Abstract
Helicobacter pylori is associated with chronic gastritis, gastric or duodenal ulcers, and gastric cancer. Since the oral cavity is the entry port and the first component of the gastrointestinal system, the oral cavity has been discussed as a potential reservoir of H. pylori. Accordingly, a potential oral-oral transmission route of H. pylori raises the question concerning whether close contact such as kissing or sharing a meal can cause the transmission of H. pylori. Therefore, this topic has been investigated in many studies, applying different techniques for detection of H. pylori from oral samples, i.e. molecular techniques, immunological or biochemical methods and traditional culture techniques. While molecular, immunological or biochemical methods usually yield high detection rates, there is no definitive evidence that H. pylori has ever been isolated from the oral cavity. The specificity of those methods may be limited due to potential cross-reactivity, especially with H. pylori-like microorganisms such as Campylobacter spp. Furthermore, the influence of gastroesophageal reflux has not been investigated so far. This review aims to summarize and critically discuss previous studies investigating the potential colonization of H. pylori in the oral cavity and suggest novel research directions for targeting this critical research question.
Introduction
Helicobacter pylori is a Gram-negative, rod-shaped, microaerophilic bacterium belonging to the order Campylobacterales (see for a field-emission scanning electron microscopy image of H. pylori) (O’Rourke Citation2001; Reshetnyak and Reshetnyak Citation2017). In most cases, this microorganism is positive for catalase, oxidase, and urease (Marshall and Goodwin Citation1987). Barry Marshall was the first to suggest a correlation between H. pylori and active chronic gastritis, duodenal ulcer, or gastric ulcer when he swallowed this bacterium bravely in a self-experiment (Marshall and Warren Citation1984), being awarded with the Nobel prize with Robin Warren in 2005. In subsequent studies, it was proven that H. pylori plays an essential role in the development of gastritis, gastroduodenal ulcers, and gastric cancer (Dixon et al. Citation1996; Luman et al. Citation1996; Yamaoka Citation2010). H. pylori is classifed as a Group 1 carcinogen for non cardia gastric carcinoma and low-grade B cell MALT gastric lymphoma by the International Agency for Research on Cancer (IARC) (IARC Monogr Eval Carcinog Risks Hum Citation2012). According to a sub-analysis from the Global Burden of Disease 2018 study, H. pylori was one of the primary causes of infection-attributable cancer cases worldwide in 2018 (Martel et al. Citation2020). Martel et al. highlighted that H. pylori may infect most adults once during their life course (Martel et al. Citation2013). Accordingly, a meta-analysis showed that about 4.4 billion individuals were infected with H. pylori worldwide in 2015 (Hooi et al. Citation2017).
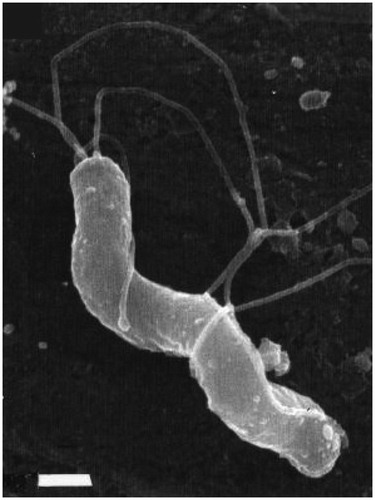
Figure 1. Morphology of H. pylori. Field-emission scanning electron microscopic (FESEM) image of a spiral-shaped H. pylori with five to seven sheathed polar flagella. The scale bar is 0.5 μm. This figure is reprinted from references (O’Rourke Citation2001; Reshetnyak and Reshetnyak Citation2017) with kind permission from the publishers.
As the oral cavity is the entry port and first component of the gastrointestinal system, researchers have also been interested in the presence of H. pylori in this niche (Singhal et al. Citation2011). There are a few studies in the literature claiming to have isolated this bacterium from dental plaque, but without any definitve proof in terms of whole genome sequencing of the given isolate or deposition in a culture collection (Agarwal and Jithendra Citation2012; Wang et al. Citation2014). Nevertheless, it was speculated that the oral cavity could also be a potential reservoir for this microorganism (Kignel et al. Citation2005; Agarwal and Jithendra Citation2012; Wang et al. Citation2014; Yee Citation2016; Urban et al. Citation2017). In 2003, Rickard et al. proposed that H. pylori was a typical colonizer of biofilms on the tooth surface in their diagrammatic representation of oral bacterial accretion on the tooth surface (Rickard et al. Citation2003). On the other hand, Bürgers et al. found no evidence for a link between oral and gastric H. pylori infections (Bürgers et al. Citation2008). In their study, the authors could not find any serum antibodies to H. pylori in patients who were found to have only oral H. pylori infections but without gastric involvement. The oral-oral transmission route is discussed as one of the most likely transmission pathways because H. pylori DNA has been found in gastric juices, vomitus, saliva, and dental plaque (Gerhard et al. Citation1999; Brown Citation2000). For instance, kissing results in an average total bacterial transfer of about 80 million bacteria in 10 s and it may lead to the exchange of H. pylori (Al-Ahmad et al. Citation2012; Kort et al. Citation2014). From this finding, the question emerges concerning whether close contact such as kissing or sharing a meal, etc. can cause the transmission of H. pylori infections. The presence of H. pylori in the oral cavity may lead to further infection in the stomach. However, it is unclear whether H. pylori can colonize in the oral cavity at all. It is also unclear whether H. pylori is permanently resident or simply temporarily present in the oral cavity.
Most studies reporting on the detection of H. pylori in samples from the oral cavity (e.g. dental plaque or saliva) have been performed by using molecular techniques, such as polymerase chain reaction (PCR) methods (Westblom and Bhatt Citation1999; Kignel et al. Citation2005). For instance, Agarwal and Jithendra found H. pylori in subgingival plaque samples of eighteen (60%) patients with confirmed gastric H. pylori infection using PCR methods, but in 15% of control patients without gastric H. pylori infection (Agarwal and Jithendra Citation2012). Immunological methods such as the Campylobacter-like organism (CLO) gel test have also been used to detect oral H. pylori. Dane and Gurbuz tested dental plaque samples from 35 patients with gastric H. pylori infection using the CLO gel test and found that 29 (82.8%) of them were oral H. pylori-positive (Dane and Gurbuz Citation2016). By contrast, when using the conventional culture technique, detection results for H. pylori have been rather contradictory (Krajden et al. Citation1989; Shankaran and Desai Citation1995; Agarwal and Jithendra Citation2012). To date, no publication has definitively demonstrated cultivation of H. pylori from any sample taken from the oral cavity (Al-Ahmad et al. Citation2010, Citation2012). Therefore, this review aims to critically discuss the evidence of a resident or transient colonization of H. pylori in the oral cavity and provide novel research directions for further studies to eliminate the confusion regarding H. pylori in the oral cavity.
Detection of H. pylori with molecular methods
Molecular methods are widely used in the detection of H. pylori from oral samples (see for a detailed overview of studies published since 2010). PCR, quantitative PCR (qPCR), nested PCR, and loop-mediated isothermal amplification (LAMP) are the most commonly used methods. Concerning the detection of H. pylori, the design of primers varies from study to study. Most of the primers designed in various studies focus on the 16S rRNA gene, 23S rRNA gene, ureA gene, cagA gene, vacA gene, babA2 gene, and glmM gene. The protein-coding genes encode components that are generally conserved between H. pylori strains and are important for the biology of the organism. For example, the ureA gene encodes for urease, which initiates urea hydrolysis to produce ammonia and neutralize gastric acid, creating a suitable pH environment for H. pylori survival and colonization (Tsuda et al. Citation1994; Dunn et al. Citation1997). Cytotoxin-associated gene A (cagA) can disrupt intracellular actin transport, stimulate inflammatory responses, and break down tight cellular junctions (Tohidpour Citation2016). CagA is encoded by the cagA gene and it is a highly immunogenic protein with an approximate weight of 140 kDa. Hatakeyama found the cagA gene in about two-thirds of H. pylori isolated from western countries (Hatakeyama Citation2004). Cytotoxin vacuolizing (VacA) protein – which causes vacuolization in epithelial cells, cell death, and the destruction of epithelial integrity – is encoded by the vacA gene and can be detected in all strains of H. pylori (Leunk et al. Citation1988; Phadnis et al. Citation1994). BabA (blood group antigen-binding adhesion) – a 78 KDa protein – is the first-identified and best-characterized adhesin of H. pylori and it is encoded by the babA2 gene (Kalali et al. Citation2014). The glmM gene – also termed ureC – encodes phosphoglucosamine mutase, a member of the enzyme superfamily that converts glucosamine 6-phosphate (GlcN-6-P) to glucosamine 1-phosphate (GlcN-1-P) (Reuse et al. Citation1997).
Table 1. Studies published since 2010 detecting oral H. pylori with molecular methods.
The 16S rRNA gene has been most widely used in studies over the past decade. When considering studies using PCR amplifying 16S rRNA genes published since 2010, the reported detection rates of H. pylori in oral samples broadly range from 5% to 78.9% when considering all experimental subjects, irrespective of gastric complaints or proven gastric H. pylori infection (Song et al. Citation1999; Silva et al. Citation2010; Agarwal and Jithendra Citation2012; Hirsch et al. Citation2012; Cai et al. Citation2014; Aksit Bıcak et al. Citation2017; Castro-Muñoz et al. Citation2017; Mendoza-Cantú et al. Citation2017; Valadan Tahbaz et al. Citation2017; Ansari et al. Citation2018; Matamala-Valdés et al. Citation2018; Wongphutorn et al. Citation2018; Šeligová et al. Citation2020). The study performed by Silva et al. was the only one to report a 0% detection rate of H. pylori from subgingival plaque samples taken from 115 patients with complaints in the upper digestive tract (Silva et al. Citation2010). Zhao et al. reported a detection rate for H. pylori of 60% (48 out of 80) in tongue samples from patients diagnosed with chronic non-atrophic gastritis using 16S rRNA gene next generation sequencing (NGS) (Zhao et al. Citation2019).
In 1993, Li et al. described primers for a 417 bp DNA fragment encoding for the ribosomal protein S20 which is the primary binding protein that bridges the 5′ domain and the 3′ minor domain of the 16S rRNA (Li et al. Citation1993). The authors reported absence of cross-reactivity when evaluating 166 non-H. pylori bacterial strains (including Campylobacter cinaedi, Campylobacter coli, Campylobacter concisus, Campylobacter cryaerophila, Campylobacter foetus, Campylobacter jejuni, Campylobacter laridis, Campylobacter sputorum subsp. bubulus, Campylobacter sputorum subsp. sputorum, Campylobacter upsaliensis) (Li et al. Citation1993). On the other hand, Šeligová et al. found these primers not to be specific as they could cross-react with Barnesiella viscericola, which can be found in the oral cavity (Šeligová et al. Citation2020). Therefore, further research needs to be done on this point.
Song et al. also used these primers targeting this 417 bp DNA fragment with nested PCR (Song et al. Citation1999). They included 40 randomly selected adult patients who visited the dental department. H. pylori’s detection rate in the oral cavity using this primer (100%) was found to be even higher than when using other 16S rRNA PCR (78.9%) (Song et al. Citation1999). In 2015, a study from Poland also used the nested PCR method amplified for the 417 bp fragment described above to study the distribution of H. pylori in the oral cavity of patients with leukoplakia and oral lichen planus (Kazanowska-Dygdała et al. Citation2016). Notably, H. pylori was only detected in samples from patients with leukoplakia or oral lichen planus but not from healthy patients. 20% of patients with leukoplakia and 23% of patients with lichen planus were found to be H. pylori-positive (Kazanowska-Dygdała et al. Citation2016). Employing the same method, Wichelhaus et al. detected oral H. pylori in 82% dental plaque samples from eleven adolescent patients who went for orthodontic therapy (Wichelhaus et al. Citation2011). As described above, the high detection rate in a healthy population raises questions about the specificity of the primers used for this 417 bp fragment (Wichelhaus et al. Citation2011). PCR (amplifying the 417 bp fragment) and nested PCR (amplifying a 109 bp fragment) were used in the study performed by Al-Ahmad et al. (Citation2012). Only one out of fifteen patients with gastric H. pylori infection were detected with oral H. pylori using PCR with both primers. The authors excluded false-negative PCR results in the study since they used a PCR inhibition control comprising a plasmid, including the target gene (Al-Ahmad et al. Citation2012).
The ureA gene is also quite commonly used in oral H. pylori research. In most of the studies using the ureA gene as an amplification target, the detection rate of H. pylori in samples from the oral cavity ranges from 15% to 50.4% when considering all experimental subjects (Song et al. Citation1999; Boyanova et al. Citation2013; Bharath et al. Citation2014; Ogaya et al. Citation2015; Medina et al. Citation2017; Nomura et al. Citation2018; Hamada et al. Citation2019; Kadota et al. Citation2019). Kadota et al. described the case of a 29-year-old woman with the chief complaint of a stomach ache. The ureA gene of H. pylori was detected in oral specimens before they started triple antibiotic therapy (potassium-competitive acid blocker, amoxicillin, and clarithromycin) for H. pylori eradication. After this treatment, the gene could no longer be detected (Kadota et al. Citation2019). By contrast, Ogaya et al. found a 0% detection rate of oral H. pylori in salivary samples from 40 Japanese children and adolescents who went for root canal treatment without any gastric complaints. However, it is unsurprising that H. pylori was not detected in those young subjects, who exhibited a high probability of not being infected with H. pylori due to the lack of any gastric complaints (Ogaya et al. Citation2015). In 2019, Hamada et al. performed ureA gene PCR on saliva specimens and extracted teeth from 87 subjects to analyse H. pylori’s distribution among these specimens (Hamada et al. Citation2019). The H. pylori-positive rate in these samples was 18.4%. Eight samples exhibited H. pylori in saliva, and thirteen samples showed H. pylori in dental plaque taken from teeth that were extracted due to dental caries or periodontal disease (Hamada et al. Citation2019).
In the study mentioned above, Ogaya et al. detected H. pylori in 15% of samples obtained from infected dental pulps (Ogaya et al. Citation2015), while Nomura et al. found H. pylori in even 38.9% of specimens from infected dental pulps using the nested PCR system targeting the ureA gene (Nomura et al. Citation2018). The difference in detection rates compared with the study by Ogaya et al. (Citation2015) may be attributed to the lower detection threshold of nested PCR as compared to conventional PCR. In another study, ureA PCR analysis was compared with the urease test for detecting H. pylori in 56 dental plaque samples from dyspeptic adult patients (Bharath et al. Citation2014). Here, a clear difference in detection efficiency was found (urease test showed a 71.4% H. pylori-positive rate, while ureA PCR showed a 33.9% positive rate) (Bharath et al. Citation2014). Besides H. pylori, other bacteria (such as Staphylococcus epidermidis, Campylobacter ureolyticus, Streptococcus salivarius, Actinomyces spp. and some strains of Haemophilus parainfluenzae) that are commonly found in the oral cavity exhibit strong urease activity (Dahlén et al. Citation2018). Since it is not clear whether the primers are specific to the H. pylori ureA gene, PCR analysis targeting this gene or testing for urease activity may lead to false-positive results due to the presence of other bacteria within the dental plaque exhibiting strong urease activity.
The glmM gene is not as commonly used for the detection of H. pylori as the ureA gene. The detection rate in studies using glmM gene PCR ranges from 5.5% to 66.7% when considering all experimental subjects (Gao et al. Citation2011; Amiri et al. Citation2015; Castro-Muñoz et al. Citation2017). Castro-Muñoz et al. investigated H. pylori’s distribution in the oral cavities of 162 healthy kindergarten children under five years of age (Castro-Muñoz et al. Citation2017). For the detection of H. pylori, they used PCR for 16S rRNA and glmM genes. As a result, 13% (21 out of 162) of the children were found H. pylori-positive with PCR for 16S rRNA, while only nine of those 16S rRNA-positive subjects were found to be H. pylori-positive with PCR for the glmM gene. The glmM detection rate in the 16S rRNA-positive samples was only 42.8% (Castro-Muñoz et al. Citation2017). Lu et al. described a similar glmM detection rate of 67.5% in 16S rRNA-positive samples (Lu et al. Citation1999). The difference in detection rate between 16S rRNA and glmM gene PCR may thus be due to differences in the specificity of the primers used (Castro-Muñoz et al. Citation2017). Without carefully analysing H. pylori-speficity of primers, a positive cross-reaction of the PCR with other H. pylori-related microorganisms such as Campylobacter spp. cannot be excluded (Al-Ahmad et al. Citation2010).
In 2015, Amiri et al. compared the efficiency of PCR and LAMP in oral H. pylori detection (Amiri et al. Citation2015). Forty-five samples of dental plaque from patients without any gastric complaints were investigated in this study. The target gene in this research was glmM. H. pylori’s detection rates in the dental plaque samples were 44% (20 out of 45) using PCR and 66.7% (30 out of 45) using LAMP, whereby in 33.3% (15 out of 45) both methods yielded positive results. The authors claimed that LAMP seems to be a more efficient method for oral H. pylori detection. However, compared with other studies using glmM gene PCR to investigate oral H. pylori in patients without gastric complaints, the detection rate of oral H. pylori in their work is relatively high. Therefore, the specificity of the primers used for detection of glmM may need further verification, as discussed above. Gao et al. also used PCR targeting the glmM gene to detect oral H. pylori in patients with gastric H. pylori infection (Gao et al. Citation2011). The H. pylori detection rate in oral samples (dental plaque, gargles, and tongue samples) was 56.9%, but only 14.6% of the glmM-positive samples were also detected as harbouring the cagA gene. The H. pylori infection rate of the oral cavity in the gastric H. pylori-infected population in this study was higher than in the non-gastric H. pylori-infected population in other studies (Gao et al. Citation2011).
The detection rate of the vacA gene in oral samples is relatively low compared with other genes, ranging from 2% to 5.5% when considering all experimental subjects (Sepúlveda et al. Citation2012; Boyanova et al. Citation2013; Valadan Tahbaz et al. Citation2017), while in a study from Thailand, the detection rate of H. pylori in saliva samples from healthy persons was 59% for the vacA gene, but still lower than the detection rate (65%) using 16S rRNA PCR (Wongphutorn et al. Citation2018). Notably, in the research conducted by Sepúlveda et al. (Sepúlveda et al. Citation2012), they did not detect the vacA and cagA genes simultaneously in any oral H. pylori-positive samples taken from patients with gastric complaints. Furthermore, it needs to be clarified whether cross-reactions with genes from other microorganisms may lead to false-positive results (Sepúlveda et al. Citation2012). Since the vacA gene cannot be found in all H. pylori-positive samples (Assumpção et al. Citation2010; Gao et al. Citation2011; Medina et al. Citation2017; Valadan Tahbaz et al. Citation2017), it may not be useful as a standard for oral H. pylori detection. Since the vacA gene is frequently but not always found in H. pylori, it can be concluded that the value of positive or negative results must be questioned when using vacA gene PCR for detection of H. pylori from oral samples.
In the studies using the cagA gene as a PCR target, detection rates of oral H. pylori range from 1.8% to 58% when considering all experimental subjects, and from 8.7% to 26.1% in gastric H. pylori-positive patients (Assumpção et al. Citation2010; Gao et al. Citation2011; Sepúlveda et al. Citation2012; Boyanova et al. Citation2013; Cai et al. Citation2014; Mendoza-Cantú et al. Citation2017; Valadan Tahbaz et al. Citation2017; Flores-Treviño et al. Citation2019). It is found that the cagA gene is not present in all H. pylori isolated from western countries. The cagA-positive H. pylori strains are more virulent than the cagA-negative strains (Hatakeyama Citation2004). Therefore, the cagA gene may not be worthwhile for oral H. pylori detection, but rather to characterize given strains in terms of whether they comprise this critical virulence factor.
The babA2 gene has only been used in two studies for oral H. pylori detection in the past decade (Medina et al. Citation2017; Valadan Tahbaz et al. Citation2017). A study from Iran (Valadan Tahbaz et al. Citation2017) reported a 5% H. pylori-positive rate in supragingival and subgingival plaque samples from 50 periodontitis patients and 50 patients without periodontal disease. They detected H. pylori with PCR using a 16S rRNA primer set and found that the babA2 gene was detected in all five 16S rRNA-positive samples (Valadan Tahbaz et al. Citation2017). On the other hand, Medina et al. reported that the ureA gene could be detected in 50.8% (31/61) of patients who attended Gastroenterology service (Medina et al. Citation2017). However, only three of those ureA-positive oral samples were seen with the babA2 gene (Medina et al. Citation2017). These findings support the notion that caution is required when babA2 PCR is conducted for detecting H. pylori because strong heterogeneity in the detection rates of the babA2 gene has been shown depending on the PCR primer sets used (Šterbenc et al. Citation2020).
Compared with other detection methods, H. pylori’s oral detection rate is relatively high when using molecular techniques. PCR is a highly sensitive technique, but the specifity of the used primers is of paramount importance. Since cross-reactions have been observed between H. pylori and different members of Campylobacter spp. that can be frequently detected in the oral cavity by using checkerboard DNA-DNA hybridization (Ximénez-Fyvie et al. Citation1999; Al-Ahmad et al. Citation2010), cross-reaction between Campylobacter spp. and H. pylori in molecular methods may cause false-positive results and explain the high detection rates. For instance, Šeligová et al. recently pointed out that many primers designed or used in previous studies had flaws. The major drawback was nonspecificity at the 3′ ends, as shown in silico by a fast search in GenBank that excluded the Helicobacter taxid (Šeligová et al. Citation2020). Another important aspect is gastroesophageal reflux, which can bring H. pylori from the stomach to the oral cavity. In western and eastern societies, the prevalence of gastroesophageal reflux disease is about 19% to 44% (Ho et al. Citation2006; Yönem et al. Citation2013). The high prevalence of gastroesophageal reflux among citizens can also influence the detection of oral H. pylori because efflux may temporarily bring H. pylori or "fragments" of H. pylori from the stomach, which may lead to positive PCR detection. Moreover, it could be speculated that DNA of H. pylori may reach the oral cavity by a hiccup and hence be detected by the PCR as a sensitive molecular method.
Detection with immunological and biochemical methods
Besides molecular methods like PCR, immunological and biochemical methods have also been described for investigating the presence of H. pylori in the oral cavity (Matamala-Valdés et al. Citation2018; Wongphutorn et al. Citation2018; Zhao et al. Citation2019). The following paragraphs summarize studies using such immunological and biochemical methods, and shows details of studies using immunological methods published in 2010 or later.
Table 2. Studies published since 2010 detecting oral H. pylori with immunological and biochemical methods.
Immunological methods used for the detection of H. pylori vary from study to study. The saliva H. pylori antigen (HPS) test and the H. pylori flagellin test (HPF) are lateral flow, immuno-chromatographic tests to detect the H. pylori urease antigen (HPS) or the flagellin antigen (HPF), respectively (Yee et al. Citation2013). The CLO test (also known as the rapid urease test) is a quick diagnostic test for H. pylori. It is designed to test H. pylori’s ability to secrete the urease enzyme, which catalyses the conversion of urea to ammonia and carbon dioxide (Dane and Gurbuz Citation2016). Indirect immunofluorescence assays (IFA) using fluorescence-labeled monoclonal antibodies (IgG anti-H. pylori antibodies) against H. pylori have also been used (Wongphutorn et al. Citation2018).
A total of 277 patients (159 with stomach pain and 118 with no stomach complaints) were included in the study by Wang et al., 201 (110 with stomach pain and 91 with no stomach complaints) of whom were tested with HPS and HPF. 70.1% (141/201) of these patients were found to be oral H. pylori-positive according to positive results in both HPS and HPF. The detection rates of oral H. pylori in the symptomatic and asymptomatic group were 87.3% and 49.5%, respectively (Wang et al. Citation2014). No cross-reactivity with Actinomyces naeslundi, Actinomyces odontolyticus, Bifidobacterium dentium, Corynebacterium matruchotii, Gemella haemolysans, Granulicatella adiacens, Streptococcus gordonii, Streptococcus salivarius, Streptococcus sanguinis, and Veillonella parvula was found using HPS and HPF. However, cross-reactivity with H. pylori-related organisms such as Campylobacter spp. that have strong potential to give false-positive results were not considered (Wang et al. Citation2014).
An anti-H. pylori antibody (a polyclonal rabbit antibody to H. pylori) was used to stain samples from 50 oral lichen planus patients for microscopic evaluation and detection of H. pylori (Hulimavu et al. Citation2014). No H. pylori was detected in both 50 tissue samples from oral lichen planus patients and ten samples from normal buccal mucosal biopsies. Since none of these patients had gastric complaints, this finding is unsurprising (Hulimavu et al. Citation2014).
Ding et al. (Citation2015) used HPS and HPF test to detect H. pylori in the oral cavity of subjects who went for an oral health examination (Ding et al. Citation2015). A total of 1050 patients were tested, 633 (60.3%) of whom were found to be H. pylori-positive. 69.5% of patients with a history of gastric ulcer carried oral H. pylori, compared with 58.3% of patients without a history of gastric ulcer. A statistically significant difference was found between these two groups. Detection rates of H. pylori in patients with caries (66.9%) and periodontal diseases (63.4%) were higher than those without oral diseases (54.1%). The authors concluded that oral H. pylori infection is closely related to periodontal diseases and caries. However, the lack of careful validation of the specificity of the tests is a shortcoming of this study (Ding et al. Citation2015). In 2016, Dane et al. tested dental plaque samples from 35 patients who had endoscopically-diagnosed cases of H. pylori-related gastritis and 35 healthy patients with the CLO gel test (Dane and Gurbuz Citation2016). Twenty-nine of 35 (82.8%) gastritis patients were found oral H. pylori-positive, while eight of 35 (22.9%) healthy patients were found to be positive. A high prevalence of H. pylori was found in dental plaque. The authors concluded that the oral cavity was a vital reservoir for H. pylori. Unfortunately, cross-reactivity to Campylobacter spp. – which are frequently isolated from dental plaque – was not evaluated in this study (Dane and Gurbuz Citation2016).
Yu et al. (Citation2017) performed a study to detect oral H. pylori infections among 4321 adults in samples from dental plaque by using HPS (Yu et al. Citation2017). They detected H. pylori in 59.6% of the young age sub-group (<45 years), while the rate of positive detection of H. pylori was 25.5% in the elder sub-group (75–89 years). One strength of this study is that researchers tested cross-reactivity of HPS with thirteen bacterial strains (Streptococcus gordonii, Streptococcus mutans, S. salivarius, S. sanguinis, Veillonella parvula, Porphyromonas gingivalis, Gemella haemolysans, Granulicatella adiacens, Campylobacter rectus, Corynebacterium matruchotii, Bifidobacterium dentium, Actinomyces naeslundii, and A. odontolyticus) and obtained a negative result (Yu et al. Citation2017). While Campylobacter rectus is known to be urease-negative (Noël et al. Citation2018), a recent study showed that the Campylobacter lari group – which is typically isolated from humans – was found to be positive for urease (Boukerb et al. Citation2019). Based on this, HPS and CLO tests that test for urease antigens may have cross-reactivity with this group of bacteria, potentially leading to false-positive detection results of H. pylori.
Wongphutorn et al. investigated H. pylori’s prevalence in 110 saliva samples from asymptomatic persons in north-eastern Thailand (Wongphutorn et al. Citation2018). Samples were tested with IFA using a mouse anti-H. pylori IgG antibody. From 110 saliva samples, 57 (51.8%) were positive according to IFA. However, cross-reactivity of IFA using this antibody (especially with H. pylori-related microorganisms) remains to be clarified in further studies (Wongphutorn et al. Citation2018).
A study from Chile focussed on oral swabs from 53 term newborns (Matamala-Valdés et al. Citation2018). They detected H. pylori with immunofluorescence using rabbit polyclonal IgG anti-H. pylori antibodies marked with FITC. Subsequently, the authors took images with fluorescence microscopy, finding only one of 53 samples to be H. pylori-positive. This result was consistent with the PCR amplified for cagA and vacA genes in this study. However, unfortunately cross-reactivity was not tested in this study (Matamala-Valdés et al. Citation2018).
In 2019, Zhao et al. performed research on samples from the gastric mucosa and tongue scrapings collected from 80 patients with chronic gastritis (Zhao et al. Citation2019). The H. pylori status was confirmed by 16S rRNA gene sequencing. The cagA status was confirmed by investigating anti-cagA immunoglobulin G (IgG) in serum. The authors found H. pylori-negative samples (32 of 80), cagA-negative H. pylori infections (13 of 80), and cagA-positive H. pylori infections (35 of 80). Notably, 27% of H. pylori infections were found to be cagA-negative (Zhao et al. Citation2019). However, as stated above, cagA is not present in all H. pylori isolated from western countries and rather serves as an indicator of the virulence of a given H. pylori strain (Hatakeyama Citation2004).
A total of 277 patients were included in the research conducted by Wang et al., with 50.9% (141 out of 277) being found oral H. pylori-positive according to positive results in both HPS and HPF (Wang et al. Citation2014). However, it is known that flagellin can also be found in the Campylobacter jejuni group (Salah Ud-Din and Roujeinikova Citation2018). Based on this, HPF – which tests for the flagellin antigen – may have cross-reactivity with this group of bacteria.
Moreover, immunofluorescence with monoclonal antibodies was used to detect oral H. pylori (Boyanova et al. Citation2013). Four monoclonal antibodies (121F3, 122Е9, 123B11, and 161F8) were used in this study. H. pylori could only be detected in the dental plaque from one out of 43 patients. 121F3 was negative in this strain, while 122Е9, 123B11, and 161F8 were positive. However, no further information on these antibodies is given, despite that there was no cross-reactivity with Campylobacter jejuni, Escherichia coli, Salmonella, Shigella, Klebsiella, Proteus, and Yersinia enterocolitica (Boyanova et al. Citation2013).
Namiot et al. collected samples of supragingival plaque from 155 patients aged 9–78 years (Namiot et al. Citation2010). They detected H. pylori with an immunological method using a kit to detect H. pylori antigens in stool samples. One hundred and one patients (65.6%) were found to be oral H. pylori-infected (Namiot et al. Citation2010). The only drawback is that no data on the specificity of this kit is given.
With immunological methods, attention should be paid to cross-reactions between H. pylori and other bacteria. Although some studies have investigated the cross-reactions between H. pylori and other bacteria (Wang et al. Citation2014), almost none of them focus on the cross-reactions between H. pylori and H. pylori-related organisms such as Campylobacter spp. In the Chinese study by Yu et al., researchers tested cross-reactivity of HPS with Campylobacter rectus and obtained negative results (Yu et al. Citation2017). Nonetheless, it is known that flagellin can also be found in the Campylobacter jejuni group (Salah Ud-Din and Roujeinikova Citation2018). Therefore, the cross-reactivity of HPF remains to be clarified. It is worth noting that some studies discovered serological detection with low specificity as common surface antigens of Campylobacter jejuni and Campylobacter coli may lead to serological cross-reactivity (Webberley et al. Citation1992; Bodhidatta et al. Citation1993; Glupczynski et al. Citation1993). Therefore, this cross-reactivity may cause false-positive results of immunological H. pylori detection in oral samples. As stated above, it has recently been shown that the Campylobacter lari group isolated from humans was positive for urease (Boukerb et al. Citation2019). Based on these findings, HPS and CLO – both of which test for the urease antigen – may cross-react with this group of bacteria. Likewise, CLO even suggests by its name that it is suitable for the detection of Campylobacter-related organisms but not specifically for H. pylori. If researchers want to use CLO in oral H. pylori detection, they need to make a rigorous assessment of CLO’s cross-reactivity. On the other hand, as highlighted above regarding DNA-based molecular methods, the impact of the high prevalence of gastroesophageal reflux among subjects on the detection of oral H. pylori cannot be ignored because efflux may temporarily bring H. pylori from the stomach, which may lead to positive immunological detection.
Detection with culture technique
When using the culture technique, H. pylori’s detection rates from oral samples are usually relatively low compared with those using the previously-mentioned detection methods. The culture technique is time- and laboratory-intensive and requires experience among the staff. Furthermore, for proofing the culture of H. pylori from an oral sample, it is crucial to provide data from whole genome sequencing of the given strain (Whittam and Bumbaugh Citation2002). Furthermore, deposition of a suchlike isolate in an international culture collection would be highly worthwhile in order to provide authenticated biological material for confirmation of the results and for further studies (Smith Citation2003). Unfortunately, no H. pylori strain isolated from the oral cavity has ever been deposited in any national or international culture collection. Accordingly, the Human Oral Microbiome Database (HOMD; www.homd.org) comprises data on the genomes of twelve H. pylori strains, none of which, however, have been isolated from the oral cavity. In the following, selected studies are described that used the culture technique for detection of H. pylori from oral samples, while summarizes the details of studies published since 2005 investigating the presence of H. pylori in the oral cavity by culture technique.
Table 3. Studies published since 2000 detecting oral H. pylori by culture techniques.
In 1989, Krajden et al. collected samples of dental plaque, saliva, and gastric biopsies from 71 patients undergoing endoscopy (Krajden et al. Citation1989). It was the first study attempting to culture H. pylori from oral samples. All such oral and gastric samples were cultured on 10% laked horse blood with Skirrow formula. The researchers successfully recovered H. pylori from 29 (40.8%) of 71 gastric biopsy samples. At the same time, no H. pylori were recovered from the salivary samples, while H. pylori could only be cultured from one of 71 dental plaque samples. Consequently, the authors did not consider saliva and dental plaque to be relevant reservoirs of this organism (Krajden et al. Citation1989). To the best knowledge of the authors, this isolate has unfortunately neither been investigated by other groups nor deposited in any culture collection.
Likewise, the detection rate of oral H. pylori in research conducted by Goosen et al. was only 3.4%. In this research, dental plaque and saliva samples were collected from 58 randomly-selected clinically healthy volunteers and cultured on brain heart infusion agar plates with 5% sheep blood. H. pylori isolates were identified with the urease test and catalase activity test, which does not rule out false-positive detection, as discussed above (Goosen et al. Citation2002).
Allaker et al. did not find any positive culture results from dental plaque samples taken from H. pylori-positive children according to gastric biopsy results, while PCR amplified for a 411 bp DNA fragment of the ureA gene exhibited a 25% H. pylori detection rate (Allaker et al. Citation2002). Their results were quite similar to those from two other studies: Teoman et al. found H. pylori in 28.3% of dental plaque samples using ureA gene PCR, although they failed to isolate H. pylori from any of those dental plaque specimens with the culture method (Teoman et al. Citation2007), while Umeda et al. made attempts to isolate H. pylori with H. pylori selective medium (specific formula unknown) from oral samples of Japanese subjects and confirmed the result by morphological observation, catalase test, oxidase test, and nested PCR. While H. pylori was identified in one sample using PCR, it could not be isolated by culture technique at all (Umeda et al. Citation2003).
In 2004, researchers tried to culture H. pylori from saliva and dental plaque samples from 100 female subjects using solid selective, enriched medium with 5% horse blood (Cześnikiewicz-Guzik et al. Citation2004; Czesnikiewicz-Guzik et al. Citation2005). The authors claimed to have successfully cultured H. pylori from 54.1% of the saliva samples and 48.3% of the supragingival plaque samples. The detection rate of oral H. pylori in gastric H. pylori-positive patients was 55.9%, and the detection rate of oral H. pylori in gastric H. pylori-negative patients was 48%. The result was confirmed by checking for bacterial urease, catalase, and oxidase activity (Cześnikiewicz-Guzik et al. Citation2004; Czesnikiewicz-Guzik et al. Citation2005). However, for instance, Brucella spp. also show a positive reaction on oxidase, catalase, and urease tests (Koestanti et al. Citation2018). Therefore, testing for urease, catalase, and oxidase may simply not be sufficiently selective to confirm the detection of H. pylori.
Wang et al. performed H. pylori culture from saliva samples (Wang et al. Citation2014; Yee Citation2016). They also tried to confirm the result with the oxidase test, catalase test, H. pylori urease test and HPF, and microscopy following Gram-staining. They claimed to have successfully cultured this bacterium from 86% of HPS and HPF positive samples (Wang et al. Citation2014). In another study, Boyanova et al. reported successful isolation of H. pylori on blood agar with Columbia agar base and 1% Isovitalex from one dental plaque sample out of 43 samples from subjects randomly selected among citizens (Boyanova et al. Citation2013).
Agarwal and Jithendra also claimed to have successfully cultured H. pylori on Columbia blood agar with 5% defibrinated sheep blood with antibiotic supplements from nine samples of subgingival plaque collected from patients with confirmed gastric H. pylori infection (Agarwal and Jithendra Citation2012).
In the study of Hirsch et al. (Citation2012), ten samples taken from root canals and corresponding supragingival plaque were collected from three children with endodontically-infected deciduous teeth (Hirsch et al. Citation2012). 16S rRNA PCR and culture methods (GC agar plates with 10% horse serum containing vancomycin, trimethoprim, nystatin, and colistin) were used to investigate the prevalence of H. pylori in those samples. With the PCR method, two samples of the root canal and four plaque samples were found to be H. pylori-positive. Interestingly, H. pylori was successfully cultured from both root canal samples with PCR-positive results, while H. pylori could not be cultured from any of the plaque samples. The authors concluded that root canals of endodontically-infected teeth – but not dental plaque – could be a reservoir for H. pylori (Hirsch et al. Citation2012). PCR (amplifying for the 417 bp fragment discussed above) and nested PCR (amplifying for a 109 bp fragment) were used in the research performed by Al-Ahmad et al. (Al-Ahmad et al. Citation2012), as mentioned above. While one out of fifteen patients with gastric H. pylori infection was detected with oral H. pylori using PCR, H. pylori could not be cultured from that sample with three different growth media (DENT agar, yeast-cysteine blood agar plates, and Columbia blood agar) (Al-Ahmad et al. Citation2012). The authors concluded that H. pylori are not residents but only transient in the oral cavity.
The methods used in previous studies for confirming H. pylori culture or isolation have been controversial. In almost all of the studies, the oxidase test, catalase test, H. pylori antigen test, or microscopy observations were used to “confirm” H. pylori isolation by culture technique. The selectivity of these methods for H. pylori detection in previous studies may be insufficient. For instance, Brucella spp. also show a positive reaction on oxidase, catalase, or urease tests (Koestanti et al. Citation2018). Therefore, potential cross-reactions may exist, and thus these methods are not sufficient to proof the isolation of H. pylori. By contrast, whole-genome sequencing of the isolates and subsequent deposition of the organisms in a culture collection is needed to confirm the H. pylori colonization of the oral cavity. Application of whole-genome sequencing on the isolated bacterial strain is crucial to prove that a given isolate is H. pylori and not another H. pylori-like microorganism. Furthermore, deposition in culture collections is crucial to provide authenticated biological material and allow other researchers to study this organism (Smith Citation2003). Until there is data from whole-genome sequencing of at least one oral H. pylori isolate and deposition of this isolate in any national or international culture collection, positive proof that the oral cavity is a reservoir for this bacterium is almost impossible.
Future research directions
Despite the large number of studies dealing with the topic of oral colonization of H. pylori as summarized above, it remains unclear whether H. pylori colonizes the oral cavity residently, transiently or even at all. Along with differences in the detection methods used in previous studies, the huge diversity in the sample types (e.g. plaque, saliva, biopsies, tongue scrapings etc.) and in the study populations (e.g. in terms of disease status, age, country, sample size) may influence the reported detection rates. As such, it makes it very difficult to accurately compare reported detection rates even when the technique is the same or similar.
One major drawback of the existing studies using molecular or immunological methods is the lack of investigating the specificity or cross-reactivity of the given methods. For instance, it has been reported that cross-reactions with H. pylori have been observed between different members of Campylobacter spp. when using molecular techniques (Ximénez-Fyvie et al. Citation1999; Al-Ahmad et al. Citation2012). Therefore, it is evident that cross-reactions between Campylobacter spp. and H. pylori may cause false-positive results when using molecular methods. Common antigens shared by Campylobacter spp. and H. pylori – which may lead to false-positive results in immunological methods – have also been found in previous research (Tanabe et al. Citation2003). Therefore, cross-reaction tests with H. pylori-like microorganisms (such as Campylobacter spp.) are needed to ensure specificity in molecular and immunological methods.
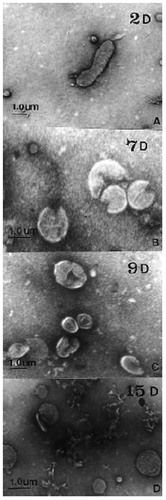
Furthermore, other points also need to be considered; for instance, a coccoid form of H. pylori has been reported in several studies (Roe et al. Citation1999; Rudnicka et al. Citation2014). It was found that the transformation of H. pylori from its spiral to its coccoid form is induced under stress conditions (such as antibiotics or environmental change; see ) (Roe et al. Citation1999) and that the coccoid form is a protection mechanism against such stress conditions (Azevedo et al. Citation2007). To prepare the coccoid state of H. pylori, H. pylori isolates were cultured in tubes containing 3 mL double distilled water with 1.5 × 108 CFU/mL bacteria under microaerophilic conditions and harvested after 1 or 2 months of incubation. The coccoid form of H. pylori was proven by using optical microscopy and LAMP. The prepared coccoid H. pylori cultures were then plated on Brucella blood agar under microaerophilic conditions at 37 °C for seven days. 90% of coccoid H. pylori could not be “resurrected” on Brucella blood agar, and only 10% of coccoid H. pylori could be re-cultured on the same agar (Chamanrokh et al. Citation2015). Likewise, Cellini et al. prepared the coccoid form H. pylori and tried to culture it on Brucella blood agar under the same conditions as described above, but they failed to “revive” H. pylori (Cellini et al. Citation1994). This may explain why H. pylori can be detected in the oral cavity by molecular methods while it hardly can be cultured. Nonetheless, it cannot be excluded that such coccoid forms of H. pylori would turn viable again when returning to the stomach. Accordingly, Cellini et al. inoculated coccoid H. pylori intragastrically in the stomachs of mice and observed alterations of the stomach tissue under transmission electron microscopy (TEM). They found a bacillary form of H. pylori in mice stomachs two weeks after inoculation, which shows successful inoculation and replication of the coccoid H. pylori. Furthermore, they observed histopathologic changes in the murine gastric mucosa after inoculation of coccoid H. pylori, albeit which were less pronounced than after inoculation of bacillary H. pylori. Interestingly, inoculation of both forms of H. pylori led to a systemic antibody response towards H. pylori in all colonized mice (Cellini et al. Citation1994). Based on this fact, H. pylori may be viable but not culturable in its coccoid form in the oral cavity, although it may be “revived” in the human stomach when swallowed. Therefore, the coccoid form of H. pylori can be considered as a “viable but not culturable” (VBNC) state of H. pylori (Rudnicka et al. Citation2014). In the VBNC state, H. pylori usually shows decreased metabolic activity and little or no ability to replicate (Roe et al. Citation1999). Most human pathogens (including H. pylori) exhibit a VBNC form, and bacteria in this state may play an essential role in recurrent and drug-resistant infections (Ozçakir Citation2007). It has been found that low levels of AI-2 (Autoinducer-2) in supragingival plaque may allow dental H. pylori to colonize the oral cavity as non-culturable forms (Krzyżek and Gościniak Citation2018). But there is still not much research on the ability of H. pylori to colonize in oral biofilms or respective in vitro models. In this light, it will be interesting to study whether H. pylori changes to its coccoid form after it encounters human saliva or typical oral bacteria and whether it can be “resurrected” again or not.
Figure 2. Morphological transformation of H. pylori from spiral to coccoid form under stress conditions. Transmission electron microscopic (TEM) images exhibiting the morphology transformation of H. pylori from its spiral to its coccoid form under stress conditions. H. pylori was cultured on Brucella blood agar with 5% horse serum and antimicrobial agents (10 mg/L vancomycin, 5 mg/L colistin, 5 mg/L trimethoprim and 5 mg/L amphotericin B) for a total of 15 days. On day 2, day 7, day 9, and day 15, its morphology was pictured by TEM: A: Day 2 (2D): bacillary form of H. pylori; B: Day 7 (7D): U-shaped form; C: Day 9 (9D): doughnut shaped form; D: Day 15 (15D): full coccoid form. The scale bar shows 1 μm. This figure is reprinted from reference (Roe et al. Citation1999) with kind permission from the publisher.
While researchers have found H. pylori in drinking water, seawater, vegetables, and animal food (Quaglia and Dambrosio Citation2018) or even on refrigerated ready-to-eat food (Poms and Tatini Citation2001), it remains unknown whether the oral cavity can be "contaminated" with H. pylori by consuming H. pylori-infected food. On the other hand, only a few studies have successfully cultured this bacterium from water or food, which means that positive results in many reports may simply reflect contamination with either dead H. pylori organisms or even naked DNA.
Likewise, positive detection of H. pylori may also be due to gastroesophageal reflux, which could bring H. pylori or “fragments” from the stomach to the oral cavity. Therefore, it may be interesting to specifically investigate a cohort of H. pylori-positive patients suffering from reflux for oral detection of H. pylori by molecular, immunological and culture-based methods. As the estimated half-life of cell-free DNA has been described to range from several minutes up two hours (Kustanovich et al. Citation2019), taking samples at least two hours after reflux might be a way to eliminate the influence of gastroesophageal reflux in such a cohort of patients.
Combining all of these aspects mentioned in the paragraphs above, depicts the potential “cycle” of H. pylori in the human body. Due to the lack of deposition of oral H. pylori strains in national or international culture collections, collaboration among researchers is particularly meaningful. Building a network of the researchers who have reported isolating H. pylori from the oral cavity would be very worthwhile to confirm these isolates as H. pylori, e.g. by whole genome sequencing approaches. Overall, future studies investigating the potential colonization of H. pylori in the oral cavity should consider some important aspects, which are outlined in . This may also help either to prove or to rule out a potential oral-to-oral transmission route of H. pylori.
Figure 3. The cycle of H. pylori in the human body. The question on whether H. pylori is resident or transient in the oral cavity (saliva, dental plaque, or tongue microbiota) remains to be clarified. H. pylori can be introduced in the oral cavity by contaminated substances including food and drinking water. There may also be a relationship between gastric and oral H. pylori infection. H. pylori can be swallowed and may subsequently cause infections in the stomach. Conversely, reflux can bring back viable organisms, DNA fragments or antigens of H. pylori to the oral cavity from the stomach of H. pylori-infected individuals. This may lead to positive results in detection with molecular methods, immunological methods or culture methods. Furthermore, H. pylori may be present in a viable, but not-culturable (VBNC) state (i.e. coccoid form).
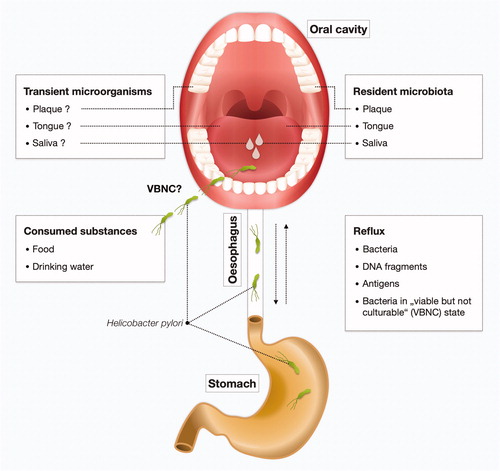
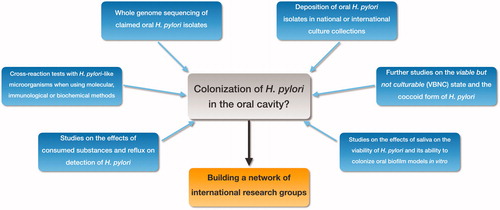
Figure 4. Future research directions. Important aspects for future studies investigating the potential colonization of H. pylori in the oral cavity: • Whole genome sequencing of oral H. pylori isolates. • Deposition of oral H. pylori in national or international culture collections. • Research on the VBNC state and the coccoid form of H. pylori. • Studies on the effects of saliva on viability of H. pylori and on the ability of H. pylori to colonize biofilms formed from oral bacteria in vitro. • Effects of consumed substances and reflux on the detection of oral H. pylori. • Cross-reaction tests with H. pylori-like microorganisms when using molecular or immunological methods. To achieve these goals, we advocate establishing a network of researchers who have reported isolating H. pylori from the oral cavity.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Agarwal S, Jithendra KD. 2012. Presence of Helicobacter pylori in subgingival plaque of periodontitis patients with and without dyspepsia, detected by polymerase chain reaction and culture. J Indian Soc Periodontol. 16(3):398–403.
- Aksit Bıcak D, Akyuz S, Kıratlı B, Usta M, Urganci N, Alev B, Yarat A, Sahin F. 2017. The investigation of Helicobacter pylori in the dental biofilm and saliva samples of children with dyspeptic complaints. BMC Oral Health. 17(1):67.
- Al-Ahmad A, Hellwig E, Wittmer A, Kist M, Waidner B. 2010. Letter to the Editor: Re: “the impact of the stone age diet on gingival conditions in the absence of oral hygiene”. J Periodontol. 81(3):338.
- Al-Ahmad A, Kürschner A, Weckesser S, Wittmer A, Rauberger H, Jakob T, Hellwig E, Kist M, Waidner B. 2012. Is Helicobacter pylori resident or transient in the human oral cavity? J Med Microbiol. 61(Pt 8):1146–1152.
- Allaker RP, Young KA, Hardie JM, Domizio P, Meadows NJ. 2002. Prevalence of Helicobacter pylori at oral and gastrointestinal sites in children: evidence for possible oral-to-oral transmission. J Med Microbiol. 51(4):312–317.
- Amiri N, Abiri R, Eyvazi M, Zolfaghari MR, Alvandi A. 2015. The frequency of Helicobacter pylori in dental plaque is possibly underestimated. Arch Oral Biol. 60(5):782–788.
- Ansari SA, Iqbal MUN, Khan TA, Kazmi SU. 2018. Association of oral Helicobacter pylori with gastric complications. Life Sci. 205:125–130.
- Assumpção MB, Martins LC, Melo Barbosa HP, Barile KAdS, Almeida SS. d, Assumpção PP, Corvelo TCdO. 2010. Helicobacter pylori in dental plaque and stomach of patients from Northern Brazil. World J Gastroenterol. 16(24):3033–3039.
- Azevedo NF, Almeida C, Cerqueira L, Dias S, Keevil CW, Vieira MJ. 2007. Coccoid form of Helicobacter pylori as a morphological manifestation of cell adaptation to the environment. Appl Environ Microbiol. 73(10):3423–3427.
- Bharath TS, Reddy MS, Dhanapal R, Raj Kumar NG, Neeladri Raju P, Saraswathi T. 2014. Molecular detection and corelation of Helicobacter pylori in dental plaque and gastric biopsies of dyspeptic patients. J Oral Maxillofac Pathol. 18(1):19–24.
- Bodhidatta L, Hoge CW, Churnratanakul S, Nirdnoy W, Sampathanukul P, Tungtaem C, Raktham S, Smith CD, Echeverria P. 1993. Diagnosis of Helicobacter pylori infection in a developing country: comparison of two ELISAs and a seroprevalence study. J Infect Dis. 168(6):1549–1553.
- Boukerb AM, Penny C, Serghine J, Walczak C, Cauchie H-M, Miller WG, Losch S, Ragimbeau C, Mossong J, Mégraud F, et al. 2019. Campylobacter armoricus sp. nov., a novel member of the Campylobacter lari group isolated from surface water and stools from humans with enteric infection. Int J Syst Evol Microbiol. 69(12):3969–3979.
- Boyanova L, Panov V, Yordanov D, Gergova G, Mitov I. 2013. Characterization of oral Helicobacter pylori strain by 4 methods. Diagn Microbiol Infect Dis. 77(4):287–288.
- Brown LM. 2000. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 22(2):283–297.
- Bürgers R, Schneider-Brachert W, Reischl U, Behr A, Hiller K-A, Lehn N, Schmalz G, Ruhl S. 2008. Helicobacter pylori in human oral cavity and stomach. Eur J Oral Sci. 116(4):297–304.
- Cai H, Li W, Shu X, Peng K, Zhang Y, Jiang M. 2014. Genetic variation of Helicobacter pylori in the oral cavity and stomach detected using thymine adenine cloning in children with chronic gastritis. Pediatr Infect Dis J. 33(1):e1–e6.
- Castro-Muñoz LJ, González-Díaz CA, Muñoz-Escobar A, Tovar-Ayona BJ, Aguilar-Anguiano LM, Vargas-Olmos R, Sánchez-Monroy V. 2017. Prevalence of Helicobacter pylori from the oral cavity of Mexican asymptomatic children under 5 years of age through PCR. Arch Oral Biol. 73:55–59.
- Cellini L, Allocati N, Angelucci D, Iezzi T, Di Campli E, Marzio L, Dainelli B. 1994. Coccoid Helicobacter pylori not culturable in vitro reverts in mice. Microbiol Immunol. 38(11):843–850.
- Chamanrokh P, Shahhosseiny MH, Mazaheri Assadi M, Nejadsattari T, Esmaili D. 2015. Three tests used to identify non-culturable form of Helicobacter pylori in water samples. Jundishapur J Microbiol. 8(4):e16811.
- Czesnikiewicz-Guzik M, Bielanski W, Guzik TJ, Loster B, Konturek SJ. 2005. Helicobacter pylori in the oral cavity and its implications for gastric infection, periodontal health, immunology and dyspepsia. J Physiol Pharmacol. 56 (Suppl 6):77–89.
- Cześnikiewicz-Guzik M, Karczewska E, Bielański W, Guzik TJ, Kapera P, Targosz A, Konturek SJ, Loster B. 2004. Association of the presence of Helicobacter pylori in the oral cavity and in the stomach. J Physiol Pharmacol. 55 (Suppl 2):105–115.
- Dahlén G, Hassan H, Blomqvist S, Carlén A. 2018. Rapid urease test (RUT) for evaluation of urease activity in oral bacteria in vitro and in supragingival dental plaque ex vivo. BMC Oral Health. 18(1):89.
- Dane A, Gurbuz T. 2016. Clinical comparative study of the effects of Helicobacter pylori colonization on oral health in children. Pak J Med Sci. 32(4):969–973.
- Ding Y-J, Yan T-L, Hu X-L, Liu J-H, Yu C-H, Li Y-M, Wang Q-Y. 2015. Association of salivary Helicobacter pylori infection with oral diseases: a cross-sectional study in a Chinese population. Int J Med Sci. 12(9):742–747.
- Dixon MF, Genta RM, Yardley JH, Correa P. 1996. Classification and grading of gastritis: the updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 20(10):1161–1181.
- Dunn BE, Cohen H, Blaser MJ. 1997. Helicobacter pylori. Clin Microbiol Rev. 10(4):720–741.
- Flores-Treviño CE, Urrutia-Baca VH, Gómez-Flores R, La Garza-Ramos M. d, Sánchez-Chaparro MM, Garza-Elizondo MA. 2019. Molecular detection of Helicobacter pylori based on the presence of cagA and vacA virulence genes in dental plaque from patients with periodontitis. J Dent Sci. 14(2):163–170.
- Gao J, Li Y, Wang Q, Qi C, Zhu S. 2011. Correlation between distribution of Helicobacter pylori in oral cavity and chronic stomach conditions. J Huazhong Univ Sci Technol. 31(3):409–412.
- Gerhard M, Lehn N, Neumayer N, Borén T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci USA. 96(22):12778–12783.
- Glupczynski Y, Goossens H, Burette A, Deprez C, van Borre C, Butzler JP. 1993. Serology in Helicobacter pylori infection. Zentralbl Bakteriol. 280(1–2):150–154.
- Goosen C, Theron J, Ntsala M, Maree FF, Olckers A, Botha SJ, Lastovica AJ, van der Merwe SW. 2002. Evaluation of a novel heminested PCR assay based on the phosphoglucosamine mutase gene for detection of Helicobacter pylori in saliva and dental plaque. J Clin Microbiol. 40(1):205–209.
- Hamada M, Nomura R, Ogaya Y, Matayoshi S, Kadota T, Morita Y, Uzawa N, Nakano K. 2019. Potential involvement of Helicobacter pylori from oral specimens in overweight body-mass index. Sci Rep. 9(1):4845.
- Hatakeyama M. 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 4(9):688–694.
- Hirsch C, Tegtmeyer N, Rohde M, Rowland M, Oyarzabal OA, Backert S. 2012. Live Helicobacter pylori in the root canal of endodontic-infected deciduous teeth. J Gastroenterol. 47(8):936–940.
- Ho KY, Cheung TK, Wong BC. 2006. Gastroesophageal reflux disease in Asian countries: disorder of nature or nurture? J Gastroenterol Hepatol. 21(9):1362–1365.
- Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, et al. 2017. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 153(2):420–429.
- Hulimavu SR, Mohanty L, Tondikulam NV, Shenoy S, Jamadar S, Bhadranna A. 2014. No evidence for Helicobacter pylori in oral lichen planus. J Oral Pathol Med. 43(8):576–578.
- IARC Monogr Eval Carcinog Risks Hum. 2012. Biological agents. Volume 100 B: a review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 100(Pt B):1–441.
- Ismail H, Morgan C, Griffiths P, Williams J, Jenkins G. 2016. A newly developed nested PCR assay for the detection of Helicobacter pylori in the oral cavity. J Clin Gastroenterol. 50(1):17–22.
- Kadota T, Ogaya Y, Hatakeyama R, Nomura R, Nakano K. 2019. Comparison of oral flora before and after triple therapy for Helicobacter pylori eradication in patient with gastric disease. Odontology. 107(2):261–267.
- Kalali B, Mejías-Luque R, Javaheri A, Gerhard M. 2014. H. pylori virulence factors: influence on immune system and pathology. Mediators Inflamm. 2014:426309.
- Kazanowska-Dygdała M, Duś I, Radwan-Oczko M. 2016. The presence of Helicobacter pylori in oral cavities of patients with leukoplakia and oral lichen planus. J Appl Oral Sci. 24(1):18–23.
- Kignel S, Almeida P. F d, André EA, Alves Mayer MP, Birman EG. 2005. Occurrence of Helicobacter pylori in dental plaque and saliva of dyspeptic patients. Oral Dis. 11(1):17–21.
- Koestanti E, Misaco W, Chusniati S, Maslachah L. 2018. Isolation and identification of brucella suis in pigs as zoonotic disease in endemic areas of east java, Indonesia. Afr J Infect Dis. 12(1 Suppl):148–151.
- Kort R, Caspers M, van de Graaf A, van Egmond W, Keijser B, Roeselers G. 2014. Shaping the oral microbiota through intimate kissing. Microbiome. 2:41.
- Krajden S, Fuksa M, Anderson J, Kempston J, Boccia A, Petrea C, Babida C, Karmali M, Penner JL. 1989. Examination of human stomach biopsies, saliva, and dental plaque for Campylobacter pylori. J Clin Microbiol. 27(6):1397–1398.
- Krzyżek P, Gościniak G. 2018. Oral Helicobacter pylori: interactions with host and microbial flora of the oral cavity. Dent Med Probl. 55(1):75–82.
- Kustanovich A, Schwartz R, Peretz T, Grinshpun A. 2019. Life and death of circulating cell-free DNA. Cancer Biol Ther. 20(8):1057–1067.
- Leunk RD, Johnson PT, David BC, Kraft WG, Morgan DR. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 26(2):93–99.
- Li C, Ferguson DA, Ha T, Chi DS, Thomas E. 1993. A highly specific and sensitive DNA probe derived from chromosomal DNA of Helicobacter pylori is useful for typing H. pylori isolates. J Clin Microbiol. 31(8):2157–2162.
- Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q, Chong SK, Lee CH. 1999. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J Clin Microbiol. 37(3):772–774.
- Luman W, Alkout AM, Blackwell CC, Weir DM, Plamer KR. 1996. Helicobacter pylori in the mouth-negative isolation from dental plaque and saliva. Eur J Gastroenterol Hepatol. 8(1):11–14.
- Marshall BJ, Goodwin CS. 1987. Notes: revised nomenclature of campylobacter pyloridis. Int J Syst Bacteriol. 37(1):68–68.
- Marshall B, Warren J. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 323(8390):1311–1315.
- Martel C. d, Forman D, Plummer M. 2013. Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am. 42(2):219–240.
- Martel C. d, Georges D, Bray F, Ferlay J, Clifford GM. 2020. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 8(2):e180–e190.
- Matamala-Valdés L, Sánchez-Alonzo K, Parra C, Sáez K, Aguayo-Reyes A, García A. 2018. Detection of intracellular Helicobacter pylori in Candida. SPP from neonate oral swabs. Rev Assoc Med Bras. 64(10):928–935.
- Medina ML, Medina MG, Merino LA. 2017. Correlation between virulence markers of Helicobacter pylori in the oral cavity and gastric biopsies. Arq Gastroenterol. 54(3):217–221.
- Mendoza-Cantú A, Urrutia-Baca VH, Urbina-Ríos CS, La Garza-Ramos M. d, García-Martínez ME, Torre-Martínez HHH. 2017. Prevalence of Helicobacter pylori vacA Genotypes and cagA gene in dental plaque of asymptomatic Mexican children. Biomed Res Int. 2017:1–10.
- Namiot DB, Leszczyńska K, Namiot Z, Chilewicz M, Bucki R, Kemona A. 2010. The occurrence of Helicobacter pylori antigens in dental plaque; an association with oral health status and oral hygiene practices. Adv Med Sci. 55(2):167–171.
- Noël A, Verroken A, Belkhir L, Rodriguez-Villalobos H. 2018. Fatal thoracic empyema involving Campylobacter rectus: a case report. Anaerobe. 49:95–98.
- Nomura R, Ogaya Y, Matayoshi S, Morita Y, Nakano K. 2018. Molecular and clinical analyses of Helicobacter pylori colonization in inflamed dental pulp. BMC Oral Health. 18(1):64.
- Ogaya Y, Nomura R, Watanabe Y, Nakano K. 2015. Detection of Helicobacter pylori DNA in inflamed dental pulp specimens from Japanese children and adolescents. J Med Microbiol. 64(Pt 1):117–123.
- O’Rourke JB. 2001. Helicobacter pylori: physiology and genetics: morphology and ultrastructure (Chapter 6). Mobley HLT, Mendz GL, Hazell SL, editors. Washington (DC): [publisher unknown].
- Ozçakir O. 2007. Viable but non-culturable form of bacteria. Mikrobiyol Bul. 41(3):477–484.
- Pataro AL, Cortelli SC, Abreu MHNG, Cortelli JR, Franco GCN, Aquino DR, Cota LOM, Costa FO. 2016. Frequency of periodontal pathogens and Helicobacter pylori in the mouths and stomachs of obese individuals submitted to bariatric surgery: a cross-sectional study. J Appl Oral Sci. 24(3):229–238.
- Phadnis SH, Ilver D, Janzon L, Normark S, Westblom TU. 1994. Pathological significance and molecular characterization of the vacuolating toxin gene of Helicobacter pylori. Infect Immun. 62(5):1557–1565.
- Poms RE, Tatini SR. 2001. Survival of Helicobacter pylori in ready-to-eat foods at 4 °C. Int J Food Microbiol. 63(3):281–286.
- Quaglia NC, Dambrosio A. 2018. Helicobacter pylori: a foodborne pathogen? World J Gastroenterol. 24(31):3472–3487.
- Reshetnyak VI, Reshetnyak TM. 2017. Significance of dormant forms of Helicobacter pylori in ulcerogenesis. World J Gastroenterol. 23(27):4867–4878.
- Reuse H. d, Labigne A, Mengin-Lecreulx D. 1997. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J Bacteriol. 179(11):3488–3493.
- Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11(2):94–100.
- Roe IH, Son SH, Oh HT, Choi J, Shin JH, Lee JH, Hah YC. 1999. Changes in the evolution of the antigenic profiles and morphology during coccoid conversion of Helicobacter pylori. Korean J Intern Med. 14(1):9–14.
- Rudnicka K, Graczykowski M, Tenderenda M, Chmiela M. 2014. [Helicobacter pylori morphological forms and their potential role in the transmission of infection]. Postepy Hig Med Dosw. 68:219–229. pol.
- Salah Ud-Din AIM, Roujeinikova A. 2018. Flagellin glycosylation with pseudaminic acid in campylobacter and helicobacter: prospects for development of novel therapeutics. Cell Mol Life Sci. 75(7):1163–1178.
- Šeligová B, Lukáč Ľ, Bábelová M, Vávrová S, Sulo P. 2020. Diagnostic reliability of nested PCR depends on the primer design and threshold abundance of Helicobacter pylori in biopsy, stool, and saliva samples. Helicobacter. 25(2):e12680
- Sepúlveda E, Moreno J, Spencer ML, Quilodrán S, Brethauer U, Briceño C, García A. 2012. [Comparison of Helicobacter pylori in oral cavity and gastric mucosa according to virulence genotype (cagA and vacA m 1)]). Rev Chilena Infectol. 29(3):278–283.
- Shankaran K, Desai HG. 1995. Helicobacter pylori in dental plaque. J Clin Gastroenterol. 21(2):82–84.
- Silva DG, Stevens RH, Macedo JMB, Albano RM, Falabella MEV, Fischer RG, Veerman EC, Tinoco EMB. 2010. Presence of Helicobacter pylori in supragingival dental plaque of individuals with periodontal disease and upper gastric diseases. Arch Oral Biol. 55(11):896–901.
- Singhal S, Dian D, Keshavarzian A, Fogg L, Fields JZ, Farhadi A. 2011. The role of oral hygiene in inflammatory bowel disease. Dig Dis Sci. 56(1):170–175.
- Smith D. 2003. Culture collections over the world. Int Microbiol. 6(2):95–100.
- Song Q, Haller B, Schmid RM, Adler G, Bode G. 1999. Helicobacter pylori in dental plaque: a comparison of different PCR primer sets. Dig Dis Sci. 44(3):479–484.
- Šterbenc A, Lunar MM, Homan M, Luzar B, Zidar N, Poljak M. 2020. Prevalence of the Helicobacter pylori babA2 Gene in Children Mainly Depends on the PCR Primer Set Used. Can J Infect Dis Med Microbiol. 2020:4080248.
- Tanabe S, Hinode D, Yokoyama M, Fukui M, Nakamura R, Yoshioka M, Grenier D, Mayrand D. 2003. Helicobacter pylori and Campylobacter rectus share a common antigen. Oral Microbiol Immunol. 18(2):79–87.
- Teoman I, Ozmeriç N, Ozcan G, Alaaddinoğlu E, Dumlu S, Akyön Y, Baloş K. 2007. Comparison of different methods to detect Helicobacter pylori in the dental plaque of dyspeptic patients. Clin Oral Investig. 11(3):201–205.
- Tohidpour A. 2016. CagA-mediated pathogenesis of Helicobacter pylori. Microb Pathog. 93:44–55.
- Tsuda M, Karita M, Morshed MG, Okita K, Nakazawa T. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 62(8):3586–3589.
- Umeda M, Kobayashi H, Takeuchi Y, Hayashi J, Morotome-Hayashi Y, Yano K, Aoki A, Ohkusa T, Ishikawa I. 2003. High prevalence of Helicobacter pylori detected by PCR in the oral cavities of periodontitis patients. J Periodontol. 74(1):129–134.
- Urban J, Koszowski R, Płachetka A, Wiczkowski A. 2017. An evaluation of selected oral health indicators and cariogenic bacteria titer in patients with Helicobacter pylori. Adv Clin Exp Med. 26(3):401–407.
- Valadan Tahbaz S, Yadegar A, Amirmozafari N, Yaghoobee S, Ehsani Ardakani MJ, Zojaji H. 2017. Occurrence of Helicobacter pylori and its major virulence genotypes in dental plaque samples of patients with chronic periodontitis in Iran. Gastroenterol Hepatol Bed Bench. 10(Suppl1):S70–S78.
- Wang XM, Yee KC, Hazeki-Taylor N, Li J, Fu HY, Huang ML, Zhang GY. 2014. Oral Helicobacter pylori, its relationship to successful eradication of gastric H. pylori and saliva culture confirmation. J Physiol Pharmacol. 65(4):559–566.
- Webberley MJ, Webberley JM, Newell DG, Lowe P, Melikian V. 1992. Seroepidemiology of Helicobacter pylori infection in vegans and meat-eaters. Epidemiol Infect. 108(3):457–462.
- Westblom TU, Bhatt BD. 1999. Diagnosis of Helicobacter pylori infection. Curr Top Microbiol Immunol. 241:215–235.
- Whittam TS, Bumbaugh AC. 2002. Inferences from whole-genome sequences of bacterial pathogens. Curr Opin Genet Dev. 12(6):719–725.
- Wichelhaus A, Brauchli L, Song Q, Adler G, Bode G. 2011. Prevalence of Helicobacter pylori in the adolescent oral cavity: dependence on orthodontic therapy, oral flora and hygiene. J Orofac Orthop. 72(3):187–195.
- Wongphutorn P, Chomvarin C, Sripa B, Namwat W, Faksri K. 2018. Detection and genotyping of Helicobacter pylori in saliva versus stool samples from asymptomatic individuals in Northeastern Thailand reveals intra-host tissue-specific H. pylori subtypes. BMC Microbiol. 18(1):10.
- Ximénez-Fyvie LA, Haffajee AD, Martin L, Tanner A, Macuch P, Socransky SS. 1999. Identification of oral Actinomyces species using DNA probes. Oral Microbiol Immunol. 14(4):257–265.
- Yamaoka Y. 2010. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 7(11):629–641.
- Yee JKC. 2016. Helicobacter pylori colonization of the oral cavity: a milestone discovery. World J Gastroenterol. 22(2):641–648.
- Yee KC, Wei MH, Yee HC, Everett KDE, Yee HP, Hazeki-Talor N. 2013. A screening trial of Helicobacter pylori-specific antigen tests in saliva to identify an oral infection. Digestion. 87(3):163–169.
- Yönem Ö, Sivri B, Özdemir L, Nadir I, Yüksel S, Uygun Y. 2013. Gastroesophageal reflux disease prevalence in the city of Sivas. Turk J Gastroenterol. 24(4):303–310.
- Yu Y, Zhao L, Wang S, Yee JK. 2017. Helicobacter pylori – Specific Antigen Tests in Saliva to identify an oral infection. Ann Clin Lab Sci. 47(3):323–327.
- Zhao Y, Gao X, Guo J, Yu D, Xiao Y, Wang H, Li Y. 2019. Helicobacter pylori infection alters gastric and tongue coating microbial communities. Helicobacter. 24(2):e12567.
