune responses can be directed against HML-2 antigens to eliminate cancer cells. The recent success of immune-mediated therapies, particularly the engineering of a chimeric antigen receptor on the surface of T cells, makes this a promising approach (Krishnamurthy et al. Citation2015; Zhou et al. Citation2015). In cases where viral assembly occurs, the use of antiretroviral drugs to inhibit productive viral replication could also be considered. Certainly, in vitro studies show that some of the reverse transcriptase and integrase inhibitors developed against HIV are also effective against HML-2, although with lower efficacy (Contreras-Galindo et al. Citation2017; Tyagi et al. Citation2017). Hence, more effective drugs need to be specifically designed that target HML-2.
10. Conclusions
It is becoming abundantly clear that HERVs may play a critical role in embryogenesis and in disease pathophysiology. Of the different HERVs in the human genome, HML-2 is distinct because of the presence of multiple nearly complete viral genomes, its high transcriptional activity and its possible non-canonical replication. Understanding the role of HERVs in cellular physiology is vital to eventually clarifying their relationship with disease pathogenesis. The mechanisms by which HERVs are regulated by the host and how they interact with other retroviruses are aspects that require more attention. At the same time, current therapeutic technologies for manipulating their expression make it possible to consider therapeutic intervention where dysregulation of HERVs is implicated.
Disclosure statement
The authors report no conflict of interests.
Additional information
Funding
References
- Aaronson SA, Todaro GJ, Scolnick EM. 1971. Induction of murine C-type viruses from clonal lines of virus-free BALB-3T3 cells. Science. 174(4005):157–159.
- Ade C, Roy-Engel AM, Deininger PL. 2013. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol. 3(6):639–645.
- Agoni L, Guha C, Lenz J. 2013. Detection of human endogenous retrovirus k (HERV-K) transcripts in human prostate cancer cell lines. Front Oncol. 3:180.
- Alfahad T, Nath A. 2013. Retroviruses and amyotrophic lateral sclerosis. Antiviral Res. 99(2):180–187.
- Andersson ML, Lindeskog M, Medstrand P, Westley B, May F, Blomberg J. 1999. Diversity of human endogenous retrovirus class II-like sequences. J Gen Virol. 80(Pt 1):255–260.
- Andersson ML, Medstrand P, Yin H, Blomberg J. 1996. Differential expression of human endogenous retroviral sequences similar to mouse mammary tumor virus in normal peripheral blood mononuclear cells. AIDS Res Hum Retroviruses. 12(9):833–840.
- Argaw-Denboba A, Balestrieri E, Serafino A, Cipriani C, Bucci I, Sorrentino R, Sciamanna I, Gambacurta A, Sinibaldi-Vallebona P, Matteucci C. 2017. HERV-K activation is strictly required to sustain CD133 + melanoma cells with stemness features. J Exp Clin Canc Res. 36:20.
- Ariza ME, Williams MV. 2011. A human endogenous retrovirus K dUTPase triggers a TH1, TH17 cytokine response: does it have a role in psoriasis? J Invest Dermatol. 131(12):2419–2427.
- Armbruester V, Sauter M, Krautkraemer E, Meese E, Kleiman A, Best B, Roemer K, Mueller-Lantzsch N. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin Cancer Res. 8(6):1800–1807.
- Armbruester V, Sauter M, Roemer K, Best B, Hahn S, Nty A, Schmid A, Philipp S, Mueller A, Mueller-Lantzsch N. 2004. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J Virol. 78(19):10310–10319.
- Aydin H, Cook JD, Lee JE. 2014. Crystal structures of beta- and gammaretrovirus fusion proteins reveal a role for electrostatic stapling in viral entry. J Virol. 88(1):143–153.
- Babaian A, Mager DL. 2016. Endogenous retroviral promoter exaptation in human cancer. Mob DNA. 7:24.
- Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A. 101(Suppl 2):14572–14579.
- Barazandeh M, Lambert SA, Albu M, Hughes TR. 2018. Comparison of ChIP-Seq data and a reference motif set for human KRAB C2H2 zinc finger proteins. G3 (Bethesda). 8(1):219–229.
- Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 9(16):861–868.
- Barmak K, Harhaj E, Grant C, Alefantis T, Wigdahl B. 2003. Human T cell leukemia virus type I-induced disease: pathways to cancer and neurodegeneration. Virology. 308(1):1–12.
- Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol. 79(19):12507–12514.
- Bentvelzen P, Daams JH, Hageman P, Calafat J. 1970. Genetic transmission of viruses that incite mammary tumor in mice. Proc Natl Acad Sci U S A. 67(1):377.
- Berkhout B, Jebbink M, Zsiros J. 1999. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J Virol. 73(3):2365–2375.
- Best S, Le Tissier PR, Stoye JP. 1997. Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol. 5(8):313–318.
- Bhardwaj N, Maldarelli F, Mellors J, Coffin JM. 2014. HIV-1 infection leads to increased transcription of human endogenous retrovirus HERV-K (HML-2) proviruses in vivo but not to increased virion production. J Virol. 88(19):11108–11120.
- Bhardwaj N, Montesion M, Roy F, Coffin JM. 2015. Differential expression of HERV-K (HML-2) proviruses in cells and virions of the teratocarcinoma cell line Tera-1. Viruses. 7(3):939–968.
- Bhat RK, Rudnick W, Antony JM, Maingat F, Ellestad KK, Wheatley BM, Tonjes RR, Power C. 2014. Human endogenous retrovirus-K(II) envelope induction protects neurons during HIV/AIDS. PLoS One. 9(7):e97984.
- Blaise S, de Parseval N, Benit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A. 100(22):13013–13018.
- Blikstad V, Benachenhou F, Sperber GO, Blomberg J. 2008. Evolution of human endogenous retroviral sequences: a conceptual account. Cell Mol Life Sci. 65(21):3348–3365.
- Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. 2010. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J Virol. 84(13):6598–6604.
- Blomberg J, Benachenhou F, Blikstad V, Sperber G, Mayer J. 2009. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene. 448(2):115–123.
- Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 74(7):3321–3329.
- Boller K, Janssen O, Schuldes H, Tonjes RR, Kurth R. 1997. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J Virol. 71(6):4581–4588.
- Boller K, Konig H, Sauter M, Muellerlantzsch N, Lower R, Lower J, Kurth R. 1993. Evidence that Herv-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus Htdv. Virology. 196(1):349–353.
- Bowen LN, Tyagi R, Li W, Alfahad T, Smith B, Wright M, Singer EJ, Nath A. 2016. HIV-associated motor neuron disease: HERV-K activation and response to antiretroviral therapy. Neurology. 87(17):1756–1762.
- Brady T, Lee YN, Ronen K, Malani N, Berry CC, Bieniasz PD, Bushman FD. 2009. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 23(5):633–642.
- Bray S, Turnbull M, Hebert S, Douville RN. 2016. Insight into the ERVK integrase – propensity for DNA damage. Front Microbiol. 7:1941.
- Brinzevich D, Young GR, Sebra R, Ayllon J, Maio SM, Deikus G, Chen BK, Fernandez-Sesma A, Simon V, Mulder LC. 2014. HIV-1 interacts with human endogenous retrovirus K (HML-2) envelopes derived from human primary lymphocytes. J Virol. 88(11):6213–6223.
- Buscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Lower J, Lower R, Kurth R, Denner J. 2006. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 16(3):223–234.
- Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 65(10):4172–4180.
- Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. 2006. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol. 80(21):10752–10762.
- Buzdin A, Ustyugova S, Khodosevich K, Mamedov I, Lebedev Y, Hunsmann G, Sverdlov E. 2003. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages. Genomics. 81(2):149–156.
- Cai BH, Chen JY, Lu MH, Chang LT, Lin HC, Chang YM, Chao CF. 2009. Functional four-base A/T gap core sequence CATTAG of P53 response elements specifically bound tetrameric P53 differently than two-base A/T gap core sequence CATG bound both dimeric and tetrameric P53. Nucleic Acids Res. 37(6):1984–1990.
- Callahan R, Chiu IM, Wong JF, Tronick SR, Roe BA, Aaronson SA, Schlom J. 1985. A new class of endogenous human retroviral genomes. Science. 228(4704):1208–1211.
- Cegolon L, Salata C, Weiderpass E, Vineis P, Palu G, Mastrangelo G. 2013. Human endogenous retroviruses and cancer prevention: evidence and prospects. BMC Cancer. 13:4.
- Chaffanet M, Popovici C, Leroux D, Jacrot M, Adelaide J, Dastugue N, Gregoire MJ, Hagemeijer A, Lafage-Pochitaloff M, Birnbaum D et al. 1998. t(6;8), t(8;9) and t(8;13) translocations associated with stem cell myeloproliferative disorders have close or identical breakpoints in chromosome region 8p11-12. Oncogene. 16(7):945–949.
- Chen T, Meng Z, Gan Y, Wang X, Xu F, Gu Y, Xu X, Tang J, Zhou H, Zhang X et al. 2013. The viral oncogene Np9 acts as a critical molecular switch for co-activating beta-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia. 27(7):1469–-1478. English.
- Cohen M, Larsson E. 1988. Human endogenous retroviruses. Bioessays. 9(6):191–196.
- Contreras-Galindo R, Almodovar-Camacho S, Gonzalez-Ramirez S, Lorenzo E, Yamamura Y. 2007. Comparative longitudinal studies of HERV-K and HIV-1 RNA titers in HIV-1-infected patients receiving successful versus unsuccessful highly active antiretroviral therapy. AIDS Res Hum Retroviruses. 23(9):1083–1086.
- Contreras-Galindo R, Contreras-Galindo A, Lorenzo E, Yamamura Y. 2006. Evidence for replication of human endogenous retroviruses type-K (HERV-K) in HIV-1 positive patients. Retrovirology. 3:S33.
- Contreras-Galindo R, Dube D, Fujinaga K, Kaplan MH, Markovitz DM. 2017. Susceptibility of human endogenous retrovirus type K to reverse transcriptase inhibitors. J Virol. 91(23):e01309-17.
- Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y et al. 2012. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol. 86(1):262–276.
- Contreras-Galindo R, Kaplan MH, Dube D, Gonzalez-Hernandez MJ, Chan S, Meng F, Dai M, Omenn GS, Gitlin SD, Markovitz DM. 2015. Human endogenous retrovirus type K (HERV-K) particles package and transmit HERV-K-related sequences. J Virol. 89(14):7187–7201.
- Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. 2007. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res Hum Retroviruses. 23(1):116–122.
- Costas J. 2001. Evolutionary dynamics of the human endogenous retrovirus family HERV-K inferred from full-length proviral genomes. J Mol Evol. 53(3):237–243.
- de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. 2003. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 77(19):10414–10422.
- Delelis O, Saib A, Sonigo P. 2003. Biphasic DNA synthesis in spumaviruses. J Virol. 77(14):8141–8146.
- Denne M, Sauter M, Armbruester V, Licht JD, Roemer K, Mueller-Lantzsch N. 2007. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J Virol. 81(11):5607–5616.
- Denner J. 1998. Immunosuppression by retroviruses: implications for xenotransplantation. Ann N Y Acad Sci. 862:75–86.
- Denner J. 2000. How does HIV induce AIDS? The virus protein hypothesis. J Human Virol. 3(2):81–82.
- Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J Virol. 79(24):15573–15577.
- Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16(12):1548–1556.
- Dickerson F, Rubalcaba E, Viscidi R, Yang S, Stallings C, Sullens A, Origoni A, Leister F, Yolken R. 2008. Polymorphisms in human endogenous retrovirus K-18 and risk of type 2 diabetes in individuals with schizophrenia. Schizophr Res. 104(1–3):121–126.
- Diem O, Schaffner M, Seifarth W, Leib-Mosch C. 2012. Influence of antipsychotic drugs on human endogenous retrovirus (HERV) transcription in brain cells. PLoS One. 7(1):e30054.
- Domansky AN, Kopantzev EP, Snezhkov EV, Lebedev YB, Leib-Mosch C, Sverdlov ED. 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472(2–3):191–195.
- Douville R, Liu J, Rothstein J, Nath A. 2011. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 69(1):141–151.
- Dube D, Contreras-Galindo R, He S, King SR, Gonzalez-Hernandez MJ, Gitlin SD, Kaplan MH, Markovitz DM. 2014. Genomic flexibility of human endogenous retrovirus type K. J Virol. 88(17):9673–9682.
- Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL. 2006. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene. 366(2):335–342.
- Ehlhardt S, Seifert M, Schneider J, Ojak A, Zang KD, Mehraein Y. 2006. Human endogenous retrovirus HERV-K(HML-2) Rec expression and transcriptional activities in normal and rheumatoid arthritis synovia. J Rheumatol. 33(1):16–23.
- Esqueda D, Xu F, Moore Y, Yang Z, Huang G, Lennon PA, Hu PC, Dong J. 2013. Lack of correlation between HERV-K expression and HIV-1 viral load in plasma specimens. Ann Clin Lab Sci. 43(2):122–125.
- Faulkner GJ, Carninci P. 2009. Altruistic functions for selfish DNA. Cell Cycle. 8(18):2895–2900.
- Feuchter A, Mager D. 1990. Functional heterogeneity of a large family of human LTR-like promoters and enhancers. Nucleic Acids Res. 18(5):1261–1270.
- Flockerzi A, Ruggieri A, Frank O, Sauter M, Maldener E, Kopper B, Wullich B, Seifarth W, Muller-Lantzsch N, Leib-Mosch C et al. 2008. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 9:354.
- Fuchs NV, Kraft M, Tondera C, Hanschmann KM, Lower J, Lower R. 2011. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J Virol. 85(7):3436–3448.
- Fuchs NV, Loewer S, Daley GQ, Izsvak Z, Lower J, Lower R. 2013. Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology. 10:115.
- Garrison KE, Jones RB, Meiklejohn DA, Anwar N, Ndhlovu LC, Chapman JM, Erickson AL, Agrawal A, Spotts G, Hecht FM et al. 2007. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 3(11):e165.
- Gautam P, Yu T, Loh YH. 2017. Regulation of ERVs in pluripotent stem cells and reprogramming. Curr Opin Genet Dev. 46:194–201.
- George M, Schwecke T, Beimforde N, Hohn O, Chudak C, Zimmermann A, Kurth R, Naumann D, Bannert N. 2011. Identification of the protease cleavage sites in a reconstituted Gag polyprotein of an HERV-K(HML-2) element. Retrovirology. 8:30.
- Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E. 2009. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J Virol. 83(12):6098–6105.
- Gonzalez-Cao M, Iduma P, Karachaliou N, Santarpia M, Blanco J, Rosell R. 2016. Human endogenous retroviruses and cancer. Cancer Biol Med. 13(4):483–488.
- Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Noel RJ, Kaplan MH, Markovitz DM. 2012. Expression of human endogenous retrovirus type K (HML-2) is activated by the TAT protein of HIV-1. J Virol. 86(15):7790–7805.
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY et al. 2015. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 522(7555):221–225.
- Guasch G, Popovici C, Mugneret F, Chaffanet M, Pontarotti P, Birnbaum D, Pebusque MJ. 2003. Endogenous retroviral sequence is fused to FGFR1 kinase in the 8p12 stem-cell myeloproliferative disorder with t(8;19)(p12;q13.3). Blood. 101(1):286–288.
- Hanke K, Chudak C, Kurth R, Bannert N. 2013. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int J Cancer. 132(3):556–567.
- Hanke K, Hohn O, Liedgens L, Fiddeke K, Wamara J, Kurth R, Bannert N. 2013. Staufen-1 interacts with the human endogenous retrovirus family HERV-K(HML-2) rec and gag proteins and increases virion production. J Virol. 87(20):11019–11030.
- Hanke K, Kramer P, Seeher S, Beimforde N, Kurth R, Bannert N. 2009. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus k and modulation of its functional activity by truncation of the cytoplasmic domain. J Virol. 83(24):12790–12800.
- Harris JM, Haynes RH, McIntosh EM. 1997. A consensus sequence for a functional human endogenous retrovirus K (HERV-K) dUTPase. Biochem Cell Biol. 75(2):143–151.
- Henzy JE, Coffin JM. 2013. Betaretroviral envelope subunits are noncovalently associated and restricted to the mammalian class. J Virol. 87(4):1937–1946.
- Herbst H, Sauter M, Mueller-Lantzsch N. 1996. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. Am J Pathol. 149(5):1727–1735.
- Hohn O, Hanke K, Bannert N. 2013. HERV-K(HML-2), the best preserved family of HERVs: endogenization, expression, and implications in health and disease. Front Oncol. 3:246.
- Holt MP, Shevach EM, Punkosdy GA. 2013. Endogenous mouse mammary tumor viruses (mtv): new roles for an old virus in cancer, infection, and immunity. Front Oncol. 3:287.
- Hughes JF, Coffin JM. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat Genet. 29(4):487–489.
- Hughes JF, Coffin JM. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc Natl Acad Sci U S A. 101(6):1668–1672.
- Huh JW, Kim DS, Kang DW, Ha HS, Ahn K, Noh YN, Min DS, Chang KT, Kim HS. 2008. Transcriptional regulation of GSDML gene by antisense-oriented HERV-H LTR element. Arch Virol. 153(6):1201–1205.
- Ikeda H, Odaka T. 1984. A cell membrane "gp70" associated with Fv-4 gene: immunological characterization, and tissue and strain distribution. Virology. 133(1):65–76.
- Ishida T, Obata Y, Ohara N, Matsushita H, Sato S, Uenaka A, Saika T, Miyamura T, Chayama K, Nakamura Y et al. 2008. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun. 8:15.
- Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF et al. 2012. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J Clin Invest. 122(12):4473–4489.
- Kammerer U, Germeyer A, Stengel S, Kapp M, Denner J. 2011. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J Reprod Immunol. 91(1–2):1–8. English.
- Kapitonov VV, Jurka J. 1999. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J Mol Evol. 48(2):248–251.
- Karamitros T, Paraskevis D, Hatzakis A, Psichogiou M, Elefsiniotis I, Hurst T, Geretti AM, Beloukas A, Frater J, Klenerman P et al. 2016. A contaminant-free assessment of Endogenous Retroviral RNA in human plasma. Sci Rep. 6:33598.
- Katoh I, Mirova A, Kurata S, Murakami Y, Horikawa K, Nakakuki N, Sakai T, Hashimoto K, Maruyama A, Yonaga T et al. 2011. Activation of the long terminal repeat of human endogenous retrovirus K by melanoma-specific transcription factor MITF-M. Neoplasia. 13(11):1081–1092.
- Kempf L, Nicodemus KK, Kolachana B, Vakkalanka R, Verchinski BA, Egan MF, Straub RE, Mattay VA, Callicott JH, Weinberger DR et al. 2008. Functional polymorphisms in PRODH are associated with risk and protection for schizophrenia and fronto-striatal structure and function. PLoS Genet. 4(11):e1000252.
- Khodosevich K, Lebedev Y, Sverdlov ED. 2004. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 32(3):e31.
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature. 453(7191):56–64.
- Kitamura Y, Ayukawa T, Ishikawa T, Kanda T, Yoshiike K. 1996. Human endogenous retrovirus K10 encodes a functional integrase. J Virol. 70(5):3302–3306.
- Knossl M, Lower R, Lower J. 1999. Expression of the human endogenous retrovirus HTDV/HERV-K is enhanced by cellular transcription factor YY1. J Virol. 73(2):1254–1261.
- Koppe B, Menendez-Arias L, Oroszlan S. 1994. Expression and purification of the mouse mammary tumor virus gag-pro transframe protein p30 and characterization of its dUTPase activity. J Virol. 68(4):2313–2319.
- Kovalskaya E, Buzdin A, Gogvadze E, Vinogradova T, Sverdlov E. 2006. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology. 346(2):373–378.
- Kramer P, Lausch V, Volkwein A, Hanke K, Hohn O, Bannert N. 2016. The human endogenous retrovirus K(HML-2) has a broad envelope-mediated cellular tropism and is prone to inhibition at a post-entry, pre-integration step. Virology. 487:121–128.
- Krishnamurthy J, Rabinovich BA, Mi T, Switzer KC, Olivares S, Maiti SN, Plummer JB, Singh H, Kumaresan PR, Huls HM et al. 2015. Genetic engineering of T cells to target HERV-K, an ancient retrovirus on melanoma. Clin Cancer Res. 21(14):3241–3251.
- Krzysztalowska-Wawrzyniak M, Ostanek M, Clark J, Binczak-Kuleta A, Ostanek L, Kaczmarczyk M, Loniewska B, Wyrwicz LS, Brzosko M, Ciechanowicz A. 2011. The distribution of human endogenous retrovirus K-113 in health and autoimmune diseases in Poland. Rheumatology (Oxford). 50(7):1310–1314.
- Kurth R, Bannert N. 2010. Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer. 126(2):306–314.
- Laderoute MP, Giulivi A, Larocque L, Bellfoy D, Hou Y, Wu HX, Fowke K, Wu J, Diaz-Mitoma F. 2007. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS. 21(18):2417–2424.
- Laderoute MP, Larocque LJ, Giulivi A, Diaz-Mitoma F. 2015. Further evidence that human endogenous retrovirus K102 is a replication competent foamy virus that may antagonize HIV-1 replication. Open AIDS J. 9:112–122.
- Langner JS, Fuchs NV, Hoffmann J, Wittmann A, Brutschy B, Lower R, Suess B. 2012. Biochemical analysis of the complex between the tetrameric export adapter protein Rec of HERV-K/HML-2 and the responsive RNA element RcRE pck30. J Virol. 86(17):9079–9087.
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK et al. 2012. Landscape of somatic retrotransposition in human cancers. Science. 337(6097):967–971.
- Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3(1):e10.
- Lemaitre C, Harper F, Pierron G, Heidmann T, Dewannieux M. 2014. The HERV-K human endogenous retrovirus envelope protein antagonizes Tetherin antiviral activity. J Virol. 88(23):13626–13637.
- Lemaitre C, Tsang J, Bireau C, Heidmann T, Dewannieux M. 2017. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 13(6):e1006451.
- Li M, Radvanyi LG, Yin B, Li J, Chivukula R, Lin K, Lu Y, Shen J, Chang DZ, Li D et al. 2017. Down-regulation of human endogenous retrovirus type K (HERV-K) viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin Cancer Res. 23:5892–5911.
- Li W, Lee MH, Henderson L, Tyagi R, Bachani M, Steiner J, Campanac E, Hoffman DA, von Geldern G, Johnson K et al. 2015. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 7(307):307ra153.
- Li XJ, Hanson C, Cmarik JL, Ruscetti S. 2009. Neurodegeneration induced by PVC-211 murine leukemia virus is associated with increased levels of vascular endothelial growth factor and macrophage inflammatory protein 1 alpha and is inhibited by blocking activation of microglia. J Virol. 83(10):4912–4922.
- Liang X, Fu C, Cui W, Ober-Blobaum JL, Zahner SP, Shrikant PA, Clausen BE, Flavell RA, Mellman I, Jiang A. 2014. Beta-catenin mediates tumor-induced immunosuppression by inhibiting cross-priming of CD8(+) T cells. J Leukoc Biol. 95(1):179–190.
- Linial ML. 1999. Foamy viruses are unconventional retroviruses. J Virol. 73(3):1747–1755.
- Longo MC, Berninger MS, Hartley JL. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain-reactions. Gene. 93(1):125–128.
- Lori F, Veronese FD, Devico AL, Lusso P, Reitz MS, Gallo RC. 1992. Viral-DNA carried by human-immunodeficiency-virus type-1 virions. J Virol. 66(8):5067–5074.
- Lower R, Lower J, Frank H, Harzmann R, Kurth R. 1984. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J Gen Virol. 65(Pt 5):887–898.
- Lower R, Lower J, Tondera-Koch C, Kurth R. 1993. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology. 192(2):501–511.
- Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol. 69(1):141–149.
- Macfarlane C, Simmonds P. 2004. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J Mol Evol. 59(5):642–656.
- Magin C, Lower R, Lower J. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol. 73(11):9496–9507.
- Magin-Lachmann C, Hahn S, Strobel H, Held U, Lower J, Lower R. 2001. Rec (formerly corf) function requires interaction with a complex, folded RNA structure within its responsive element rather than binding to a discrete specific binding site. J Virol. 75(21):10359–10371.
- Manghera M, Douville RN. 2013. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology. 10:16.
- Manghera M, Ferguson J, Douville R. 2015. ERVK polyprotein processing and reverse transcriptase expression in human cell line models of neurological disease. Viruses. 7(1):320–332.
- Manghera M, Ferguson-Parry J, Lin R, Douville RN. 2016. NF-kappaB and IRF1 induce endogenous retrovirus k expression via interferon-stimulated response elements in its 5' long terminal repeat. J Virol. 90(20):9338–9349.
- Marchi E, Kanapin A, Magiorkinis G, Belshaw R. 2014. Unfixed endogenous retroviral insertions in the human population. J Virol. 88(17):9529–9537.
- Marguerat S, Wang WY, Todd JA, Conrad B. 2004. Association of human endogenous retrovirus K-18 polymorphisms with type 1 diabetes. Diabetes. 53(3):852–854.
- Martin MA, Bryan T, Rasheed S, Khan AS. 1981. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc Natl Acad Sci U S A. 78(8):4892–4896.
- Mayer J, Ehlhardt S, Seifert M, Sauter M, Muller-Lantzsch N, Mehraein Y, Zang KD, Meese E. 2004. Human endogenous retrovirus HERV-K(HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology. 322(1):190–198.
- Mayer J, Meese EU. 2003. Presence of dUTPase in the various human endogenous retrovirus K (HERV-K) families. J Mol Evol. 57(6):642–649.
- Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 21(3):257–258.
- McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 36(6):344–355.
- McCormick AL, Brown RH, Jr., Cudkowicz ME, Al-Chalabi A, Garson JA. 2008. Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology. 70(4):278–283.
- McIntosh EM, Haynes RH. 1996. HIV and human endogenous retroviruses: an hypothesis with therapeutic implications. Acta Biochim Pol. 43(4):583–592.
- Medstrand P, Blomberg J. 1993. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 67(11):6778–6787.
- Medstrand P, Mager DL. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 72(12):9782–9787.
- Medstrand P, Mager DL, Yin H, Dietrich U, Blomberg J. 1997. Structure and genomic organization of a novel human endogenous retrovirus family: HERV-K (HML-6). J Gen Virol. 78(Pt 7):1731–1744.
- Michaud HA, de Mulder M, SenGupta D, Deeks SG, Martin JN, Pilcher CD, Hecht FM, Sacha JB, Nixon DF. 2014. Trans-activation, post-transcriptional maturation, and induction of antibodies to HERV-K (HML-2) envelope transmembrane protein in HIV-1 infection. Retrovirology. 11:10.
- Moelling K, Broecker F. 2015. The reverse transcriptase-RNase H: from viruses to antiviral defense. Ann N Y Acad Sci. 1341:126–135.
- Monde K, Contreras-Galindo R, Kaplan MH, Markovitz DM, Ono A. 2012. Human endogenous retrovirus K Gag coassembles with HIV-1 Gag and reduces the release efficiency and infectivity of HIV-1. J Virol. 86(20):11194–11208.
- Monde K, Terasawa H, Nakano Y, Soheilian F, Nagashima K, Maeda Y, Ono A. 2017. Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity. Retrovirology. 14(1):27.
- Morozov VA, Dao Thi VL, Denner J. 2013. The transmembrane protein of the human endogenous retrovirus–K (HERV-K) modulates cytokine release and gene expression. PLoS One. 8(8):e70399.
- Moyes D, Griffiths DJ, Venables PJ. 2007. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 23(7):326–333.
- Mueller-Lantzsch N, Sauter M, Weiskircher A, Kramer K, Best B, Buck M, Grasser F. 1993. Human endogenous retroviral element K10 (HERV-K10) encodes a full-length gag homologous 73-kDa protein and a functional protease. AIDS Res Hum Retroviruses. 9(4):343–350.
- Muradrasoli S, Forsman A, Hu L, Blikstad V, Blomberg J. 2006. Development of real-time PCRs for detection and quantitation of human MMTV-like (HML) sequences HML expression in human tissues. J Virol Methods. 136(1–2):83–92.
- Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Fodinger D, Seppele H, Schanab O, Magin-Lachmann C et al. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 63(24):8735–8741.
- Ono M. 1986. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 58(3):937–944.
- Ono M, Yasunaga T, Miyata T, Ushikubo H. 1986. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 60(2):589–598.
- Oricchio E, Sciamanna I, Beraldi R, Tolstonog GV, Schumann GG, Spadafora C. 2007. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 26(29):4226–4233.
- Ormsby CE, Sengupta D, Tandon R, Deeks SG, Martin JN, Jones RB, Ostrowski MA, Garrison KE, Vazquez-Perez JA, Reyes-Teran G et al. 2012. Human endogenous retrovirus expression is inversely associated with chronic immune activation in HIV-1 infection. PLoS One. 7(8):e41021.
- Otowa T, Tochigi M, Rogers M, Umekage T, Kato N, Sasaki T. 2006. Insertional polymorphism of endogenous retrovirus HERV-K115 in schizophrenia. Neurosci Lett. 408(3):226–229.
- Pani MA, Wood JP, Bieda K, Toenjes RR, Usadel KH, Badenhoop K. 2002. The variable endogenous retroviral insertion in the human complement C4 gene: a transmission study in type I diabetes mellitus. Hum Immunol. 63(6):481–484.
- Perot P, Mugnier N, Montgiraud C, Gimenez J, Jaillard M, Bonnaud B, Mallet F. 2012. Microarray-based sketches of the HERV transcriptome landscape. PLoS One. 7(6):e40194.
- Ponferrada VG, Mauck BS, Wooley DP. 2003. The envelope glycoprotein of human endogenous retrovirus HERV-W induces cellular resistance to spleen necrosis virus. Arch Virol. 148(4):659–675.
- Reis BS, Jungbluth AA, Frosina D, Holz M, Ritter E, Nakayama E, Ishida T, Obata Y, Carver B, Scher H et al. 2013. Prostate cancer progression correlates with increased humoral immune response to a human endogenous retrovirus GAG protein. Clin Cancer Res. 19(22):6112–6125.
- Rethwilm A. 2003. The replication strategy of foamy viruses. Curr Top Microbiol Immunol. 277:1–26.
- Reus K, Mayer J, Sauter M, Scherer D, Muller-Lantzsch N, Meese E. 2001. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics. 72(3):314–320.
- Reus K, Mayer J, Sauter M, Zischler H, Muller-Lantzsch N, Meese E. 2001. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J Virol. 75(19):8917–8926.
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G et al. 2015. Integrative clinical genomics of advanced prostate cancer. Cell. 161(5):1215–1228.
- Robinson LR, Whelan SP. 2016. Infectious entry pathway mediated by the human endogenous retrovirus K envelope protein. J Virol. 90(7):3640–3649.
- Romano CM, Ramalho RF, Zanotto PM. 2006. Tempo and mode of ERV-K evolution in human and chimpanzee genomes. Arch Virol. 151(11):2215–2228.
- Ruggieri A, Maldener E, Sauter M, Mueller-Lantzsch N, Meese E, Fackler OT, Mayer J. 2009. Human endogenous retrovirus HERV-K(HML-2) encodes a stable signal peptide with biological properties distinct from Rec. Retrovirology. 6:17.
- Schmitt K, Heyne K, Roemer K, Meese E, Mayer J. 2015. HERV-K(HML-2) rec and np9 transcripts not restricted to disease but present in many normal human tissues. Mobile DNA-Uk. 6:4.
- Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. 2013. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol. 5(2):307–328.
- Schommer S, Sauter M, Krausslich HG, Best B, Mueller-Lantzsch N. 1996. Characterization of the human endogenous retrovirus K proteinase. J Gen Virol. 77(Pt 2):375–379.
- Schulte AM, Lai S, Kurtz A, Czubayko F, Riegel AT, Wellstein A. 1996. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc Natl Acad Sci U S A. 93(25):14759–14764.
- Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol. 79(1):341–352.
- SenGupta D, Tandon R, Vieira RG, Ndhlovu LC, Lown-Hecht R, Ormsby CE, Loh L, Jones RB, Garrison KE, Martin JN et al. 2011. Strong human endogenous retrovirus-specific T cell responses are associated with control of HIV-1 in chronic infection. J Virol. 85(14):6977–6985.
- Steele AJ, Al-Chalabi A, Ferrante K, Cudkowicz ME, Brown RH, Jr., Garson JA. 2005. Detection of serum reverse transcriptase activity in patients with ALS and unaffected blood relatives. Neurology. 64(3):454–458.
- Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 8:90.
- Sugimoto J, Matsuura N, Kinjo Y, Takasu N, Oda T, Jinno Y. 2001. Transcriptionally active HERV-K genes: identification, isolation, and chromosomal mapping. Genomics. 72(2):137–144.
- Suntsova M, Gogvadze EV, Salozhin S, Gaifullin N, Eroshkin F, Dmitriev SE, Martynova N, Kulikov K, Malakhova G, Tukhbatova G et al. 2013. Human-specific endogenous retroviral insert serves as an enhancer for the schizophrenia-linked gene PRODH. P Natl Acad Sci U S A. 110(48):19472–19477.
- Suryawanshi A, Manicassamy S. 2015. Tumors induce immune tolerance through activation of beta-catenin/TCF4 signaling in dendritic cells: a novel therapeutic target for cancer immunotherapy. Oncoimmunology. 4(12):e1052932.
- Tandon R, SenGupta D, Ndhlovu LC, Vieira RG, Jones RB, York VA, Vieira VA, Sharp ER, Wiznia AA, Ostrowski MA et al. 2011. Identification of human endogenous retrovirus-specific T cell responses in vertically HIV-1-infected subjects. J Virol. 85(21):11526–11531.
- Terry SN, Manganaro L, Cuesta-Dominguez A, Brinzevich D, Simon V, Mulder LCF. 2017. Expression of HERV-K108 envelope interferes with HIV-1 production. Virology. 509:52–59.
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B et al. 2007. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 448(7153):595–599.
- Tonjes RR, Czauderna F, Kurth R. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J Virol. 73(11):9187–9195.
- Trono D. 1992. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 66(8):4893–4900.
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 11(19):1531–1535.
- Tyagi R, Li W, Parades D, Bianchet MA, Nath A. 2017. Inhibition of human endogenous retrovirus-K by antiretroviral drugs. Retrovirology. 14(1):21.
- van der Kuyl AC. 2012. HIV infection and HERV expression: a review. Retrovirology. 9:6.
- Vertessy BG, Toth J. 2009. Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc Chem Res. 42(1):97–106.
- Vincendeau M, Gottesdorfer I, Schreml JM, Wetie AG, Mayer J, Greenwood AD, Helfer M, Kramer S, Seifarth W, Hadian K et al. 2015. Modulation of human endogenous retrovirus (HERV) transcription during persistent and de novo HIV-1 infection. Retrovirology. 12:27.
- Viola MV, Frazier M, White L, Brody J, Spiegelman S. 1975. RNA-instructed DNA polymerase activity in a cytoplasmic particulate fraction in brains from Guamanian patients. J Exp Med. 142(2):483–494.
- Vogetseder W, Dumfahrt A, Mayersbach P, Schonitzer D, Dierich MP. 1993. Antibodies in human sera recognizing a recombinant outer membrane protein encoded by the envelope gene of the human endogenous retrovirus K. AIDS Res Hum Retroviruses. 9(7):687–694.
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 104(47):18613–18618.
- Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. 2007. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 120(1):81–90.
- Watanabe T. 2017. Adult T-cell leukemia: molecular basis for clonal expansion and transformation of HTLV-1-infected T cells. Blood. 129(9):1071–1081.
- Weiss RA. 1969a. The host range of Bryan strain Rous sarcoma virus synthesized in the absence of helper virus. J Gen Virol. 5:511–528.
- Weiss RA. 1969b. Interference and neutralization studies with Bryan strain Rous sarcoma virus synthesized in the absence of helper virus. J Gen Virol. 5:529–539.
- Weiss RA. 2006. The discovery of endogenous retroviruses. Retrovirology. 3:67.
- Whitelaw E, Martin DIK. 2001. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 27(4):361–365.
- Wildschutte JH, Ram D, Subramanian R, Stevens VL, Coffin JM. 2014. The distribution of insertionally polymorphic endogenous retroviruses in breast cancer patients and cancer-free controls. Retrovirology. 11:62.
- Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. 2016. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci U S A. 113(16):E2326–E2334.
- Wilkinson D, Mager D, Leong JC. 1994. Endogenous human retroviruses. In: Levy J, editor. The retroviridae. New York: Plenum Press; p. 465–535.
- Wodrich H, Krausslich HG. 2001. Nucleocytoplasmic RNA transport in retroviral replication. Results Probl Cell Differ. 34:197–217.
- Wolf G, Yang P, Fuchtbauer AC, Fuichtbauer EM, Silva AM, Park C, Wu WR, Nielsen AL, Pedersen FS, Macfarlan TS. 2015. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Gene Dev. 29(5):538–554.
- Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP et al. 2015. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 163(1):230–245.
- Yin H, Medstrand P, Kristofferson A, Dietrich U, Aman P, Blomberg J. 1999. Characterization of human MMTV-like (HML) elements similar to a sequence that was highly expressed in a human breast cancer: further definition of the HML-6 group. Virology. 256(1):22–35.
- Yu SF, Baldwin DN, Gwynn SR, Yendapalli S, Linial ML. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 271(5255):1579–1582.
- Yu SF, Sullivan MD, Linial ML. 1999. Evidence that the human foamy virus genome is DNA. J Virol. 73(2):1565–1572.
- Zhou F, Krishnamurthy J, Wei Y, Li M, Hunt K, Johanning GL, Cooper LJ, Wang-Johanning F. 2015. Chimeric antigen receptor T cells targeting HERV-K inhibit breast cancer and its metastasis through downregulation of Ras. Oncoimmunology. 4(11):e1047582.
