Abstract
Group A Streptococcus (GAS) is a major human pathogen, causing diseases ranging from mild superficial infections of the skin and pharyngeal epithelium to severe systemic and invasive diseases. Moreover, post infection auto-immune sequelae arise by a yet not fully understood mechanism. The ability of GAS to cause a wide variety of infections is linked to the expression of a large set of virulence factors and their transcriptional regulation in response to various physiological environments. The use of transcriptomics, among others -omics technologies, in addition to traditional molecular methods, has led to a better understanding of GAS pathogenesis and host adaptation mechanisms. This review focusing on bacterial transcriptomic provides new insight into gene-expression patterns in vitro, ex vivo and in vivo with an emphasis on metabolic shifts, virulence genes expression and transcriptional regulators role.
Introduction
Group A Streptococcus (GAS) is a major cause of morbidity globally and causes more than 500.000 deaths per year. GAS engenders a broad spectrum of diseases ranging from uncomplicated pharyngitis and skin infections to life-threatening invasive illnesses as well as non-suppurative immune-mediated sequelae, acute rheumatic fever, and glomerulonephritis (Barnett et al. Citation2018). GAS can either rapidly destroy tissues in their environment during purulent infections or show little macroscopic effects when asymptomatically colonizing the host tonsils or skin. Carriers without diseases symptoms represent the main reservoir of GAS in the population.
GAS has been shown to adhere to, internalize into, and persist within human cells (Courtney et al. Citation2002; Kreikemeyer et al. Citation2004). Its success as an opportunistic pathogen is due to a complex interplay between host factors and secreted or surface-bound bacterial factors. This remarkable set of factors includes adhesins, lytic enzymes, proteases, cytotoxins, DNases, superantigens and immune-protective proteins (Cunningham Citation2000). Expression of these virulence factors is regulated by the action of at least 13 two-components systems (TCS) and 30 stand-alone transcriptional regulators, integrating environmental information in and around GAS (Kreikemeyer et al. Citation2003).
Virulence factors are not expressed at the same rate and/or time during infection across the many different M-types of GAS (> 220), defining a specific ‘virulence factor profile’ for each M-type. However, to date the link between virulence factor profile and clinical phenotype is still poorly understood.
Much uncertainty still exists regarding the molecular mechanisms responsible for the many GAS clinical manifestations in general and for the switch between localized and systemic infections in particular. Severe invasive infections (iGAS) such as necrotizing fasciitis (NF) and streptococcal toxic shock syndrome (STSS) are rapidly progressing diseases and are associated with high mortality rates (32 to 44%) in Europe (Lamagni et al. Citation2008). Whilst all GAS M-types can cause severe diseases, some emm-types are more frequently recovered from iGAS, such as M1, M3, M28, M89 and M12 (Gherardi et al. Citation2018). Among these strains, invasive phenotypes have been associated with bacteriophages-encoded specific virulence factors, like DNases and superantigens (Barnett et al. Citation2018; McShan and Nguyen Citation2022). However, genomic comparison between ‘aggressive’ and ‘mild’ strains does not always show differences in genes content, highlighting the limits of genomics studies.
More recently, SNPs in transcriptional regulators or promoter regions of virulence genes have been shown to impact virulence by enhancing virulence genes expression. For example, SNPs in the nga-ifs-slo promoter region lead to an increase in their transcription and a high mortality rate in a mouse model (Kachroo et al. Citation2019). These genes encode a cytolysin (SLO for Streptolysin O) and a glycolytic enzyme (NAD-glycohydrolase encoded by the nga gene) which, after translocation through SLO, can lead to cell death (Madden et al. Citation2001; Bricker et al. Citation2002). Intrabacterial toxicity is prevented by an endogenous inhibitor of NAD, called Ifs. Lot of the SNPs described in iGAS strains are located in the covRS genes encoding the most characterized TCS in GAS.
Pathogens behavior is well-known to be strongly dependent on the environment, i.e. the human host probably being the most important one for GAS. Within the host, GAS progresses through diverse microenvironments which could be nutrient (carbohydrates/amino acids) limited or rich, anaerobic or aerobic, with different temperatures, osmolarity and pH. Each of these microenvironments challenges GAS with various stress conditions (oxidative stress …) and obligates it to continuously adapt its metabolism (Slade Citation1954; Davies et al. Citation1965). In parallel, GAS has also to coordinate expression of its virulence factors plethora, targeting diverse host proteins. Interestingly, transcriptome analyses of GAS grown in various in vitro and in vivo models, including primate and non-primate animal models, have unequivocally established that GAS metabolism and virulence are closely linked.
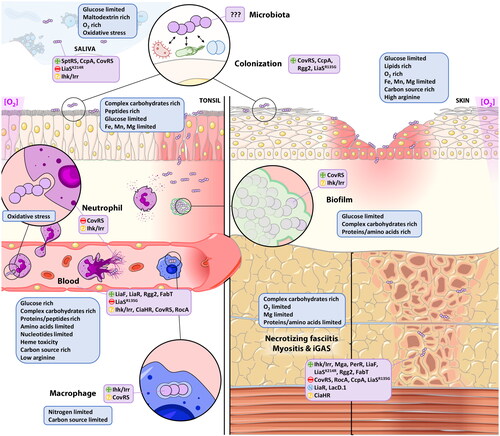
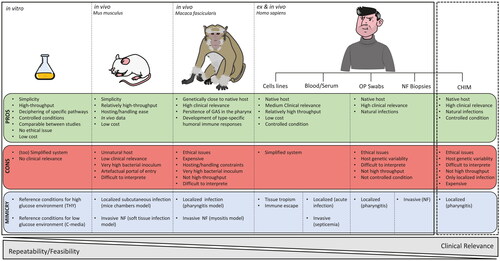
Deciphering GAS metabolic reprogramming and virulence expression within the host requires the use of well-defined and controlled environment like in vitro media but also more complex in vivo settings (). Currently, more than 2000 GAS genomes are available in public dataset. Moreover, RNAseq technology has become affordable and has largely replaced the microarray techniques to study bacterial transcriptome. In this review, we present and discuss current knowledge about the effect of different in vitro, ex vivo or in vivo environments on the GAS metabolism, expression of virulence factors and of phage genes, focusing on recent RNAseq analysis as data before 2010 have been smartly reviewed elsewhere (Fiedler et al. Citation2010).
Figure 1. Models used to study GAS transcriptome. Advantages (PROS) and disadvantages (CONS) of current models (plain lines) used for GAS transcriptomic studies as well as the clinical manifestations there are reproducing (MIMICRY). Dash lines highlight that the more clinically relevant model for GAS transcriptomic studies is the challenge human infection model (CHIM). OP: oro-pharyngeal, NF: necrotizing fasciitis.
Environmental impact on metabolic reprogramming
GAS is a lactic acid bacterium, preferentially uses glucose as a primary source of energy, generating lactic acid and auxotrophic for multiple amino acids. When glucose is present in scarce amount, like in raw saliva or on the skin, GAS switches to complex carbohydrates use (Pancholi and Caparon Citation2016). The metabolic and virulence genes differentially regulated in the different conditions discussed in this review are summarized in Table S1. The function of all cited genes is described in Table S2.
Transcriptomic in laboratory rich media
Studies using laboratory media (Todd-Hewitt supplemented with yeast extract and C-medium) have provided with important information regarding GAS metabolism and diet adaption, notably regarding the impact of sugar versus amino acids/peptides rich media (Table S1 and ). THY (glucose rich, 0.5%) and C-medium (glucose limited, 0.05%) may reflect different niches for GAS growth like blood/serum (high glucose) or saliva/lower respiratory tract/deep tissues (glucose limited). It has also permitted to decipher the expression of GAS genes during its different growth phases (Figure S1). Indeed, the metabolic and replicative state of GAS in the different growth phases somewhat reflects the infection steps. Broadly speaking, exponential phase (EP) is characterized by actively dividing bacteria tempting to invade a host, while stationary phase (SP) bacteria represent settled cells into a host and responding to starvation. Non-optimal growth conditions can also lead to biofilm formation (Yin et al. Citation2019).
By contrast to other bacteria, GAS is lacking a classical sigma factor (σs) to regulate the stationary phase (usually encoded by rpoS or alternative genes) (Wood et al. Citation2009). The role of this transcriptional factor is to aid in survival and improve resistance to stressful conditions (Schellhorn Citation2020) by regulating the expression of an array of ‘late’ genes. To decipher how this regulation is managed by GAS in absence of any σs, growth phases transcriptomic studies have been conducted using microarray. Most of these data arise from bacteria cultured in THY media supplemented with yeast extract, incubated at 37 °C with 5% CO2. THY is a medium rich in carbohydrates which can reflect, in term of sugars use, what GAS encounters at skin and muscles in acute infection. Transcriptomic analyses have been performed in different growth phases (Figure S1). During the EP, GAS uses glucose as a preferred carbon source and consequently produces lactate (Beyer-Sehlmeyer et al. Citation2005). Then, GAS enters in a transient phase in which it modifies its metabolism to adapt to the new environment lacking glucose (Beyer-Sehlmeyer et al. Citation2005). Nearly 80% of the genes are expressed during this phase by contrast to only 56% in EP. The need for proteins synthesis is fulfilled by the upregulation of ribosomes. In this phase, expression of stress response and nutrients transporters genes increased concomitantly with virulence factors genes expression (Beyer-Sehlmeyer et al. Citation2005). Stationary phase in THY allows to study GAS adaptation if carbon sources are scarce. The need of amino acid residues is met by upregulating peptide permeases, peptidases, and transporters. Lactate, produced in EP, is oxidized and results in the formation of pyruvate and acetyl-CoA, further leading to adenosine triphosphate (ATP), acetate accumulation and consequently acidification of the medium.
Overall, 87% of the genes are expressed during the SP suggesting a low redundancy on the (small) genome of GAS. However, longer half-life of some mRNAs can also explain this difference with the EP (Barnett et al. Citation2007). Two genes (spym49s0045 and c0766) were shown to be steadily expressed during all phases and should be considered as good reference genes in RT-qPCR experiments (Beyer-Sehlmeyer et al. Citation2005).
Differences in transcriptome in EP and early stationary phase (ESP) in THY have also been observed between different M-types as shown for M1 and M3 (Calfee et al. Citation2018). The chosen M-types are from same M-clade (Y) but from different M-cluster (AC3 vs AC5) and FCT-type (2 vs 3). In this RNAseq study, the observed differences in expression mainly concerned virulence genes (). Especially, the pilus expression was downregulated in M3 strains due to SNPs in nra and a decrease expression of nra compared to the M1 strain. This low pilus expression was correlated to the poor capacity of M3 isolates to adhere to cells but their increased capacity to survive in blood. The low pilus expression in M3, and maybe other M-types, raises the question of their use as a candidate for a global vaccine (Loh et al. Citation2021). However, expression of the pilus by M3 strains have never been monitored in vivo.
Table 1. Expression of virulence genes in different conditions is highlighted by blue (down), red (up) or green boxes (both) compared to a control condition and at least in one serotype.
Differences in transcriptome between strains from the same M-type was also investigated by RNAseq using a collection of 50 clonally related progeny of M28 strains (Kachroo et al. Citation2019). Three subclades were defined based on SNPs in the core genome. The differences between the 3 subclades are limited to 0.9 to 2% of the genes, that are differentially expressed in EP, except 2 outliers harboring a CovS mutation. Around one third of these genes are localized in a Horizontal Gene Transfer (HGT) region of 28 kb, including the nga-ifs-slo operon. In these 50 strains, one third expresses the R28 encoding gene at low level due to insertion of a T in the intergenic region between R28 and the adjacent gene. R28 is an important protein for adhesion to cells (Weckel et al. Citation2018) and has also been regarded as a potential vaccine antigen (Stålhammar-Carlemalm et al. Citation1999). In ESP, the differences between the 3 subclades increased by 9 times, probably related to the fact that most of the genome is expressed during this phase (80% against 56%).
Principal Component Analysis (PCA) on the transcriptome of 442 M28 isolates defines two transcriptomic profiles, with 142 differentially expressed genes, defined as 2 clusters (A and B). All cluster B strains have a significantly greater number of differentially expressed genes (DEGs) than cluster A and are all covR or covS mutants. However, some CovRS mutants present a wild-type-like transcriptome and belong to cluster A, confirming that all polymorphisms are not equivalent. Interestingly, no simple correlation between transcriptomic changes and genome-to-genome genetic distance can be found (Kachroo et al. Citation2019). Another study has compared the transcriptome of iGAS M89 isolates belonging to 3 distinct phylogenetic clades with strains from clade 3 emerging in the US (Turner et al. Citation2015; Zhu et al. Citation2015). Clade 3 isolates expressed high level of slo and nga (Beres et al. Citation2016). Main differences in transcriptomes of the 3 clades strains were also seen in ESP (Beres et al. Citation2016). If 11% of the gene content was reshaped between clade 1 and 2 strains explaining the emergence of clade 2, only 1% of the gene content differs between clade 2 and 3. However, at the transcriptomic level, 4 to 11% of genes were differentially expressed between these two clades. As for M28, 28 upregulated genes were in the HGT region including the nga-ifs-slo operon but also the speC/spd1 encoding phages.
Even if progress has been made in understanding phases growth regulation in GAS, identifying which signals are sensed by the bacteria and how these signals are transmitted remain to be investigated. Cell density (measured by QS molecules), by-products accumulation (due to bacterial multiplication), nutrient consumption and cell cycle status, should all together be integrated by the bacteria to properly adapt to various phases of infections.
In vitro, most of the studies are performed at 37 °C, global human body temperature. However, the mean mucosal temperature roughly ranged between 30 to 34 °C and the skin temperature, depending on the body sites, between 29 and 35 °C (Lindemann et al. Citation2002; Lee et al. Citation2019). Importantly, the comparison of a M1 transcriptome by microarray between 29 °C and 37 °C have highlighted that 9% of the genes are differentially expressed (Smoot et al. Citation2001). Sixty-three genes are upregulated at 29 °C compared to 37 °C, including a high number of cell-wall associated and phages-associated proteins, suggesting a very different extracellular proteome of GAS at 29 °C compared to 37 °C. iGAS are associated with fever, during which free iron is extremely limited to prevent bacterial multiplication (Kluger and Rothenburg Citation1979; Bullen Citation1981; Green et al. Citation1981). Up-regulation of genes related to iron transport and acquisition, including hemolysins like SLO, were observed at 40 °C (Smoot et al. Citation2001).
GAS can form biofilm-like bacterial communities and notably into human soft tissues (Lembke et al. Citation2006; Siemens et al. Citation2016). A first study using microarray showed that biofilms formed at 23 °C in a carbon poor-media (C-media) provokes change in 25% of the gene expression upon adaptation to biofilm lifestyle (Cho and Caparon Citation2005). However, scarce data are available on expression profile of GAS genes in biofilms. One RNAseq study with the M1T1 clone has determined the transcriptome changes between 6 h, 16 h, 6 days and 10 days in biofilms in a flow reactor (Freiberg et al., Citation2016). Compared to a planktonic culture in diluted THY, 55% of the genes were differentially expressed in biofilms. Among them the TCS ihk/irr and the isp (immunogenic secreted protein) gene were upregulated at all time points. Superantigens, destructive enzymes (including SpeB), and virulence factors against the adaptative immune response were also upregulated () (Freiberg et al., Citation2016).
Transcriptomic in animal models
In vivo studies in animal models, even if less artificial than laboratory media/conditions, have the limitation of the host specificity (). Mouse models have been exploited to understand GAS transcriptomic changes within a host. In these models, bacteria present a metabolic shift to adapt to different sources of carbon and probably to microbiota competition.
The transcriptome studies in mice were nearly all performed by microarray and are briefly outlined hereafter. Aziz et al. (Citation2010) have first studied the GAS transcriptome in a mice chamber model, in which bacteria are confined into a Teflon-FEP tissue chamber reflecting a subcutaneous localized infection (). The well-known M1T1 was used to infect mice tissues and a microarray analysis was performed after 24h. They mainly observed a metabolic shift by increasing sucrose and dipeptides transporters showing a switch in diet. Downregulation of the arginine deiminase arcA challenges its use as vaccine antigen (Henningham et al. Citation2012). In terms of virulence, only a few genes were up-regulated in vivo including the M protein, the streptodornases, SLO and NAD () (Aziz et al. Citation2010) but this result is challenged by the low sensitivity of microarray techniques.
In soft tissues infections, GAS encounters a low glucose, peptides rich environment in an anaerobic atmosphere. A mice model of necrotizing fasciitis with the M1T1 strain (Hirose et al. Citation2019) using RNAseq has shown that the difference between in vitro (THY) and in vivo expression data concerns around 28% of the genome. No differentially expressed genes (DEGs) were observed between the 3 time points (24-48-96h). By contrast to the mice model chamber, the major differences observed were the upregulation of virulence genes with the top 4 being: speB encoding the cysteine protease, sagA the SLS precursor, the DNase gene spd and spi, the SpeB-inhibitor (FC > 6-9) (). Surprisingly, grab was downregulated in all time points despite its function in virulence, by binding of alpha-2-macroglobulin (A2M), as well as an anti-GRAB antibody response after natural infection, have been well described (Rasmussen et al. Citation1999; Akesson et al. Citation2004). The absence or poor conservation of host binding partners like A2M (pairwise identity of 72% between Homo sapiens and Mus musculus) could be a limit of non-natural host model. Carbohydrate uptake is also favored in vivo by increasing several PTS and ABC transporters. Arginine (arc operon) and histidine (hutDGHIU, ftcD, fchA, fhs.2) metabolisms, available into the host tissues and for which GAS is auxotrophic (among others) were also upregulated, probably to sustain DNA/RNA synthesis and for energy production (Hirose et al. Citation2019). Genes for metal acquisition, like iron (siaABC, shr, shp, and hupYZ) (Bates et al. Citation2003; Cook et al. Citation2019), manganese (mtsAB) (Turner et al. Citation2019) and zinc (adcRCB-adcA) (Ong et al. Citation2015; Sanson et al. Citation2015) were also upregulated (Hirose et al. Citation2019). As observed previously, genes involved in division were rather downregulated showing that bacteria are probably hardly dividing, suggesting either a stationary phase- or a biofilm-like behavior. Genes encoding enzymes to counter oxidative stress (SodA, GpoA) were curiously downregulated.
As mice are quite genetically distant from the native host of GAS, models were developed in non-human primates (NHPs) (). In a microarray study involving 20 NHPs, Virtaneva et al. studied M1 GAS gene transcript changes that occurred during a 84-days infection cycle involving the initial colonization step of the upper respiratory tract, the following clinical pharyngitis, and the subsequent asymptomatic carriage (Virtaneva et al. Citation2005). By analyzing blood, saliva and swabs samples, they showed that, during the first days of colonization (1-4d), the carbohydrates metabolism and genes that counteract immune defenses (streptodornases, scpA, superantigens, …) were upregulated. During the acute phase (5-23d) bacteria were expressing the M, Sic, SpeA, Mac and SpeB virulence factors to evade immune response (). Finally, the asymptomatic phase was characterized by the expression of the myosin cross-reactive antigen and cardiolipin synthase, associated with ARF.
In a more severe infection model in NHPs, Kachroo et al. described the transcriptome of NHPs and GAS during necrotizing fasciitis (NF)/myositis. Biopsies were analyzed after 24h of infection by dual-RNAseq. As for the mice model, the DEGs were around 30% of the genome between in vitro (ME/ESP in THY) and in vivo conditions (Kachroo et al. Citation2020). The upregulated genes are mainly stress response genes, transcriptional regulators, and virulence genes (). For example, 14 genes and/or operons involved in capsule synthesis, cytotoxicity, surface-associated proteins/adhesins, and immune evasion were highly (>5-fold) upregulated in vivo, including the sagA-I operon (encoding the streptolysin SLS), slr, lmb (laminin binding protein), htpA (histidine triad protein), hasABC operon (capsule), speA2, nga/ifs/slo, sclA, sic (complement inhibitor), spd3 and spnA (DNases), spyA, mac (IgG degrading enzyme), isp (immunogenic secreted protein), speJ, and the streptin lantibiotic (srt locus). Similarly, genes that are involved in alternative carbon sources use and metal ion homeostasis including regulation of iron, zinc, and manganese were upregulated as predicted. This study allows the identification of 5 genes specific to NF: inlA (encoding a putative internalin), isp, ihk/irr (TCS), ciaHR (TCS) and dahA (defence against host protein A).
Transcriptomic in human samples
To be more clinically relevant, study of GAS transcriptome in ex vivo human samples have also been published (Table S1 and ). First, since GAS survival in blood is one of the most frequently used functional tests for vaccine efficacy, its transcriptome in human blood has been described (Graham et al. Citation2005). M1 GAS in ESP phase was incubated for 30, 60 and 90 min in human heparinized blood and compared with an in vitro (THY) condition using microarray. An extensive remodeling of the transcriptome has been observed with 75% of the genome differentially expressed, including 97 regulatory genes with different patterns of expression throughout blood exposure time. GAS responds to blood exposure by coordinating expression of metabolic, regulatory and virulence genes (). Blood is low in free amino acids and being auxotrophic for 15 of them, GAS must obtain amino acids by proteolysis, transport and catabolism of oligopeptides. Overall, during exposure to blood, GAS downregulates biosynthesis of cell envelope, cofactors (folate/chromate) and glycolysis but firstly upregulates alternative carbohydrates pathways and then transport and catabolism of oligopeptides (pepF, pepB, opp operon). In parallel, GAS adapts its arginine (arc operon) and serine (sdhAB) metabolisms. Regarding the virulence profile, mga transcripts increased after blood exposure with a peak at 30 min resulting in upregulation of emm and sic. Other virulence factors such as hasABC, isp, sag operon (SLS), mac, scpA, slo and superantigens (speA, speJ, speG and smeZ) were also upregulated. speB and crgR (cathelicidin resistance gene regulator) were both downregulated, probably to enhance bacterial resistance to phagocytosis and host anti-microbial peptides (AMPs) (Thänert et al. Citation2019).
As for the NHPs, pharyngitis (Livezey et al. Citation2011) and necrotizing fasciitis (Thänert et al. Citation2019) have also been investigated. A microarray study on samples from pharyngitis cases separated the eight responsible M-types strains in two distinct expression clusters. Both clusters showed an upregulation of carbohydrates metabolism and iron uptake. The oxidative (per) and peroxide (sodA, dpr) stress responses were also uniformly upregulated. However, only the strains from cluster 1 (M28-M2-M1) showed an upregulation of the sag operon, the mga regulator, three lantibiotic immunity and a bacteriocin-like genes. Strains from cluster 2 (M4-M12-M68-M3-M102) presented an upregulation of amino acids starvation response genes (opp, luxS, clp) not observed in cluster 1. Adhesion molecules also presented a different pattern between clusters as prtF2 was upregulated in C1 strains and sfb1 and emm in C2. This discrepancy warns us that the expression of vaccine targets could be different for each (group of) strain(s). More recently, biopsies from human necrotizing fasciitis have been studied by the dual-RNAseq technique (Thänert et al. Citation2019). In monomicrobial (GAS) infection, complex carbohydrates like lactose, trehalose and galactose use and transport were upregulated. Expression of adhesion molecules (prtF1, prtF2, dfbII, sfbX), immune evasion factors (ska, sic), proteolytic enzymes (speB, scpA) and toxins (speA, speC, speG, smeZ) was upregulated. Interestingly, the adhesion to the extracellular matrix (ECM) and entry into cells by binding to fibronectin seem a crucial step in NF development in human.
Overall, transcriptomic data from in vitro, ex vivo and in vivo studies have helped in understanding metabolism and diet switches that GAS can perform (Table S1 and ). However, comparing these data also highlight some limits. For example, the lack of stress enzymes expression (like sodA) in a mice model contrast with their high expression in human blood, human pharyngitis, and NF. The same observation is made about the grab gene expression, showing the limit of using a non-natural host. The host specificity is probably due to some specific human proteins binding to GAS partners but could also imply specific signals from the host and/or its microbiota. The recently developed GAS Controlled Human Infection Model (CHIM) may provide interesting data regarding some of those issues (Osowicki et al. Citation2021) (). Moreover, in a vaccine research context, in vivo data in the natural host, seem to be required to assess the expression of vaccine antigen targets (Box 1). Finally, even in well-controlled laboratory conditions, the impact of the same condition on different M-types or even different strains from the same M-type can diverge. This prompts us to be extremely prudent with generalization of such data.
Box 1Future key research directions.
Study the expression of all vaccine targets in relevant in vivo conditions (human or NHP host) and in different clinical manifestations
Transcriptomic study of metabolic genes knock-out to decipher links between metabolism and virulence
ChIPseq experiments to identify direct regulon of each TCS/regulators
Transcriptomic study of GAS retrieved from CHIM and in human carriage
Single-cell and in situ transcriptomic studies
Transcriptomic study in co-culture with microbiota to understand relationship between GAS and other bacteria at mucosal surfaces
Use of genetically distant GAS strains, expressing different virulence factors
Perform more Dual-RNAseq studies in the natural host with different GAS strains
Develop new tools to analyze all the generated data
Validate microarray data obtained in vitro and ex vivo by RNAseq
Main regulatory pathways in GAS
Because GAS can cause various inflammatory infections, it implies that GAS has developed a high capability to mount an appropriate response to oxidative stress, to maintain metal homeostasis and to adapt its metabolism to nutrient deprived environments (). Adaptation is mainly regulated by two components systems (TCSs) and stand-alone regulators (reviewed in Siemens and Lütticken (Citation2021). The TCSs generally consist of a sensory and a regulatory component. The sensor component is typically a transmembrane histidine kinase that recognizes one or more environmental signals, and the response regulators are cytoplasmic transcriptional factors that may either repress or activate transcription of target genes (Vega et al. Citation2016). Until now, 13 different TCSs have been mapped in GAS (Ferretti et al. Citation2001; Beres et al. Citation2002; Smoot et al. Citation2002; Vega et al. Citation2016; Siemens and Lütticken Citation2021) as well as 30 stand-alone regulators increasing the level of complexity in gene expression regulation (reviewed in Siemens and Lütticken Citation2021).
Virulence regulation
CovR/S: the master regulator
The discovery that spontaneous mutations in CovRS are selected in vivo (Aziz et al. Citation2004; Sumby et al. Citation2006; Walker et al. Citation2007; Bao et al. Citation2015) or in epidemiologically successful strains (Beres et al. Citation2017) have urged researchers to identify its complete regulon in different strains and conditions. Several mutants of CovS have been identified in hyperinvasive strains from different emm-types (M1, M3, M23, M53, M81) (Cole et al. Citation2006; Miyoshi-Akiyama et al. Citation2006; Mayfield et al. Citation2014; Bao et al. Citation2015). This invasiveness has been attributed to the lack of SpeB secretion and the upregulation of virulence factors such as Sda1, a NETs (Neutrophils Extracellular Traps) degrading DNase, and the capsule (Aziz et al. Citation2004; Engleberg et al. Citation2004; Cole et al. Citation2006; Walker et al. Citation2007; Görke and Stülke Citation2008). The in vivo selection of CovRS variants seems however a rare phenomenon, i.e. concerning only 0.2% to 0.4% of the GAS population in a mouse model, leading to the replacement of the WT population after 7 days (Kazmi et al. Citation2001; Hirose et al. Citation2019).
Transcriptomic analysis of covS mutants (spontaneous mutation abolishing their function or isogenic gene knock-out) showed that if only 6% to 18% of the genes were controlled by CovRS in vitro (THY) (Bao et al. Citation2015; Horstmann et al. Citation2015) around 25% were differentially expressed in vivo (Aziz et al. Citation2010), which, again, emphasizes the importance of in vivo studies to understand GAS regulation. In a M23 strain, CovRS has been described as a negative regulator of 34 out of the 43 genes of the M23 virulome (including hasABC, slo, nga, mga, spyA and prtS) (Bao et al. Citation2015), mainly in EP or ESP. Moreover, CovS was found to be a positive regulator of speB but also of grab, encoding a protein known to protect surface proteins from SpeB proteolysis in blood (Rasmussen et al. Citation1999). Interestingly, spd1 and spd3 were also negatively regulated, in contrast to sda1 in M1T1, suggesting distinct roles of these DNases in GAS virulence. Several genes are not regulated by CovRS in the M23 strain including the superantigens smeZ, speG, but also spd, ska, sclA, hylA, and mac (Bao et al. Citation2015). However, the virulome genes regulated by CovRS diverge from strain to strain. For example, ska is not regulated by CovRS in M23 (Bao et al. Citation2015) but well in M1 (Graham et al. Citation2002), although the techniques used are quite different. In acapsular M4 strain, CovS mutant shows a higher mga expression, which is not observed in M1 and M3 strains (Galloway-Peña et al. Citation2018). In M3 strain, CovRS represses the pilus expression, by contrast to its role in M1 (Calfee et al. Citation2018) or other M-types (Roshika et al. Citation2022). Even in the same M-type, CovRS shows difference in its regulon. Indeed, transcriptomic studies of CovS spontaneous mutants in M28 strains from phylogenetically defined subclade A and B (Kachroo et al. Citation2019) showed that 142 genes were differentially expressed.
Even if CovRS is defined as a negative regulator of virulence, it also regulates carbohydrates and nitrogen metabolism, cell division (parE-parC), and other stand-alone transcriptional regulators making it a ‘master regulator of GAS genes expression’ (Roberts et al. Citation2007). Depending on the strain and the environmental conditions, up to 25% of the genome could be CovRS-dependent. Among the regulated genes, only a small part has a binding motif for CovR (8%) (Bao et al. Citation2015), showing that CovRS regulation is mainly indirect. Indeed, CovRS is known to regulate at least 26 regulators (Graham et al. Citation2002) among them rivR (Roberts et al. Citation2007), a member of the RofA-like protein type (RALP) family (Roberts et al. Citation2007) or some TCS, like ihk/irr (Graham et al. Citation2002).
However, these data have been challenged by recent data using both RNAseq and CovR ChIPseq. Indeed, Horstmann et al. found that CovR binds the promoter of 9 transcriptional regulators with 6 regulators being differentially expressed in the covR-D53A mutant (Horstmann et al. Citation2022). By comparing RNAseq in the M1T1 854 covR deletion mutant and CovR ChIPseq, Finn et al. highlighted that only half of the genes differentially regulated in the covR mutant had CovR bound to their promoter. They further demonstrated that several CovR-targeted regulators (ralp4/rivR, a marR-like regulator and ropB) were responsible for part of the CovR-indirect effect (Finn et al. Citation2021). By contrast, the Horstmann work with the M1T1 MGAS2221 found that CovR repression was direct, since 20 out of 23 CovR-repressed genes had CovR bound to their promoter. Besides, the promoters of only 5 out of 17 CovR-activated genes were bound by CovR. Intriguingly, they did not find CovR binding to the promoter of the three transcriptional regulators mentioned above, but to the rgg4 regulator and the trxTSR TCS system (Horstmann et al. Citation2022). A third CovR ChIPseq analysis in a M3 background confirmed a direct regulation for nearly all CovR-regulated virulence genes, like for the MGAS2221 data (Horstmann et al. Citation2022; Citation2023). Moreover, they identified several highly variable regions like the nga-slo operon, spyA gene, and sag operon that are targeted by CovR, which could explain the strain-dependent impact of covS inactivation on GAS virulence (Horstmann et al. Citation2023).
The CovRS regulon seems slightly impacted by the growth phase (Bao et al. Citation2015). Several ex vivo (human blood, macrophages) (Graham et al. Citation2005; Hertzén et al. Citation2012) and in vivo microarray studies (mice chamber model, pharyngitis in NHPs) (Virtaneva et al. Citation2005; Aziz et al. Citation2010) have shown that CovRS is implicated in virulence modulation in more physiological conditions (Virtaneva et al. Citation2003; Graham et al. Citation2005; Aziz et al. Citation2010) but with some discrepancies. Signals regulating the expression and activity of CovRS in vivo are not known but the Mg++ concentration is enhancing CovR phosphorylation in vitro (Gryllos et al. Citation2003; Horstmann et al. Citation2015; Citation2018; Finn et al. Citation2021). This effect seems however dependent of the M-type (Horstmann et al. Citation2015). Data in blood, where the Mg++ concentration is high, shows that covR expression is also upregulated (Graham et al. Citation2005). The defensin LL37, secreted by the immune system in response to infection, has been shown to relieve CovR-mediated repression by direct binding to CovR. However, once again, this effect is dependent of the strain (Tran-Winkler et al. Citation2011; Horstmann et al. Citation2015; Finn et al. Citation2021).
CovRS mutants, by their invasiveness, their in vivo selection and their out-competition with wild type (WT) strains are of significant concern for the scientific community (Aziz et al. Citation2010). Fortunately, data obtained in an improved murine model of nasopharyngeal colonization showed that inactivation of covRS in a M75 strain was detrimental for colonization and shedding (Alam et al. Citation2013). Moreover, the M1T1 variant has shown to be less persistent in saliva (Treviño et al. Citation2009). Decreased adherence to epithelial cells and mouse skin, as well as lower biofilms formation, have been observed for an isogenic covS mutant (Hollands et al. Citation2010). These data predict a lower ability of the covRS mutants to disseminate in the human population and explain why they have not been fixed in the GAS population. However, these results have been recently challenged by the identification of a CovR mutation (S130N) in M89 which presents a higher fitness in saliva than its ancestor strains (Beres et al. Citation2016). Both strains are separated by more than 20 years which suggest that GAS might be evolving to counterbalance the negative effect of covRS mutations.
Inactivation of the master regulator CovRS is an immediate advantage to counteract the strong selective pressure of the innate immune system, and particularly neutrophils which clear GAS from infected tissues (Li et al. Citation2013; Citation2014; Liu et al. Citation2015). Yet, the host genetics and infection sites where covRS mutants are selected, the differences in virulence factors repertoires, regulatory regions and regulator networks of the emm-types involved could explain the discrepancies between strains (Wilkening and Federle Citation2017).
RocA: positive regulator and accessory protein for CovRS
Whole genomes sequencing data highlighted a non-sense mutation at the beginning of the rocA gene in all sequenced M18 strains. This SNP results in a hyper-encapsulation and extends the carriage in the mouse nasopharynx (Lynskey et al. Citation2013). Knock-out mutants of rocA in diverse M-types (M1, M3, M6, M14, M18 and M89) have an increased virulence phenotype due to an increased expression of CovRS-dependent virulence factors (Biswas and Scott Citation2003; Lynskey et al. Citation2015; Miller et al. Citation2015; Zhu et al. Citation2016; Feng et al. Citation2017; Jain et al. Citation2017). RocA is natively inactivated in all M3 strains (stop insertion at position 410) resulting also in a hyper-encapsulated phenotype (Lynskey et al. Citation2013; Citation2015; Miller et al. Citation2015). However, the enhanced activity of CovR in a M28 rocA mutant is mainly due to an increase in its phosphorylation state rather than of its expression (Bernard et al. Citation2018). The mechanism of CovRS regulation by RocA has been recently identified by three independent groups (Lynskey et al. Citation2019; Bernard et al. Citation2020; Jain et al. Citation2020). RocA, forming dimers by its cytoplasmic part, interact directly with CovS at the streptococcal membranes and inhibit its phosphatase activity leading to an increase in phosphorylation of CovR (Chiang-Ni et al. Citation2020). In M3, the observed polymorphisms of rocA disrupt the dimerization step, resulting in protein aggregates formation and inability of RocA to interact with CovS (Lynskey et al. Citation2019; Bernard et al. Citation2020).
Interestingly, rocA in M28 strains isolated from iGAS is highly polymorphic (Bernard et al. Citation2019), suggesting an emm-type specific function of RocA in M28. To understand this phenomenon, Bernard et al. had studied the transcriptome of a rocA deletion mutant (ΔrocA) in the M28 background. When compared to the WT strain in EP and ESP in THY, the ΔrocA mutant has a markedly different transcriptome with 427 (25,8%) and 323 (19,5%) differentially expressed genes, respectively. Of these genes, 109 were common between both phases. RocA regulates (directly or not) 41 transcriptional regulators (TRs) in M28, among which two are dedicated to virulence; CovRS and Mga (multiple virulence gene regulator of GAS). Consequently, the isogenic ΔrocA mutant produces increased amount of SPN and SLO and has increased SOF, NADase, SLO and SSE secreted activity (Bernard et al. Citation2018). However, the increased transcripts of speB do not correlate with an increase in proteolytic activity (Bernard et al. Citation2019). The other TRs regulated by RocA in M28 are mainly linked to stress response including ciaHR, nrdR and spxA2 (Ben Zakour et al. Citation2015; Zhang et al. Citation2015; Port et al. Citation2017) as well as the arcABCD operon involved in acidic environment response (Cusumano and Caparon Citation2015). This correlates with a growth advantage of the ΔrocA mutant in acidic condition like encountered in purulent lesions.
Comparing the transcriptome of forty-eight M28 clinical isolates to WT strains or ΔrocA deletion mutants divides strains into two main phenotypes: one subtly altered ‘WT-like’ or a substantially altered ‘ΔrocA-like’ (Bernard et al. Citation2019). However, each polymorphism in rocA had a unique effect on GAS transcriptome. All these data explain that rocA polymorphisms are selected during human invasive infection with M28. In vivo, the ΔrocA mutant has also an increased virulence in a mouse model of NF and also in a non-human primate model (Bernard et al. Citation2018; Citation2019).
As observed for other regulators, the RocA regulon differs between M-types. In M28, the ΔrocA mutant and the polymorphic rocA alleles result in a decrease of ska transcripts in contrast to other M-types (Miller et al. Citation2015; Sarkar and Sumby Citation2017; Sarkar et al. Citation2018). However, the molecular mechanism of regulation through a direct interaction between RocA and CovS seems conserved between M-types (Lynskey et al. Citation2019; Bernard et al. Citation2020).
Scarce data are available considering the triggering and regulation of rocA expression. In M1, rocA is upregulated in human blood at all time points tested (30, 60 and 90 min) but covRS is upregulated only after 30 min and goes back to basal level for the later time points (Graham et al. Citation2005) suggesting that another negative regulator of covRS is acting in vivo. Much remains to be determined in the complex regulation of CovRS by RocA, especially at the transcriptional level since RocA does not possess a canonical DNA binding domain.
Ihk/Irr and the immune evasion
The regulon of the ihk/irr TCS has been investigated by several microarray studies (THY, H2O2, neutrophils and macrophages contact, blood, saliva, mouse NF model) (Graham et al. Citation2005; Shelburne et al. Citation2005; Grifantini et al. Citation2011; Hertzén et al. Citation2012). A large set of differentially expressed genes are implicated in cell wall synthesis suggesting a role of Ihk/Irr in regulating this process (Voyich et al. Citation2004; Grifantini et al. Citation2011; Hertzén et al. Citation2012). Ihk/Irr also regulates the expression of oxidative stress genes (Voyich et al. Citation2004; Grifantini et al. Citation2011) and protects against killing by H2O2.
Expression of ihk/irr is triggered by H2O2 and neutrophils primary granules exposition by up to 2-fold and up to 20-50-fold, in both M1 and M18 strains respectively (Voyich et al. Citation2004). Upregulation of ihk/irr is observed when GAS is in contact with human polymorphonuclear neutrophils (PMNs) (Voyich et al. Citation2003). At early stage of human macrophages infection (after 2h of macrophages invasion), the ihk/irr genes are upregulated up to 8-fold (Hertzén et al. Citation2012). They found that the TCS has a positive impact on bacterial survival intracellularly and/or on the bacterial release. In blood, GAS is also able to enter and survive in phagocytes by avoiding maturation of the phagolysosomes using the M protein (Staali et al. Citation2006). Graham et al. in 2005 have shown that the ihk/irr genes are also upregulated after 60 min in an ex-vivo culture in whole human blood (Graham et al. Citation2005).
Oxidative stress encountered during a 16h culture in human saliva also enhances ihk/irr expression (Shelburne et al. Citation2005). More recently, the role of Ihk/Irr have been described in a model of necrotizing fasciitis (Kachroo et al. Citation2020) using dual-RNAseq and TraDIS (genome-wide transposon-directed insertion site sequencing). They showed that the TCS is expressed 11-fold more in vivo than in vitro and the Δirr mutant is less virulent in a mouse model of NF. This is in line with data showing that irr gene is upregulated (2 to 20-fold) in human pharyngitis caused by different M-types (Voyich et al. Citation2003).
The role of this TCS is clearly involved in the response to oxidative stress encountered in or at the vicinity of innate immune cells like PMNs and macrophages, regardless of the tissue. As the cell wall synthesis proteins seem also implicated in this process, we could postulate that reshaping the cell wall inside immune cells could be a mechanism of immune evasion. However, most of the data come from microarray studies, except for one study in NHPs. Further RNAseq studies are needed to understand the role of this TCS.
Links between virulence and metabolism
CcpA and the carbohydrates use
Carbon catabolite repression is a global process used by bacteria to prioritize the utilization of favorable energy sources like glucose and CcpA is the key mediator of this repression. It acts as a transcriptional repressor in Gram-positive bacteria and its repression activity is facilitated by its interaction with HPrSer46∼P (Görke and Stülke Citation2008; Shelburne et al. Citation2008). Of note is that CcpA activity is also regulated by glucose-6-phosphate, fructose-1,6-bisphosphate and NADP in Bacillus (Gösseringer et al. Citation1997; Kim et al. Citation1998). To identify the CcpA regulon, DebRoy et al. used RNAseq in a M1T1 isolate (MGAS2221) and introduced ChIPseq for the first time in GAS (DebRoy et al. Citation2021). They found that 514 genes (31%) were differentially regulated in the ΔccpA strain with more upregulated (361) than downregulated (153) genes. ChIPseq analysis highlighted that only 105 genes (6%) were the direct targets of CcpA. Moreover, using a CcpAV301A mutant (unable to interact with the phosphorylated PTS protein HPr), they further dissociated the HPr-dependent/independent regulons (not discussed here). Amongst the genes differentially regulated, they found genes encoding 13/14 of the phosphotransferase systems (PTS) and 4 ABC carbohydrate transporters. Two PTS were activated indirectly by CcpA and the 15 other transport systems were repressed (7 directly and 8 indirectly), which is consistent with CcpA role in carbon source acquisition and utilization. Amongst virulence genes, speB, grab and endoS were found to be activated by CcpA while speA2, sic, spd, ska, nga-slo, spyCEP and sagA (SLS operon) were repressed. ChIPseq analysis further highlighted that only speB, spyCEP and sagA were the direct targets of CcpA (DebRoy et al. Citation2021). They proposed that the indirect effect of ccpA deletion on the expression of virulence genes was not due to a direct effect of CcpA on different regulators since only 4 out of the 21 differentially expressed regulators were directly targeted by CcpA and none of them are known or expected to regulate virulence genes.
RT-qPCR analysis of the M1T1 and its ΔccpA derivative previously highlighted its role in sagA repression. Functional analysis showed that the ΔccpA strain is hypervirulent during systemic infection in mice. This phenotype was abolished by the inactivation of the sag (SLS) operon, confirming the RNAseq data (Kinkel and McIver Citation2008). RT-qPCR on both strains grown in saliva, which is a glucose limited environment, confirmed that CcpA upregulates the expression of speB, which inactivates several virulence factors. Besides, the ΔccpA mutant was impaired in saliva persistence and mice oropharynx colonization compared to the parent M1T1 strain (Shelburne et al. Citation2008). Finally, another study found that the ΔccpA mutant was attenuated during soft tissue infection (Kietzman and Caparon Citation2011).
The study of CcpA role in different M-types (M1, M3 and M28) grown in THY to the EP phase has shown that even if a core CcpA regulon exists, the role of CcpA presents some specificity across M-types (DebRoy et al. Citation2016). RNAseq analysis identified five COGs consistently enriched in all 3 mutants, among which carbohydrates metabolism and transport, and the sag (SLS) operon. However, the 3 mutants present diverse phenotypes in terms of growth, colony size, spyCEP and endoS expression, and virulence in vivo (DebRoy et al. Citation2016).
Glucose-rich and glucose-limited environments are encountered by GAS during infection (). This is consistently reflected by the different behaviors of the ccpA deletion mutants according to infection sites. CcpA repression should logically be relieved when GAS is in a low glucose environment, like in deep tissues infection. Indeed, expression of ccpA itself is upregulated in vivo in a mouse model of necrotizing fasciitis (Hirose et al. Citation2019). RNAseq of GAS grown in C medium has shown that low glucose/high peptides environment allow the differential expression of 496 genes, among which 24% are CcpA-dependent (Valdes et al. Citation2018). Late logarithmic growth in C medium reflects, in part, the CcpA regulon, in absence of glucose. Finally, it has been shown that CcpA can override the CovR-mediated virulence downregulation in vivo (Shelburne et al. Citation2010).
Mga and the surface proteins
Mga (multiple gene regulator of GAS) is a DNA binding protein, first identified as the regulator of the emm gene (Caparon and Scott Citation1987). Two alleles of mga (mga-1 and mga-2) have been described and belong to the clade X and Y strains (Hollingshead et al. Citation1993; Flores et al. Citation2013; Sanderson-Smith et al. Citation2014) which suggest a potential role in tissue tropism (Hondorp and McIver Citation2007). Comparison of the transcriptome between strains harboring mga-1 (M1 and M6) or mga-2 (M4) has shown that the numbers of DEGs in M1/M4 (200 genes) and M6 (37 genes) were very different (Ribardo et al. Citation2004). However, the M6 strain used in this study (M6 JRA) was totally atypical compared to other M6, and results could not be generalized to other M6. Comparing the M1 and M4 mga regulon shows a lot of common regulated genes (Ribardo et al. Citation2004). Overall, Mga regulates approximately 10% of the GAS genome during exponential phase of growth in rich medium (THY), including transcription of several sugar transport and utilization operons (Ribardo and McIver Citation2006). Polymorphism in mga-2 allele has been observed in a collection of 800 epidemic M59 isolates (Sanson et al. Citation2015). Deletion of mga in a M59 strain showed that 7% of the genome was differentially expressed in THY. Among them, 74 genes were upregulated and 70 downregulated, the latter corresponding to virulence factors, nutrient metabolism, and transport.
Mga is the best carbohydrate-responsive regulator described to date and its mechanism of action has been extensively reviewed in Vega et al. in 2016 (Vega et al. Citation2016). Its function is mainly linked to colonization and invasion by GAS and mga is mainly expressed in EP and ESP and nutrient rich environment (Beyer-Sehlmeyer et al. Citation2005). Indeed, the ‘core mga regulon’ is composed of mga itself, the emm and emm-like genes, sic, fba and scpA, encoding proteins involved in adhesion or immune evasion (Smeesters et al. Citation2010; Frost et al. Citation2018). Mga also regulates genes located outside the mga locus like speB, sclA (encoding a fibronectin binding protein), sof and sfbX (Fiedler et al. Citation2010). However, the mga dependent expression of speB seems different between M-types; speB expression is activated by Mga in M1 and M49 but not in M4 and M6 (Ribardo and McIver Citation2006). Mga also represses the uptake of alternative sugars and activates iron acquisition (sia operon), fatty acid metabolism (fab operon) and the expression of salA (lantibiotic). The Mga regulon and the carbohydrates utilization operon are both expressed in vivo during the acute phase of pharyngitis in an NHP model (Virtaneva et al. Citation2005). All mga mutants generated to date present a decrease in virulence in vivo (Kihlberg et al. Citation1995; Terao et al. Citation2001).
Recently, the impact of sugar availability on the mga mutant has been further studied (Valdes et al. Citation2018). The transcriptome of a mga mutant (M1T1 5448) was compared between growths in THY and C media (Valdes et al. Citation2018). The study was performed on bacteria in late exponential phase, when mga is the most expressed. Only few overlaps exist (only 16 genes) between genes regulated by Mga in both conditions other than virulence genes. 135 genes (44 repressed, 91 activated) were shown to be regulated by Mga in a glucose limited environment. Among the activated genes, we found phage-encoded streptodornases (spd3 and sdaD2), the streptolysin S (sag operon), the streptolysin O (slo) and the streptokinase (ska), which were not part of the Mga regulon in THY. By contrast, Mga did not regulate speB in C medium. They observed a broader induction of known virulence factors in a Δmga background in C medium than in THY, suggesting that glucose (presence or absence) impacts the GAS Mga regulon. As the level of mga transcripts was not significantly different between both conditions, the change in Mga regulon seems rather linked to post-transcriptional modifications (Hondorp and McIver Citation2007; Hondorp et al. Citation2013; Valdes et al. Citation2018). Mga, even if auto-regulated, is also repressed by RopB/Rgg (Federle Citation2012), CcpA in late exponential phase (Almengor et al. Citation2007) and by the RALPs (RivR, Nra, RALP3 and RofA) (Beckert et al. Citation2001; Chaussee et al. Citation2002). Mga, in turn, represses CcpA in the exponential phase of growth (Almengor et al. Citation2007).
LiaFSR
Interestingly, a naturally occurring mutation in LiaS (R135G) has been found in a M3 GAS isolated from a carrier after an acute pharyngitis (Flores et al. Citation2015). LiaS is part of a three-component system (3CS), called LiaFSR (Lipid II interacting antibiotic), first described in Bacillus subtilis (Jordan et al. Citation2006). LiaFSR is specific to Firmicutes and has a role in stress envelope response (Jordan et al. Citation2007). LiaS is the sensor kinase, LiaR the response regulator and LiaF is a membrane-bound regulator of LiaS and consequently of LiaR. In Streptococcus, LiaF and LiaS are associated with the ExPortal (Lin et al. Citation2020) (whose integrity is compromised in presence of antimicrobials or human antimicrobial peptides like the human neutrophil peptide 1 (hNP1)).
The LiaSR135G strain shows a better ability to colonize and persist in the mouse nasopharynx as well as an increased adherence on human cell lines. By contrast, the strain is less virulent in a mouse model of iGAS and survives less ex vivo in human blood (Flores et al. Citation2015). RNAseq analysis of this carrier strain (MGAS10870) compared to the infecting parental M3 strain in EP or ESP showed that 6.6% and 7.6% of the genes have differential expression levels. Among them, they observed a decrease in mga transcripts, like previously described in a carrier strain (Flores et al. Citation2013), and an increase in speB transcripts. However as previously mentioned, an increase in speB transcripts is not always correlated with an increase in SpeB activity (Bernard et al. Citation2019). The decrease in mga transcripts and its regulated products in the carrier strain suggests that M (and M-like proteins) might not be expressed in the first steps of colonization. The dlt operon, allowing the synthesis of D-ala-LTA, is also down-regulated in the carrier strain, potentially explaining its susceptibility to antimicrobials. No impact on capsule expression was observed, in contrast to what has been observed in rocA variant allowing a longer time of carriage in mice nasopharynx (Jain et al. Citation2017).
By contrast, a naturally occurring amino acid replacement in LiaS (K214R) was positively correlated with iGAS in some Clade 3 M89 strain (Beres et al. Citation2016). In this strain, the upregulation of virulence traits like the sag operon encoding the SLS in exponential phase and the speG toxins in stationary phase was observed (Beres et al. Citation2016). However, the strain has been shown to have a better fitness in ex vivo human saliva, which reflects, at least partially, the environment of GAS colonization. This contrasts with previous data on CovRS mutation associated with invasive phenotype but less able to spread between hosts (Treviño et al. Citation2009).
Recently, an in depth understanding of the LiaFSR TCS has been published by Sanson et al. (Citation2021). They showed that LiaFSR positively regulates the transcription of spxA2 (suppressor of ClpP and ClpX) which is similar to RNA polymerase binders. In the liaR isogenic mutant, the level of speB is increased by 432-fold but is not associated by an increase in its regulator ropB. The speB transcripts increase has been linked to the absence of SpxA2 which has an inhibitory effect on RopB binding to the speB promoter (Port et al. Citation2017). The opposite phenotype (decrease of speB transcripts) was observed with the previously described carrier strain, suggesting that the R135G mutation activates LiaR. Pilus and grab genes, implicated in GAS adhesion, were also increased in the ΔliaR mutant.
The ΔliaR mutant also shows a decrease of hasABC, spyCEP, mac-1, ska and ideS expression, suggesting a link between LiaFSR and the CovRS system. Indeed, SpxA2 is also responsible for a decrease in CovR phosphorylation, leading to upregulation of its regulon. Surprisingly, the ΔliaR mutant was not impaired in a mouse model of NF but well in a human whole blood survival test. This discrepancy between natural host and animal model could be attributed to the lack of expression of specific host proteins (like hNP1) in mice.
MtsR and the iron homeostasis
The carrier state remains an interesting enigma in GAS and probably results from several distinct pathways. Some single mutations affecting capsule production (Flores et al. Citation2014), the stand-alone regulator Mga or surface protein SclA have been associated with the carrier state (Flores et al. Citation2013; Citation2015).
A recent study has highlighted the role of another regulator, MtsR in a mouse model of vaginal carriage (Cook et al. Citation2019). In these conditions of iron limitation, the mtsR regulon, including sia (iron acquisition locus), metal transporters and hupY (heme binding), is upregulated due to impaired binding of MtsR (Cook et al. Citation2019). As this regulator was not differentially expressed in a carrier strain recovered from human (Flores et al. Citation2015), it could be tissues or host specific and should be investigated in depth. Microarray analysis of GAS M49 WT and ΔmtsR mutant grown to mid-logarithmic phase in iron-complete medium identified 64 differentially regulated genes (44 up- and 20 down-regulated) (Toukoki et al. Citation2010). Genes activated by MtsR are involved in nucleotide metabolism and transport, cell envelope and regulation. Besides the sia operon and PerR-regulated pmtA, repressed genes are involved in protein synthesis and amino acid metabolism. However, in contrast to M1 and M3 strains, mts transcripts (Mn/Fe transport) are not significantly affected by the absence of MtsR although the protein was able to bind the mtsR/A intergenic region. Finally, the ska virulence gene is repressed while mga and emm genes are directly activated by MtsR in the M49 strain, suggesting a dual function for the MtsR regulator. By contrast, to avoid iron poisoning, GAS encodes an Fe(II) efflux pump, called PmtA (Turner et al. Citation2017; VanderWal et al. Citation2017). PmtA, whose expression is regulated by PerR, is crucial for GAS resistance to oxidative stress and survival in mice. RNAseq data in presence of excess iron showed that GAS upregulated pmtA and pathways allowing energy generation during stress like man, fru, arcBCD and sdhAB operons (VanderWal et al. Citation2017). Fatty acid metabolism as well as DNA and amino acid metabolism were downregulated, suggesting a change in membrane permeability and a decrease in macromolecules synthesis.
The homeostasis of other metals such as Mn and Zn is also crucial for GAS survival in the host. The two efflux pumps MntE (Mn) and CzcD (Zn) have been identified and help GAS to remove intracellular excess of metals and to resist to oxidative stress in macrophages and neutrophils (Turner et al. Citation2015; Makthal et al. Citation2020; Aikawa et al. Citation2023). By contrast, GAS growth needs Zn and Mn, which can be scarce inside the host, notably by their binding to calprotectin (CP). RNAseq study of GAS transcriptome in presence of CP showed that the AdcR regulon responsible for Zn import and scavenging is upregulated as well as pmtA, hasABC and the dipeptide uptake system (dppABDCE). CP downregulates carbohydrates metabolism (lac.1 operon), the virulence factors grab, cfa and cbp, as well as the copper regulator, copY (Makthal et al. Citation2020).
A role of SptRS in human saliva has also been described (Shelburne et al. Citation2005). This TCS is important for GAS growth in saliva since the survival of the ΔsptR mutant is impaired. The TCS is expressed only in stationary phase (M1) and in clinical strains of different emm-types such as M1, M2, M4, M5 and M6. SptRS positively regulates carbohydrates utilization, and virulence genes such as spd1, spd3, sic, hasA, sagA, speA and lmb.
Understanding expression profiles in carrier state, inside the nasopharynx and at the skin surface (), seems to be a priority to develop relevant vaccine candidates against GAS (Box 1). For example, mga downregulation in carrier state could impair the indirect effect of an emm-based vaccine on carriage. The only RNAseq study on GAS carriage was done in a mouse model of vaginal colonization (Cook et al. Citation2019). This model, even if interesting regarding the percentage (11-20%) of vulvovaginitis in prepubertal girls due to GAS (Donald et al. Citation1991; Stricker et al. Citation2003), is relatively far from the more common GAS niches like the tonsils.
Other players in metabolism and virulence
LacD.1, even if part of the lac1 operon, is not used to metabolize galactose (done by LacD.2) but has regulatory properties, notably on speB (Loughman and Caparon Citation2006). LacD.1 was identified as a catabolism regulator, responding to glucose but with opposing activities to CcpA (Kietzman and Caparon Citation2010). Its regulatory activity is independent from its enzymatic activity, but substrate binding is necessary for functionality (Loughman and Caparon Citation2006). In deep tissue like environments, LacD.1 seems to increase the fitness of GAS (Cusumano and Caparon Citation2013). However, in mice subcutaneous infections, the lacD.1 mutant induce similar lesions than the WT strain (Kietzman and Caparon Citation2010). The transcriptome analysis of a lacD.1 mutant in M14 has shown that LacD.1 represses speB while CcpA induces speB expression and that this co-regulation is performed at different infection phases (Kietzman and Caparon Citation2011).
Even if the link between virulence and metabolism is obvious, only scarce data are available on transcriptome of metabolic gene mutants. As high rates of polymorphisms in the fabT gene were observed in M1 isolates, Eraso et al. studied the transcriptome of a knock-out mutant of fabT compared to its parental M1 strain (MGAS2221) in EP and SP in THY, at 35° and 40 °C (Eraso et al. Citation2016). In all tested conditions, FabT represses the fab operon responsible for fatty acids synthesis and activates the expression of the lac operon and other genes implicated in sugar fermentation. The difference between the mutant and the WT was much more visible in SP and mainly concerns genes related to metabolism (glycolytic pathways, pyruvate hub, purin/pyrimidin synthesis). Interestingly, some virulence genes were either upregulated (graB, mga, sag operon) or downregulated (covS, sic). The fabT mutant is less virulent in vivo (necrotizing fasciitis) and survives less in blood but presents an increased resistance to polymyxin B. Finally, the fab operon is downregulated in a model of vaginal carriage (Cook et al. Citation2019).
Recently, the arginine deiminase (ADI) pathway (arcABCD operon) has been shown to be important in skin colonization where glucose is limited (Hirose et al. Citation2021). This pathway, found in many bacteria, provides acid stress protection and supplements energy production during glucose starvation (Abdelal Citation1979; Cotter and Hill Citation2003). The ADI pathway is under the negative control of CcpA, CovRS and Rgg (Dmitriev et al. Citation2006; Shelburne et al. Citation2010), which are also virulence regulators. Recently, the role of the ADI pathway in virulence has been elucidated (Hirose et al. Citation2021). In low glucose but high arginine environment, like on the human skin (Sylvestre et al. Citation2010; Kubo et al. Citation2013), GAS expresses more virulence genes such as slo, the sag operon, nga and spd3 and downregulates covS. The arcA mutant is less cytotoxic for keratinocytes ex vivo and less virulent in a mouse model of skin infection. However, this mutant has a WT phenotype in a systemic mouse infection or in ex vivo human blood growth. These data elegantly show that arginine, present in some specific sites of the host, allows cytolysins expression and virulence. This effect is dependent of a fully functional ADI pathway and not solely on arginine presence. It could be interesting to test the expression/activity of the ADI pathway between strains with a skin tropism as D4 strains (Sanderson-Smith et al. Citation2014).
Additionally, the global regulator of the stringent response (amino acid and nitrogen starvation), CodY, was also shown to positively influence the expression of the sag operon (SLS) and other virulence factors relative to nutritional status (Malke et al. Citation2006). In M49, CodY and CovRS regulate several virulence genes in common (ska, has operon, …) but with an opposite effect as CodY is a positive regulator while CovRS is a negative regulator of virulence (Kreth et al. Citation2011).
Four RALPS have been identified in GAS: RofA (Ralp1), Nra (Ralp2), Ralp3 and RivR (Ralp4), for which previous data has already been extensively reviewed (Vega et al. Citation2016). Recent data has shown that Nra is a positive regulator of pilus expression (Calfee et al. Citation2018). Ralp3 has been shown to regulate around 3,5% of the gene content in M49, with a negative regulation on CcpA and some stress response genes and a positive effect on lactose and fructose operon expression as well as the mga regulon (Siemens et al. Citation2012).
In the Rgg family, the role of Rgg1, also known as RopB, and Rgg4 (ComR) has been extensively reviewed elsewhere (Vega et al. Citation2016). Briefly, Rgg1 regulates genes for carbohydrates use, induction of phages and repression of nga-ifs-slo operon in SP (Beyer-Sehlmeyer et al. Citation2005; Dmitriev et al. Citation2006). Rgg2 (MutR) and Rgg3 mutants, implicated in GAS quorum sensing, have been more recently characterized (Zutkis et al. Citation2014; Rued et al. Citation2022). The rgg2 mutant in a M1 strain shows 155 DEGs with the upregulation of some virulence genes like scpA, slo and nga. However, the mutant is less virulent in mice, less able to survive in blood and to adhere to cells (Zutkis et al. Citation2014). Recently, a rgg3 mutant has been shown to have a huge impact on RNA transcripts and proteome in presence of the short hydrophobic peptide (SHP) pheromone. The changes represent mainly a stress response due to SHP excess but also involving genes not directly regulated by Rgg2/3.
Transcriptional regulation of phage genes
Genomic and functional studies have shown that GAS can contain up to 7 temperate phages (prophages) in its genome. These temperate phages can either integrate in the host genome (lysogeny) or enter in a lytic cycle, releasing new progeny after lysis of the bacterial host. GAS prophages carry and mobilize several virulence factors, such as DNases and superantigens, which significantly interfere with the human immune system (Commons et al. Citation2014; Remmington and Turner Citation2018; McShan and Nguyen Citation2022). They were found to play an important role in the expansion of the different pandemic lineages that have emerged since the mid-1980s (reviewed in Barnett et al. Citation2018). The study of the conditions and pathways that lead to the induction and mobilization of temperate phages is therefore of high importance to better understand the epidemiology of GAS. Although phage-associated virulence genes expression is routinely analyzed in transcriptomic studies, only few studies have addressed the expression of GAS phage encoded genes. Prophages generally enter in lytic cycle in response to different types of signals and damages that are considered as detrimental for the survival of their host (and so the prophage) (reviewed in Nanda et al. (Citation2015). Interestingly, Broudy and colleagues found that some human-derived soluble factors increase the production of GAS phages in two M6 and M76 strains and promote the transfer of the M76 phage to a M12 strain (Broudy et al. Citation2001; Broudy and Fischetti Citation2003). Additionally, the two M3 phages φ315.3 and φ315.4 were found to be induced in presence of hydrogen peroxide and human cells, respectively (Banks et al. Citation2003).
Transcriptomic analysis of a M23 strain in EP and ESP growth phases highlighted differences in gene expression of two out of the four M23 temperate phages, with 14% and 24% of their genes activated during ESP, respectively (Bao et al. Citation2015). Additionally, ex vivo studies with macrophages and PMNs highlighted the upregulation of phage-encoded genes from two M1T1 strains (Voyich et al. Citation2003; Hertzén et al. Citation2012).
The transcriptomic analysis from mutants of TCS and stand-alone regulators has also highlighted their roles in GAS prophages biology. In the M1T1 ΔcovR mutant grown to EP and ESP in rich medium (THY), an increase in the number of upregulated phages genes was observed compared to the WT strain (Graham et al. Citation2002). Similar results were obtained for the 4 prophages of the M23 strain ΔcovS mutant in ESP (Bao et al. Citation2015). The role of CovRS in repressing phages induction was further observed with the animal passage M1T1 strain (Aziz et al. Citation2010). The phage φNZ131.1 (M49 strain) is spontaneously induced during the EP in THY medium. Interestingly, 19 out of 21 differentially expressed genes were less induced in the Δrgg1 (ΔropB) mutant in contrast to the integrase gene. Prophage induction was further confirmed by excised vs. lysogenized- specific PCR (Dmitriev et al. Citation2006). Since the Rgg family (including RopB) proteins are bona fide quorum sensing regulators (Jimenez and Federle Citation2014; Makthal et al. Citation2016), it suggests that this phage could sense cell population density in a RopB-dependent manner. Microarray analysis of the Δnra mutant (FCT-region regulator) and ΔciaH mutants also highlighted differential regulation of phage genes in EP, ESP and SP, and ESP, respectively, compared to the WT M49 strain (Kreikemeyer et al. Citation2007; Riani et al. Citation2007).
Microarray analysis of throat swabs samples from human pharyngitis due to different M-types (M3, M4, M12, M68 and M102) identified 35 phage genes amongst the top200 upregulated genes suggesting their induction during the onset of the disease (Livezey et al. Citation2011). Similar results were previously obtained in a NHP model of pharyngitis with the M1T1 strain MGAS5005 (Virtaneva et al. Citation2005). During both the colonization and acute phases, the phage gene category (notably structural genes from φ5005.1, .2 and .3) was enriched when CFUs were high, suggesting a relationship between high cell densities and prophage gene expression as observed for the M23 strain in vitro (Virtaneva et al. Citation2005; Bao et al. Citation2015). Moreover, there was a positive correlation between phages copy number and expression of their associated virulence factors (Virtaneva et al. Citation2005). In a necrotizing fasciitis model in mice, contrasting results were obtained. Basal expression of genes of the three phages of the M1T1 5448 strain was observed in THY, suggesting spontaneous induction. However, phages genes involved in lysogeny and lytic cycle were up- and down-regulated during necrotizing fasciitis compared to exponential growth in THY, respectively (Hirose et al. Citation2019).
Finally, GAS also carries satellite prophages, known as SpyCI (S. pyogenes Chromosomal Island). The best characterized SpyCI (SpyCIM1) is found as an episome during exponential growth in rich medium and induces a mutator-phenotype during stationary phase by actively integrating in the DNA mismatch repair MMR operon. Transcriptomic analysis of a ΔSpyCIM1 strain (M1) highlighted an additional role in the regulation of virulence genes. Indeed, the emm, hasABC, nga and slo genes are upregulated by SpyCIM1 independently of its role in MMR regulation during the EP in rich medium (THY). Moreover, the expression of speB is >140-fold upregulated in presence of SPyCIM1 in the late log phase at 39 °C, a condition mimicking growth in a febrile human (Hendrickson et al. Citation2015). The FCT-region regulator Nra was found to repress a SpyCI element during growth of a M49 strain in EP, ESP and SP (Hendrickson et al. Citation2015). Importantly, a mice chamber model identified several SpyCIM1 putative transcriptional regulators as differentially regulated compared to rich medium (Aziz et al. Citation2010), suggesting a role for these elements in virulence.
Altogether, these results suggest that GAS prophages respond in different ways according to the sites of infection, like the throat and deep tissue. From the phage point of view, induction could be due to damage by the host immune system and an attempt to escape from dying bacteria, like in macrophages and PMNs. Alternatively, the phage could sense bacterial and/or environmental cues favorable for their dissemination, the throat being more promising to find new bacterial host than deep tissue. From the bacterial side, phages induction may contribute to an increase in both phage DNA and expression of virulence genes. Their secretion and/or release after completion of the lytic cycle will then contribute to the infection process. The exact mechanisms that govern GAS prophages induction in vivo are not well understood and require further studies. Additionally, the effect of prophages genes expression on GAS transcriptome, like observed for satellite phages, remains to be determined. The intricate nature of GAS regulatory networks may, however, complicate the study of these biological processes.
Conclusion and perspectives
In the last 10 years, RNAseq technology has largely contributed to the understanding of GAS virulence and metabolism. However, a global understanding of GAS regulation remains a challenge despite the huge quantity of generated data notably because the diversity of environmental conditions in those experimental procedures renders the comparison between studies difficult ( and ). Indeed, numerous differences at the technical (difference in yeast extract percentage in THY, presence or not of CO2, differences in OD regarded as exponential, stationary, or transitional phase…) and analysis (threshold, mapping) points of view could have a huge impact on transcriptomic results. Moreover, data coming from previous studies using microarrays are difficult to compare with current RNAseq data. The use of different strains, even belonging to the same M type, has also shown discordant results. Moreover, regulon identification must be done in the adequate condition, i.e. condition in which the regulator is important. For example, we have seen that the PerR regulon changed completely between basic and stress condition (6 and 232 DEGs respectively) (Brenot et al. Citation2005; Grifantini et al. Citation2011). Another understated aspect of genes regulation is the direct and indirect effect of regulators. The GAS community has recently implemented ChIPseq analyses for CcpA (with HPr∼P) and CovR regulators. These first studies have highlighted that they directly control only 21% and 48% of the differentially regulated genes in the ΔccpA and ΔcovR strain, respectively.
Overall, all these studies seem to be like pieces of a puzzle that cannot yet be entirely assembled. This lack of standardization and crosstalk between research teams hinder scientific progress in understanding the GAS transcriptomic landscape. Initiatives such as transcriptomic compendium of Salmonella RNAseq data performed under 22 infection-relevant conditions (Kröger et al. Citation2013) or PATHOGENEX, an RNA atlas of human pathogen in stress conditions (Avican et al. Citation2021), should be considered in the future (Box 1). Moreover, an integrated analysis of ChIPseq and RNAseq data is still missing. Hirose et al. recently explored the reanalysis of 116 publicly available RNAseq datasets of M1 strains grown in THY medium with an independent component analysis (ICA)-based framework (Hirose et al. Citation2023). They identified 42 iModulons, i.e. independently modulated sets of genes. Comparison between in vitro and in vivo RNAseq datasets allowed them to estimate the stress, like carbohydrate depletion, to which GAS is exposed during necrotizing fasciitis (Hirose et al. Citation2023). In E. coli, the analysis of more than 2600 RNAseq datasets obtained from different teams highlighted the overall similarity of iModulons structure (Lamoureux et al. Citation2023), which could smooth out differences between GAS laboratories.
Animal models which are undoubtedly useful to demonstrate virulence can also bias our interpretation of important genes in vivo (). For example, results obtained in mice versus human show a lot of discrepancies questioning the utility of such models. Ex vivo models with human cells and tissues and, ideally, human samples or models should preferentially be used, especially to conclude on vaccine/therapeutic target expression (Box 1). However, the use of more relevant animal model such as NHPs, has clearly helped in understanding GAS virulence (). In the next few years, transcriptomic data coming from GAS strain used in the human challenge model (Osowicki et al. Citation2019) should become a huge source of clinically relevant information ( and Box 1).
Dual-RNAseq has been successfully applied to GAS infection in non-human primates (Kachroo et al. Citation2020), mouse (Wilkening et al. Citation2023) and human NF biopsies (Thänert et al. Citation2019; Jahagirdar et al. Citation2022). Although out of the scope of this review, this systems biology approach will increase our understanding of the host-GAS interactions, especially if it is coupled with functional studies of deletion mutants (Kachroo et al. Citation2020).
On the other hand, even if uniformity could be reached by some efforts, it seems that, as usual, there is GAS emm-type/strain specificity, especially regarding the regulation by TCS and stand-alone transcriptional factors. Understanding the regulome of each TCS/stand-alone regulator and the cross talk between all these systems in several physiologically relevant conditions is the major challenge for the future. TCS and stand-alone regulators should therefore be investigated in depth by using multiple knock-mutants. Moreover, ChIPseq data need to be generated to decouple direct and indirect targets of regulators. These experimental works could be combined with in silico analysis of intergenic regions in TCS, which could help predict differences between M-types and strains (Buckley et al. Citation2018) (Box 1).
A next breakthrough in the understanding of GAS behavior, especially in precious human samples, could be the analysis of specific pattern of genes expression by single-cell RNAseq (Kuchina et al. Citation2021) and in situ sequencing (Ke et al. Citation2013) (Box 1). Moreover, transcriptomic studies should be coupled to proteomic since RNA and protein data are not always correlated (Chaussee et al. Citation2008; Freiberg et al., Citation2016) and that protein isoforms and/or proteins degradation are also important actors of the regulation (Aziz et al. Citation2004; Cain et al., Citation2014).
Supplemental Material
Download Zip (58.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abdelal AT. 1979. Arginine catabolism by microorganisms. Annu Rev Microbiol. 33(1):139–168. doi:10.1146/annurev.mi.33.100179.001035.
- Aikawa C, Shimizu A, Nakakido M, Murase K, Nozawa T, Tsumoto K, Nakagawa I. 2023. Group A Streptococcus cation diffusion facilitator proteins contribute to immune evasion by regulating intracellular metal concentrations. Biochem Biophys Res Commun. 676:141–148. doi:10.1016/j.bbrc.2023.07.052.
- Akesson P, Rasmussen M, Mascini E, von Pawel-Rammingen U, Janulczyk R, Collin M, Olsen A, Mattsson E, Olsson ML, Bjorck L, et al. 2004. Low antibody levels against cell wall-attached proteins of Streptococcus pyogenes predispose for severe invasive disease. J Infect Dis. 189(5):797–804. doi:10.1086/381982.
- Alam FM, Turner CE, Smith K, Wiles S, Sriskandan S. 2013. Inactivation of the CovR/S virulence regulator impairs infection in an improved murine model of Streptococcus pyogenes naso-pharyngeal infection. PLoS One. 8(4):e61655. doi:10.1371/journal.pone.0061655.
- Almengor AC, Kinkel TL, Day SJ, McIver KS. 2007. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the Group A Streptococcus. J Bacteriol. 189(23):8405–8416. doi:10.1128/JB.01038-07.
- Avican K, Aldahdooh J, Togninalli M, Mahmud AKMF, Tang J, Borgwardt KM, Rhen M, Fällman M. 2021. RNA atlas of human bacterial pathogens uncovers stress dynamics linked to infection. Nat Commun. 12(1):3282. doi:10.1038/s41467-021-23588-w.
- Aziz RK, Kansal R, Aronow BJ, Taylor WL, Rowe SL, Kubal M, Chhatwal GS, Walker MJ, Kotb M. 2010. Microevolution of group A streptococci in vivo: capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence. PLoS One. 5(4):e9798. doi:10.1371/journal.pone.0009798.
- Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol. 51(1):123–134. doi:10.1046/j.1365-2958.2003.03797.x.
- Banks DJ, Lei B, Musser JM. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect Immun. 71(12):7079–7086. doi:10.1128/IAI.71.12.7079-7086.2003.
- Bao Y-J, Liang Z, Mayfield JA, Lee SW, Ploplis VA, Castellino FJ. 2015. CovRS-regulated transcriptome analysis of a hypervirulent M23 strain of group a Streptococcus pyogenes provides new insights into virulence determinants. J Bacteriol. 197(19):3191–3205. doi:10.1128/JB.00511-15.
- Barnett TC, Bowen AC, Carapetis JR. 2018. The fall and rise of Group A Streptococcus diseases. Epidemiol Infect. 147:e4. doi:10.1017/S0950268818002285.
- Barnett TC, Bugrysheva JV, Scott JR. 2007. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J Bacteriol. 189(5):1866–1873. doi:10.1128/JB.01658-06.
- Bates CS, Montañez GE, Woods CR, Vincent RM, Eichenbaum Z. 2003. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect Immun. 71(3):1042–1055. doi:10.1128/IAI.71.3.1042-1055.2003.
- Beckert S, Kreikemeyer B, Podbielski A. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect Immun. 69(1):534–537. doi:10.1128/IAI.69.1.534-537.2001.
- Ben Zakour NL, Davies MR, You Y, Chen JHK, Forde BM, Stanton-Cook M, Yang R, Cui Y, Barnett TC, Venturini C, et al. 2015. Transfer of scarlet fever-associated elements into the group A Streptococcus M1T1 clone. Sci Rep. 5(1):15877. doi:10.1038/srep15877.
- Beres SB, Kachroo P, Nasser W, Olsen RJ, Zhu L, Flores AR, de la Riva I, Paez-Mayorga J, Jimenez FE, Cantu C, et al. 2016. Transcriptome remodeling contributes to epidemic disease caused by the human pathogen Streptococcus pyogenes. MBio. 7(3):e00403–16. doi:10.1128/mBio.00403-16.
- Beres SB, Olsen RJ, Ojeda Saavedra M, Ure R, Reynolds A, Lindsay DSJ, Smith AJ, Musser JM. 2017. Genome sequence analysis of emm89 Streptococcus pyogenes strains causing infections in Scotland, 2010-2016. J Med Microbiol. 66(12):1765–1773. doi:10.1099/jmm.0.000622.
- Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu M-Y, Smoot JC, Porcella SF, Parkins LD, et al. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci U S A. 99(15):10078–10083. doi:10.1073/pnas.152298499.
- Bernard PE, Duarte A, Bogdanov M, Musser JM, Olsen RJ. 2020. Single amino acid replacements in RocA disrupt protein-protein interactions to alter the molecular pathogenesis of group A Streptococcus. Infect Immun. 88(11):e00386–20. doi:10.1128/IAI.00386-20.
- Bernard PE, Kachroo P, Eraso JM, Zhu L, Madry JE, Linson SE, Ojeda Saavedra M, Cantu C, Musser JM, Olsen RJ, et al. 2019. Polymorphisms in regulator of Cov contribute to the molecular pathogenesis of serotype M28 group A Streptococcus. Am J Pathol. 189(10):2002–2018. doi:10.1016/j.ajpath.2019.06.009.
- Bernard PE, Kachroo P, Zhu L, Beres SB, Eraso JM, Kajani Z, Long SW, Musser JM, Olsen RJ. 2018. RocA has serotype-specific gene regulatory and pathogenesis activities in serotype M28 group A Streptococcus. Infect Immun. 86(11):e00467–18. doi:10.1128/IAI.00467-18.
- Beyer-Sehlmeyer G, Kreikemeyer B, Hörster A, Podbielski A. 2005. Analysis of the growth phase-associated transcriptome of Streptococcus pyogenes. Int J Med Microbiol. 295(3):161–177. doi:10.1016/j.ijmm.2005.02.010.
- Biswas I, Scott JR. 2003. Identification of rocA, a positive regulator of covR expression in the group A Streptococcus. J Bacteriol. 185(10):3081–3090. doi:10.1128/JB.185.10.3081-3090.2003.
- Brenot A, King KY, Caparon MG. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol Microbiol. 55(1):221–234. doi:10.1111/j.1365-2958.2004.04370.x.
- Bricker AL, Cywes C, Ashbaugh CD, Wessels MR. 2002. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol Microbiol. 44(1):257–269. doi:10.1046/j.1365-2958.2002.02876.x.
- Broudy TB, Fischetti VA. 2003. In vivo lysogenic conversion of Tox(-) Streptococcus pyogenes to Tox(+) with Lysogenic streptococci or free phage. Infect Immun. 71(7):3782–3786. doi:10.1128/IAI.71.7.3782-3786.2003.
- Broudy TB, Pancholi V, Fischetti VA. 2001. Induction of lysogenic bacteriophage and phage-associated toxin from group a streptococci during coculture with human pharyngeal cells. Infect Immun. 69(3):1440–1443. doi:10.1128/IAI.69.3.1440-1443.2001.
- Buckley SJ, Timms P, Davies MR, McMillan DJ. 2018. In silico characterisation of the two-component system regulators of Streptococcus pyogenes. PLoS One. 13(6):e0199163. doi:10.1371/journal.pone.0199163.
- Bullen JJ. 1981. The significance of iron in infection. Rev Infect Dis. 3(6):1127–1138. doi:10.1093/clinids/3.6.1127.
- Cain JA, Solis N, Cordwell SJ. 2014. Beyond gene expression: the impact of protein post-translational modifications in bacteria. J Proteomics. 97:265–286. doi:10.1016/j.jprot.2013.08.012.
- Calfee G, Danger JL, Jain I, Miller EW, Sarkar P, Tjaden B, Kreikemeyer B, Sumby P. 2018. Identification and characterization of serotype-specific variation in group A Streptococcus pilus expression. Infect Immun. 86(2):e00792–17. doi:10.1128/IAI.00792-17.
- Caparon MG, Scott JR. 1987. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci U S A. 84(23):8677–8681. doi:10.1073/pnas.84.23.8677.
- Chaussee MA, McDowell EJ, Chaussee MS. 2008. Proteomic analysis of proteins secreted by Streptococcus pyogenes. Methods Mol Biol. 431:15–24.
- Chaussee MS, Sylva GL, Sturdevant DE, Smoot LM, Graham MR, Watson RO, Musser JM. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect Immun. 70(2):762–770. doi:10.1128/IAI.70.2.762-770.2002.
- Chiang-Ni C, Chiou H-J, Tseng H-C, Hsu C-Y, Chiu C-H. 2020. RocA regulates phosphatase activity of virulence sensor CovS of group A Streptococcus in growth phase- and pH-dependent manners. mSphere. 5(3):e00361–20. doi:10.1128/mSphere.00361-20.
- Cho KH, Caparon MG. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol Microbiol. 57(6):1545–1556. doi:10.1111/j.1365-2958.2005.04786.x.
- Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, et al. 2006. Trigger for group A streptococcal M1T1 invasive disease. Faseb J. 20(10):1745–1747. doi:10.1096/fj.06-5804fje.
- Commons RJ, Smeesters PR, Proft T, Fraser JD, Robins-Browne R, Curtis N. 2014. Streptococcal superantigens: categorization and clinical associations. Trends Mol Med. 20(1):48–62. doi:10.1016/j.molmed.2013.10.004.
- Cook LCC, Chatterjee N, Li Y, Andrade J, Federle MJ, Eichenbaum Z. 2019. Transcriptomic analysis of Streptococcus pyogenes colonizing the vaginal mucosa identifies hupY, an MtsR-regulated adhesin involved in heme utilization. mBio. 10(3):e00848–19. doi:10.1128/mBio.00848-19.
- Cotter PD, Hill C. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev. 67(3):429–453, table of contents. doi:10.1128/MMBR.67.3.429-453.2003.
- Courtney HS, Hasty DL, Dale JB. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann Med. 34(2):77–87. doi:10.1080/07853890252953464.
- Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 13(3):470–511. doi:10.1128/CMR.13.3.470.
- Cusumano Z, Caparon M. 2013. Adaptive evolution of the Streptococcus pyogenes regulatory aldolase LacD.1. J Bacteriol. 195(6):1294–1304. doi:10.1128/JB.01997-12.
- Cusumano ZT, Caparon MG. 2015. Citrulline protects Streptococcus pyogenes from acid stress using the arginine deiminase pathway and the F1Fo-ATPase. J Bacteriol. 197(7):1288–1296. doi:10.1128/JB.02517-14.
- Davies HC, Karush F, Rudd JH. 1965. Effect of amino acids on steady-state growth of a group A hemolytic Streptococcus. J Bacteriol. 89(2):421–427. doi:10.1128/jb.89.2.421-427.1965.
- DebRoy S, Aliaga-Tobar V, Galvez G, Arora S, Liang X, Horstmann N, Maracaja-Coutinho V, Latorre M, Hook M, Flores AR, et al. 2021. Genome-wide analysis of in vivo CcpA binding with and without its key co-factor HPr in the major human pathogen group A Streptococcus. Mol Microbiol. 115(6):1207–1228. doi:10.1111/mmi.14667.
- DebRoy S, Saldaña M, Travisany D, Montano A, Galloway-Peña J, Horstmann N, Yao H, González M, Maass A, Latorre M, et al. 2016. A multi-serotype approach clarifies the catabolite control protein A regulon in the major human pathogen group a Streptococcus. Sci Rep. 6(1):32442. doi:10.1038/srep32442.
- Dmitriev AV, McDowell EJ, Kappeler KV, Chaussee MA, Rieck LD, Chaussee MS. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J Bacteriol. 188(20):7230–7241. doi:10.1128/JB.00877-06.
- Donald FE, Slack RC, Colman G. 1991. Streptococcus pyogenes vulvovaginitis in children in Nottingham. Epidemiol Infect. 106(3):459–465. doi:10.1017/s0950268800067509.
- Engleberg NC, Heath A, Vardaman K, DiRita VJ. 2004. Contribution of CsrR-regulated virulence factors to the progress and outcome of murine skin infections by Streptococcus pyogenes. Infect Immun. 72(2):623–628. doi:10.1128/IAI.72.2.623-628.2004.
- Eraso JM, Olsen RJ, Beres SB, Kachroo P, Porter AR, Nasser W, Bernard PE, DeLeo FR, Musser JM. 2016. Genomic landscape of intrahost variation in group A Streptococcus: repeated and abundant mutational inactivation of the fabt gene encoding a regulator of fatty acid synthesis. Infect Immun. 84(12):3268–3281. doi:10.1128/IAI.00608-16.
- Federle M. 2012. Pathogenic streptococci speak, but what are they saying? Virulence. 3(1):92–94. doi:10.4161/viru.3.1.18652.
- Feng W, Minor D, Liu M, Li J, Ishaq SL, Yeoman C, Lei B. 2017. Null mutations of group A Streptococcus orphan kinase RocA: selection in mouse infection and comparison with CovS mutations in alteration of in vitro and in vivo protease SpeB expression and virulence. Infect Immun. 85(1):e00790–16. doi:10.1128/IAI.00790-16.
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, et al. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A. 98(8):4658–4663. doi:10.1073/pnas.071559398.
- Fiedler T, Kreikemeyer B, Sugareva V, Redanz S, Arlt R, Standar K, Podbielski A. 2010. Impact of the Streptococcus pyogenes Mga regulator on human matrix protein binding and interaction with eukaryotic cells. Int J Med Microbiol. 300(4):248–258. doi:10.1016/j.ijmm.2009.07.004.
- Fiedler T, Sugareva V, Patenge N, Kreikemeyer B. 2010. Insights into Streptococcus pyogenes pathogenesis from transcriptome studies. Future Microbiol. 5(11):1675–1694. doi:10.2217/fmb.10.128.
- Finn MB, Ramsey KM, Dove SL, Wessels MR. 2021. Identification of group A Streptococcus genes directly regulated by CsrRS and novel intermediate regulators. mBio. 12(4):e0164221. doi:10.1128/mBio.01642-21.
- Flores AR, Jewell BE, Olsen RJ, Shelburne SA, Fittipaldi N, Beres SB, Musser JM. 2014. Asymptomatic carriage of group A Streptococcus is associated with elimination of capsule production. Infect Immun. 82(9):3958–3967. doi:10.1128/IAI.01788-14.
- Flores AR, Jewell BE, Versalovic EM, Olsen RJ, Bachert BA, Lukomski S, Musser JM. 2015. Natural variant of collagen-like protein a in serotype M3 group a Streptococcus increases adherence and decreases invasive potential. Infect Immun. 83(3):1122–1129. doi:10.1128/IAI.02860-14.
- Flores AR, Jewell BE, Yelamanchili D, Olsen RJ, Musser JM. 2015. A single amino acid replacement in the sensor kinase LiaS contributes to a carrier phenotype in group A Streptococcus. Infect Immun. 83(11):4237–4246. doi:10.1128/IAI.00656-15.
- Flores AR, Olsen RJ, Wunsche A, Kumaraswami M, Shelburne SA, Carroll RK, Musser JM. 2013. Natural variation in the promoter of the gene encoding the Mga regulator alters host-pathogen interactions in group a Streptococcus carrier strains. Infect Immun. 81(11):4128–4138. doi:10.1128/IAI.00405-13.
- Freiberg JA, Le Breton Y, Tran BQ, Scott AJ, Harro JM, Ernst RK, Goo YA, Mongodin EF, Goodlett DR, McIver KS, et al. 2016. global analysis and comparison of the transcriptomes and proteomes of group A Streptococcus biofilms. mSystems. 1(6):e00149–16. doi:10.1128/mSystems.00149-16.
- Freiberg JA, Le Breton Y, Tran BQ, Scott AJ, Harro JM, Ernst RK, Goo YA, Mongodin EF, Goodlett DR, McIver KS, et al. 2016. Global analysis and comparison of the transcriptomes and proteomes of group A. mSystems. 1(6):e00149–16. doi:10.1128/mSystems.00149-16.
- Frost HR, Sanderson-Smith M, Walker M, Botteaux A, Smeesters PR. 2018. Group A streptococcal M-like proteins: from pathogenesis to vaccine potential. FEMS Microbiol Rev. 42(2):193–204. doi:10.1093/femsre/fux057.
- Galloway-Peña J, DebRoy S, Brumlow C, Li X, Tran TT, Horstmann N, Yao H, Chen K, Wang F, Pan B-F, et al. 2018. Hypervirulent group A Streptococcus emergence in an acaspular background is associated with marked remodeling of the bacterial cell surface. PLoS One. 13(12):e0207897. doi:10.1371/journal.pone.0207897.
- Gherardi G, Vitali LA, Creti R. 2018. Prevalent emm types among invasive GAS in Europe and North America since year 2000. Front Public Health. 6:59. doi:10.3389/fpubh.2018.00059.
- Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 6(8):613–624. doi:10.1038/nrmicro1932.
- Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. 1997. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 266(4):665–676. doi:10.1006/jmbi.1996.0820.
- Graham MR, Smoot LM, Migliaccio CAL, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM, et al. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A. 99(21):13855–13860. doi:10.1073/pnas.202353699.
- Graham MR, Virtaneva K, Porcella SF, Barry WT, Gowen BB, Johnson CR, Wright FA, Musser JM. 2005. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 166(2):455–465. doi:10.1016/S0002-9440(10)62268-7.
- Green R, Miller J, Crosby W. 1981. Enhancement of iron chelation by desferrioxamine entrapped in red blood cell ghosts. Blood. 57(5):866–872. doi:10.1182/blood.V57.5.866.bloodjournal575866.
- Grifantini R, Toukoki C, Colaprico A, Gryllos I. 2011. Peroxide stimulon and role of PerR in group A Streptococcus. J Bacteriol. 193(23):6539–6551. doi:10.1128/JB.05924-11.
- Gryllos I, Levin JC, Wessels MR. 2003. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+. Proc Natl Acad Sci U S A. 100(7):4227–4232. doi:10.1073/pnas.0636231100.
- Hendrickson C, Euler CW, Nguyen SV, Rahman M, McCullor KA, King CJ, Fischetti VA, McShan WM. 2015. Elimination of chromosomal island SpyCIM1 from Streptococcus pyogenes strain SF370 reverses the mutator phenotype and alters global transcription. PLoS One. 10(12):e0145884. doi:10.1371/journal.pone.0145884.
- Henningham A, Chiarot E, Gillen CM, Cole JN, Rohde M, Fulde M, Ramachandran V, Cork AJ, Hartas J, Magor G, et al. 2012. Conserved anchorless surface proteins as group A streptococcal vaccine candidates. J Mol Med (Berl). 90(10):1197–1207. doi:10.1007/s00109-012-0897-9.
- Hertzén E, Johansson L, Kansal R, Hecht A, Dahesh S, Janos M, Nizet V, Kotb M, Norrby-Teglund A. 2012. Intracellular Streptococcus pyogenes in human macrophages display an altered gene expression profile. PLoS One. 7(4):e35218. doi:10.1371/journal.pone.0035218.
- Hirose Y, Poudel S, Sastry AV, Rychel K, Lamoureux CR, Szubin R, Zielinski DC, Lim HG, Menon ND, Bergsten H, et al. 2023. Elucidation of independently modulated genes in Streptococcus pyogenes reveals carbon sources that control its expression of hemolytic toxins. mSystems. 8(3):e0024723. doi:10.1128/msystems.00247-23.
- Hirose Y, Yamaguchi M, Okuzaki D, Motooka D, Hamamoto H, Hanada T, Sumitomo T, Nakata M, Kawabata S. 2019. Streptococcus pyogenes transcriptome changes in the inflammatory environment of necrotizing fasciitis. Appl Environ Microbiol. 85(21):e01428–19. doi:10.1128/AEM.01428-19.
- Hirose Y, Yamaguchi M, Sumitomo T, Nakata M, Hanada T, Okuzaki D, Motooka D, Mori Y, Kawasaki H, Coady A, et al. 2021. Streptococcus pyogenes upregulates arginine catabolism to exert its pathogenesis on the skin surface. Cell Rep. 34(13):108924. doi:10.1016/j.celrep.2021.108924.
- Hollands A, Pence MA, Timmer AM, Osvath SR, Turnbull L, Whitchurch CB, Walker MJ, Nizet V. 2010. Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A Streptococcus M1T1 clone. J Infect Dis. 202(1):11–19. doi:10.1086/653124.
- Hollingshead SK, Readdy TL, Yung DL, Bessen DE. 1993. Structural heterogeneity of the emm gene cluster in group A streptococci. Mol Microbiol. 8(4):707–717. doi:10.1111/j.1365-2958.1993.tb01614.x.
- Hondorp ER, Hou SC, Hause LL, Gera K, Lee C-E, McIver KS. 2013. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A Streptococcus. Mol Microbiol. 88(6):1176–1193. doi:10.1111/mmi.12250.
- Hondorp ER, McIver KS. 2007. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 66(5):1056–1065. doi:10.1111/j.1365-2958.2007.06006.x.
- Horstmann N, Myers KS, Tran CN, Flores AR, Shelburne Iii SA. 2022. CovS inactivation reduces CovR promoter binding at diverse virulence factor encoding genes in group A Streptococcus. PLoS Pathog. 18(2):e1010341. doi:10.1371/journal.ppat.1010341.
- Horstmann N, Sahasrabhojane P, Saldaña M, Ajami NJ, Flores AR, Sumby P, Liu C-G, Yao H, Su X, Thompson E, et al. 2015. Characterization of the effect of the histidine kinase CovS on response regulator phosphorylation in group A Streptococcus. Infect Immun. 83(3):1068–1077. doi:10.1128/IAI.02659-14.
- Horstmann N, Tran CN, Brumlow C, DebRoy S, Yao H, Nogueras Gonzalez G, Makthal N, Kumaraswami M, Shelburne SA. 2018. Phosphatase activity of the control of virulence sensor kinase CovS is critical for the pathogenesis of group A Streptococcus. PLoS Pathog. 14(10):e1007354. doi:10.1371/journal.ppat.1007354.
- Horstmann N, Tran CN, Flores AR, Shelburne SA. 2023. Hyperphosphorylation of the group A streptococcal control of virulence regulator increases promoter occupancy specifically at virulence factor-encoding genes. J Bacteriol. 205(6):e0011823. doi:10.1128/jb.00118-23.
- Jahagirdar S, Morris L, Benis N, Oppegaard O, Svenson M, Hyldegaard O, Skrede S, Norrby-Teglund A, Martins Dos Santos VAP, Saccenti E, et al. 2022. Analysis of host-pathogen gene association networks reveals patient-specific response to streptococcal and polymicrobial necrotising soft tissue infections. BMC Med. 20(1):173. doi:10.1186/s12916-022-02355-8.
- Jain I, Danger JL, Burgess C, Uppal T, Sumby P. 2020. The group A Streptococcus accessory protein RocA: regulatory activity, interacting partners and influence on disease potential. Mol Microbiol. 113(1):190–207. doi:10.1111/mmi.14410.
- Jain I, Miller EW, Danger JL, Pflughoeft KJ, Sumby P. 2017. RocA is an accessory protein to the virulence-regulating CovRS two-component system in group A Streptococcus. Infect Immun. 85(11):e00274–17. doi:10.1128/IAI.00274-17.
- Jimenez JC, Federle MJ. 2014. Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol. 4:127. doi:10.3389/fcimb.2014.00127.
- Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol. 188(14):5153–5166. doi:10.1128/JB.00310-06.
- Jordan S, Rietkötter E, Strauch MA, Kalamorz F, Butcher BG, Helmann JD, Mascher T. 2007. LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology (Reading). 153(Pt 8):2530–2540. doi:10.1099/mic.0.2007/006817-0.
- Kachroo P, Eraso JM, Beres SB, Olsen RJ, Zhu L, Nasser W, Bernard PE, Cantu CC, Saavedra MO, Arredondo MJ, et al. 2019. Integrated analysis of population genomics, transcriptomics and virulence provides novel insights into Streptococcus pyogenes pathogenesis. Nat Genet. 51(3):548–559. doi:10.1038/s41588-018-0343-1.
- Kachroo P, Eraso JM, Olsen RJ, Zhu L, Kubiak SL, Pruitt L, Yerramilli P, Cantu CC, Ojeda Saavedra M, Pensar J, et al. 2020. New pathogenesis mechanisms and translational leads identified by multidimensional analysis of necrotizing myositis in primates. mBio. 11(1):e03363–19. doi:10.1128/mBio.03363-19.
- Kazmi SU, Kansal R, Aziz RK, Hooshdaran M, Norrby-Teglund A, Low DE, Halim AB, Kotb M. 2001. Reciprocal, temporal expression of SpeA and SpeB by invasive M1T1 group a streptococcal isolates in vivo. Infect Immun. 69(8):4988–4995. doi:10.1128/IAI.69.8.4988-4995.2001.
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. 2013. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 10(9):857–860. doi:10.1038/nmeth.2563.
- Kietzman CC, Caparon MG. 2010. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect Immun. 78(1):241–252. doi:10.1128/IAI.00746-09.
- Kietzman CC, Caparon MG. 2011. Distinct time-resolved roles for two catabolite-sensing pathways during Streptococcus pyogenes infection. Infect Immun. 79(2):812–821. doi:10.1128/IAI.01026-10.
- Kihlberg BM, Cooney J, Caparon MG, Olsén A, Björck L. 1995. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in Mga. Microb Pathog. 19(5):299–315. doi:10.1016/s0882-4010(96)80003-9.
- Kim JH, Voskuil MI, Chambliss GH. 1998. NADP, corepressor for the Bacillus catabolite control protein CcpA. Proc Natl Acad Sci U S A. 95(16):9590–9595. doi:10.1073/pnas.95.16.9590.
- Kinkel TL, McIver KS. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect Immun. 76(8):3451–3463. doi:10.1128/IAI.00343-08.
- Kluger MJ, Rothenburg BA. 1979. Fever and reduced iron: their interaction as a host defense response to bacterial infection. Science. 203(4378):374–376. doi:10.1126/science.760197.
- Kreikemeyer B, Klenk M, Podbielski A. 2004. The intracellular status of Streptococcus pyogenes: role of extracellular matrix-binding proteins and their regulation. Int J Med Microbiol. 294(2-3):177–188. doi:10.1016/j.ijmm.2004.06.017.
- Kreikemeyer B, McIver KS, Podbielski A. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11(5):224–232. doi:10.1016/s0966-842x(03)00098-2.
- Kreikemeyer B, Nakata M, Köller T, Hildisch H, Kourakos V, Standar K, Kawabata S, Glocker MO, Podbielski A. 2007. The Streptococcus pyogenes serotype M49 Nra-Ralp3 transcriptional regulatory network and its control of virulence factor expression from the novel eno ralp3 epf sagA pathogenicity region. Infect Immun. 75(12):5698–5710. doi:10.1128/IAI.00175-07.
- Kreth J, Chen Z, Ferretti J, Malke H. 2011. Counteractive balancing of transcriptome expression involving CodY and CovRS in Streptococcus pyogenes. J Bacteriol. 193(16):4153–4165. doi:10.1128/JB.00061-11.
- Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, et al. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe. 14(6):683–695. doi:10.1016/j.chom.2013.11.010.
- Kubo A, Ishizaki I, Kubo A, Kawasaki H, Nagao K, Ohashi Y, Amagai M. 2013. The stratum corneum comprises three layers with distinct metal-ion barrier properties. Sci Rep. 3(1):1731. doi:10.1038/srep01731.
- Kuchina A, Brettner LM, Paleologu L, Roco CM, Rosenberg AB, Carignano A, Kibler R, Hirano M, DePaolo RW, Seelig G, et al. 2021. Microbial single-cell RNA sequencing by split-pool barcoding. Science. 371(6531):eaba5257. doi:10.1126/science.aba5257.
- Lamagni TL, Darenberg J, Luca-Harari B, Siljander T, Efstratiou A, Henriques-Normark B, Vuopio-Varkila J, Bouvet A, Creti R, Ekelund K, et al. 2008. Epidemiology of severe Streptococcus pyogenes disease in Europe. J Clin Microbiol. 46(7):2359–2367. doi:10.1128/JCM.00422-08.
- Lamoureux CR, Decker KT, Sastry AV, Rychel K, Gao Y, McConn JL, Zielinski DC, Palsson BO. 2023. A multi-scale expression and regulation knowledge base for Escherichia coli. Nucleic Acids Res. 51(19):10176–10193. doi:10.1093/nar/gkad750.
- Lee CM, Jin S-P, Doh EJ, Lee DH, Chung JH. 2019. Regional variation of human skin surface temperature. Ann Dermatol. 31(3):349–352. doi:10.5021/ad.2019.31.3.349.
- Lembke C, Podbielski A, Hidalgo-Grass C, Jonas L, Hanski E, Kreikemeyer B. 2006. Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl Environ Microbiol. 72(4):2864–2875. doi:10.1128/AEM.72.4.2864-2875.2006.
- Li J, Liu G, Feng W, Zhou Y, Liu M, Wiley JA, Lei B. 2014. Neutrophils select hypervirulent CovRS mutants of M1T1 group A Streptococcus during subcutaneous infection of mice. Infect Immun. 82(4):1579–1590. doi:10.1128/IAI.01458-13.
- Lindemann J, Leiacker R, Rettinger G, Keck T. 2002. Nasal mucosal temperature during respiration. Clin Otolaryngol Allied Sci. 27(3):135–139. doi:10.1046/j.1365-2273.2002.00544.x.
- Lin Y, Sanson MA, Vega LA, Shah B, Regmi S, Cubria MB, Flores AR. 2020. ExPortal and the LiaFSR regulatory system coordinate the response to cell membrane stress in Streptococcus pyogenes. mBio. 11(5):e01804–20. doi:10.1128/mBio.01804-20.
- Liu G, Feng W, Li D, Liu M, Nelson DC, Lei B. 2015. The Mga regulon but not deoxyribonuclease Sda1 of invasive M1T1 group A Streptococcus contributes to in vivo selection of CovRS mutations and resistance to innate immune killing mechanisms. Infect Immun. 83(11):4293–4303. doi:10.1128/IAI.00857-15.
- Livezey J, Perez L, Suciu D, Yu X, Robinson B, Bush D, Merrill G. 2011. Analysis of group A Streptococcus gene expression in humans with pharyngitis using a microarray. J Med Microbiol. 60(Pt 12):1725–1733. doi:10.1099/jmm.0.022939-0.
- Li J, Zhu H, Feng W, Liu M, Song Y, Zhang X, Zhou Y, Bei W, Lei B. 2013. Regulation of inhibition of neutrophil infiltration by the two-component regulatory system CovRS in subcutaneous murine infection with group A streptococcus. Infect Immun. 81(3):974–983. doi:10.1128/IAI.01218-12.
- Loh JMS, Rivera-Hernandez T, McGregor R, Khemlani AHJ, Tay ML, Cork AJ, M Raynes J, Moreland NJ, Walker MJ, Proft T, et al. 2021. A multivalent T-antigen-based vaccine for Group A Streptococcus. Sci Rep. 11(1):4353. doi:10.1038/s41598-021-83673-4.
- Loughman JA, Caparon MG. 2006. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. Embo J. 25(22):5414–5422. doi:10.1038/sj.emboj.7601393.
- Lynskey NN, Goulding D, Gierula M, Turner CE, Dougan G, Edwards RJ, Sriskandan S. 2013. RocA truncation underpins hyper-encapsulation, carriage longevity and transmissibility of serotype M18 group A streptococci. PLoS Pathog. 9(12):e1003842. doi:10.1371/journal.ppat.1003842.
- Lynskey NN, Turner CE, Heng LS, Sriskandan S. 2015. A truncation in the regulator RocA underlies heightened capsule expression in serotype M3 group A streptococci. Infect Immun. 83(4):1732–1733. doi:10.1128/IAI.02892-14.
- Lynskey NN, Velarde JJ, Finn MB, Dove SL, Wessels MR. 2019. RocA binds CsrS to modulate CsrRS-mediated gene regulation in group A Streptococcus. mBio. 10(4):e01495–19. doi:10.1128/mBio.01495-19.
- Madden JC, Ruiz N, Caparon M. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 104(1):143–152. doi:10.1016/s0092-8674(01)00198-2.
- Makthal N, Do H, Wendel BM, Olsen RJ, Helmann JD, Musser JM, Kumaraswami M. 2020. Group A Streptococcus AdcR regulon participates in bacterial defense against host-mediated zinc sequestration and contributes to virulence. Infect Immun. 88(8):e00097–20. doi:10.1128/IAI.00097-20.
- Makthal N, Gavagan M, Do H, Olsen RJ, Musser JM, Kumaraswami M. 2016. Structural and functional analysis of RopB: a major virulence regulator in Streptococcus pyogenes. Mol Microbiol. 99(6):1119–1133. doi:10.1111/mmi.13294.
- Malke H, Steiner K, McShan WM, Ferretti JJ. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int J Med Microbiol. 296(4-5):259–275. doi:10.1016/j.ijmm.2005.11.008.
- Mayfield JA, Liang Z, Agrahari G, Lee SW, Donahue DL, Ploplis VA, Castellino FJ. 2014. Mutations in the control of virulence sensor gene from Streptococcus pyogenes after infection in mice lead to clonal bacterial variants with altered gene regulatory activity and virulence. PLoS One. 9(6):e100698. doi:10.1371/journal.pone.0100698.
- McShan WM, Nguyen SV. 2022. The bacteriophages of Streptococcus pyogenes. In: JJ Ferretti, DL Stevens, VA Fischetti, editors. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City (OK): University of Oklahoma Health Sciences Center.
- Miller EW, Danger JL, Ramalinga AB, Horstmann N, Shelburne SA, Sumby P. 2015. Regulatory rewiring confers serotype-specific hyper-virulence in the human pathogen group A Streptococcus. Mol Microbiol. 98(3):473–489. doi:10.1111/mmi.13136.
- Miyoshi-Akiyama T, Ikebe T, Watanabe H, Uchiyama T, Kirikae T, Kawamura Y. 2006. Use of DNA arrays to identify a mutation in the negative regulator, csrR, responsible for the high virulence of a naturally occurring type M3 group A Streptococcus clinical isolate. J Infect Dis. 193(12):1677–1684. doi:10.1086/504263.
- Nanda AM, Thormann K, Frunzke J. 2015. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J Bacteriol. 197(3):410–419. doi:10.1128/JB.02230-14.
- Ong CL, Walker MJ, McEwan AG. 2015. Zinc disrupts central carbon metabolism and capsule biosynthesis in Streptococcus pyogenes. Sci Rep. 5(1):10799. doi:10.1038/srep10799.
- Osowicki J, Azzopardi KI, Baker C, Waddington CS, Pandey M, Schuster T, Grobler A, Cheng AC, Pollard AJ, McCarthy JS, et al. 2019. Controlled human infection for vaccination against Streptococcus pyogenes (CHIVAS): Establishing a group A Streptococcus pharyngitis human infection study. Vaccine. 37(26):3485–3494. doi:10.1016/j.vaccine.2019.03.059.
- Osowicki J, Azzopardi KI, Fabri L, Frost HR, Rivera-Hernandez T, Neeland MR, Whitcombe AL, Grobler A, Gutman SJ, Baker C, et al. 2021. A controlled human infection model of Streptococcus pyogenes pharyngitis (CHIVAS-M75): an observational, dose-finding study. Lancet Microbe. 2(7):e291–e299. doi:10.1016/S2666-5247(20)30240-8.
- Pancholi V, Caparon M. 2016. Streptococcus pyogenes metabolism. In: JJ Ferretti, DL Stevens, VA Fischetti, editors. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City (OK): University of Oklahoma Health Sciences Center.
- Port GC, Cusumano ZT, Tumminello PR, Caparon MG. 2017. SpxA1 and SpxA2 act coordinately to fine-tune stress responses and virulence in Streptococcus pyogenes. mBio. 8(2):e00288–17. doi:10.1128/mBio.00288-17.
- Rasmussen M, Müller HP, Björck L. 1999. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial surface by binding alpha2-macroglobulin. J Biol Chem. 274(22):15336–15344. doi:10.1074/jbc.274.22.15336.
- Remmington A, Turner CE. 2018. The DNases of pathogenic Lancefield streptococci. Microbiology (Reading). 164(3):242–250. doi:10.1099/mic.0.000612.
- Riani C, Standar K, Srimuang S, Lembke C, Kreikemeyer B, Podbielski A. 2007. Transcriptome analyses extend understanding of Streptococcus pyogenes regulatory mechanisms and behavior toward immunomodulatory substances. Int J Med Microbiol. 297(7-8):513–523. doi:10.1016/j.ijmm.2007.04.005.
- Ribardo DA, Lambert TJ, McIver KS. 2004. Role of Streptococcus pyogenes two-component response regulators in the temporal control of Mga and the Mga-regulated virulence gene emm. Infect Immun. 72(6):3668–3673. doi:10.1128/IAI.72.6.3668-3673.2004.
- Ribardo DA, McIver KS. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A Streptococcus. Mol Microbiol. 62(2):491–508. doi:10.1111/j.1365-2958.2006.05381.x.
- Roberts SA, Churchward GG, Scott JR. 2007. Unraveling the regulatory network in Streptococcus pyogenes: the global response regulator CovR represses rivR directly. J Bacteriol. 189(4):1459–1463. doi:10.1128/JB.01026-06.
- Roshika R, Jain I, Glenaldo T, Sickler T, Musser JM, Sumby P. 2022. Characterization of M-type-specific pilus expression in group A Streptococcus. J Bacteriol. 204(11):e0027022. doi:10.1128/jb.00270-22.
- Rued BE, Anderson CM, Federle MJ. 2022. The proteomic and transcriptomic landscapes altered by Rgg2/3 activity in Streptococcus pyogenes. J Bacteriol. 204(11):e0017522. doi:10.1128/jb.00175-22.
- Sanderson-Smith M, De Oliveira DMP, Guglielmini J, McMillan DJ, Vu T, Holien JK, Henningham A, Steer AC, Bessen DE, Dale JB, et al. 2014. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 210(8):1325–1338. doi:10.1093/infdis/jiu260.
- Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. 2015. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 43(1):418–432. doi:10.1093/nar/gku1304.
- Sanson M, O’Neill BE, Kachroo P, Anderson JR, Flores AR, Valson C, Cantu CC, Makthal N, Karmonik C, Fittipaldi N, et al. 2015. A naturally occurring single amino acid replacement in multiple gene regulator of group A Streptococcus significantly increases virulence. Am J Pathol. 185(2):462–471. doi:10.1016/j.ajpath.2014.10.018.
- Sanson MA, Vega LA, Shah B, Regmi S, Cubria MB, Horstmann N, Shelburne SA, Flores AR. 2021. The LiaFSR transcriptome reveals an interconnected regulatory network in group A Streptococcus. Infect Immun. 89(11):e0021521. doi:10.1128/IAI.00215-21.
- Sarkar P, Danger JL, Jain I, Meadows LA, Beam C, Medicielo J, Burgess C, Musser JM, Sumby P. 2018. Phenotypic variation in the group A Streptococcus due to natural mutation of the accessory protein-encoding gene rocA. mSphere. 3(5):e00519–18. doi:10.1128/mSphere.00519-18.
- Sarkar P, Sumby P. 2017. Regulatory gene mutation: a driving force behind group A Streptococcus strain- and serotype-specific variation. Mol Microbiol. 103(4):576–589. doi:10.1111/mmi.13584.
- Schellhorn HE. 2020. Function, evolution, and composition of the RpoS regulon in Escherichia coli. Front Microbiol. 11:560099. doi:10.3389/fmicb.2020.560099.
- Shelburne SA, Granville C, Tokuyama M, Sitkiewicz I, Patel P, Musser JM.3rd 2005. Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun. 73(8):4723–4731., doi: 10.1128/IAI.73.8.4723-4731.2005.
- Shelburne SA, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 105(5):1698–1703. doi:10.1073/pnas.0711767105.
- Shelburne SA, Olsen RJ, Suber B, Sahasrabhojane P, Sumby P, Brennan RG, Musser JM. 2010. A combination of independent transcriptional regulators shapes bacterial virulence gene expression during infection. PLoS Pathog. 6(3):e1000817. doi:10.1371/journal.ppat.1000817.
- Shelburne SA, Sumby P, Sitkiewicz I, Granville C, DeLeo FR, Musser JM3rd. 2005. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci U S A. 102(44):16037–16042., doi: 10.1073/pnas.0505839102.
- Siemens N, Chakrakodi B, Shambat SM, Morgan M, Bergsten H, Hyldegaard O, Skrede S, Arnell P, Madsen MB, Johansson L, et al. 2016. Biofilm in group A streptococcal necrotizing soft tissue infections. JCI Insight. 1(10):e87882. doi:10.1172/jci.insight.87882.
- Siemens N, Fiedler T, Normann J, Klein J, Münch R, Patenge N, Kreikemeyer B. 2012. Effects of the ERES pathogenicity region regulator Ralp3 on Streptococcus pyogenes serotype M49 virulence factor expression. J Bacteriol. 194(14):3618–3626. doi:10.1128/JB.00227-12.
- Siemens N, Lütticken R. 2021. Streptococcus pyogenes ("group A Streptococcus"), a highly adapted human pathogen-potential implications of its virulence regulation for epidemiology and disease management. Pathogens. 10(6):776. doi:10.3390/pathogens10060776.
- Slade HD. 1954. The effect of fluoride on the arsenolysis of citrulline by soluble enzymes of streptococci. Biochim Biophys Acta. 15(3):411–414. doi:10.1016/0006-3002(54)90044-x.
- Smeesters PR, McMillan DJ, Sriprakash KS. 2010. The streptococcal M protein: a highly versatile molecule. Trends Microbiol. 18(6):275–282. doi:10.1016/j.tim.2010.02.007.
- Smoot JC, Korgenski EK, Daly JA, Veasy LG, Musser JM. 2002. Molecular analysis of group A Streptococcus type emm18 isolates temporally associated with acute rheumatic fever outbreaks in Salt Lake City, Utah. J Clin Microbiol. 40(5):1805–1810. doi:10.1128/JCM.40.5.1805-1810.2002.
- Smoot LM, Smoot JC, Graham MR, Somerville GA, Sturdevant DE, Migliaccio CA, Sylva GL, Musser JM. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc Natl Acad Sci U S A. 98(18):10416–10421. doi:10.1073/pnas.191267598.
- Staali L, Bauer S, Mörgelin M, Björck L, Tapper H. 2006. Streptococcus pyogenes bacteria modulate membrane traffic in human neutrophils and selectively inhibit azurophilic granule fusion with phagosomes. Cell Microbiol. 8(4):690–703. doi:10.1111/j.1462-5822.2005.00662.x.
- Stålhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. 1999. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol Microbiol. 33(1):208–219. doi:10.1046/j.1365-2958.1999.01470.x.
- Stricker T, Navratil F, Sennhauser FH. 2003. Vulvovaginitis in prepubertal girls. Arch Dis Child. 88(4):324–326. doi:10.1136/adc.88.4.324.
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2(1):e5. doi:10.1371/journal.ppat.0020005.
- Sylvestre J-P, Bouissou CC, Guy RH, Delgado-Charro MB. 2010. Extraction and quantification of amino acids in human stratum corneum in vivo. Br J Dermatol. 163(3):458–465. doi:10.1111/j.1365-2133.2010.09805.x.
- Terao Y, Kawabata S, Kunitomo E, Murakami J, Nakagawa I, Hamada S. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol Microbiol. 42(1):75–86. doi:10.1046/j.1365-2958.2001.02579.x.
- Thänert R, Itzek A, Hoßmann J, Hamisch D, Madsen MB, Hyldegaard O, Skrede S, Bruun T, Norrby-Teglund A, Oppegaard O, et al. 2019. Molecular profiling of tissue biopsies reveals unique signatures associated with streptococcal necrotizing soft tissue infections. Nat Commun. 10(1):3846. doi:10.1038/s41467-019-11722-8.
- Toukoki C, Gold KM, McIver KS, Eichenbaum Z. 2010. MtsR is a dual regulator that controls virulence genes and metabolic functions in addition to metal homeostasis in the group A Streptococcus. Mol Microbiol. 76(4):971–989. doi:10.1111/j.1365-2958.2010.07157.x.
- Tran-Winkler HJ, Love JF, Gryllos I, Wessels MR. 2011. Signal transduction through CsrRS confers an invasive phenotype in group A Streptococcus. PLoS Pathog. 7(10):e1002361. doi:10.1371/journal.ppat.1002361.
- Treviño J, Perez N, Ramirez-Peña E, Liu Z, Shelburne SA, Musser JM, Sumby P. 2009. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun. 77(8):3141–3149. doi:10.1128/IAI.01560-08.
- Turner CE, Abbott J, Lamagni T, Holden MTG, David S, Jones MD, Game L, Efstratiou A, Sriskandan S. 2015. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. MBio. 6(4):e00622. doi:10.1128/mBio.00622-15.
- Turner AG, Djoko KY, Ong C-LY, Barnett TC, Walker MJ, McEwan AG. 2019. Group A Streptococcus co-ordinates manganese import and iron efflux in response to hydrogen peroxide stress. Biochem J. 476(3):595–611. doi:10.1042/BCJ20180902.
- Turner AG, Ong C-LY, Djoko KY, West NP, Davies MR, McEwan AG, Walker MJ. 2017. The PerR-regulated P1B-4-type ATPase (PmtA) acts as a ferrous iron efflux pump in Streptococcus pyogenes. Infect Immun. 85(6):e00140–17. doi:10.1128/IAI.00140-17.
- Turner AG, Ong C-LY, Gillen CM, Davies MR, West NP, McEwan AG, Walker MJ. 2015. Manganese homeostasis in group A Streptococcus is critical for resistance to oxidative stress and virulence. mBio. 6(2):e00278–15. doi:10.1128/mBio.00278-15.
- Valdes KM, Sundar GS, Belew AT, Islam E, El-Sayed NM, Le Breton Y, McIver KS. 2018. Glucose levels alter the Mga virulence regulon in the group A Streptococcus. Sci Rep. 8(1):4971. doi:10.1038/s41598-018-23366-7.
- VanderWal AR, Makthal N, Pinochet-Barros A, Helmann JD, Olsen RJ, Kumaraswami M. 2017. Iron efflux by PmtA is critical for oxidative stress resistance and contributes significantly to group A Streptococcus virulence. Infect Immun. 85(6):e00091–17. doi:10.1128/IAI.00091-17.
- Vega LA, Malke H, McIver KS. 2016. Virulence-related transcriptional regulators of Streptococcus pyogenes. In : JJ Ferretti, DL Stevens, VA Fischetti, editors. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City (OK): University of Oklahoma Health Sciences Center.
- Virtaneva K, Graham MR, Porcella SF, Hoe NP, Su H, Graviss EA, Gardner TJ, Allison JE, Lemon WJ, Bailey JR, et al. 2003. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 71(4):2199–2207. doi:10.1128/IAI.71.4.2199-2207.2003.
- Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, Parkins LD, Romero RA, Corn GJ, et al. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci U S A. 102(25):9014–9019. doi:10.1073/pnas.0503671102.
- Voyich JM, Braughton KR, Sturdevant DE, Vuong C, Kobayashi SD, Porcella SF, Otto M, Musser JM, DeLeo FR. 2004. Engagement of the pathogen survival response used by group A Streptococcus to avert destruction by innate host defense. J Immunol. 173(2):1194–1201. doi:10.4049/jimmunol.173.2.1194.
- Voyich JM, Sturdevant DE, Braughton KR, Kobayashi SD, Lei B, Virtaneva K, Dorward DW, Musser JM, DeLeo FR. 2003. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc Natl Acad Sci U S A. 100(4):1996–2001. doi:10.1073/pnas.0337370100.
- Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, et al. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 13(8):981–985. doi:10.1038/nm1612.
- Weckel A, Ahamada D, Bellais S, Méhats C, Plainvert C, Longo M, Poyart C, Fouet A. 2018. The N-terminal domain of the R28 protein promotes emm28 group A Streptococcus adhesion to host cells via direct binding to three integrins. J Biol Chem. 293(41):16006–16018. doi:10.1074/jbc.RA118.004134.
- Wilkening RV, Federle MJ. 2017. Evolutionary constraints shaping Streptococcus pyogenes-host interactions. Trends Microbiol. 25(7):562–572. doi:10.1016/j.tim.2017.01.007.
- Wilkening RV, Langouët-Astrié C, Severn MM, Federle MJ, Horswill AR. 2023. Identifying genetic determinants of Streptococcus pyogenes-host interactions in a murine intact skin infection model. Cell Rep. 42(11):113332. doi:10.1016/j.celrep.2023.113332.
- Wood DN, Weinstein KE, Podbielski A, Kreikemeyer B, Gaughan JP, Valentine S, Buttaro BA. 2009. Generation of metabolically diverse strains of Streptococcus pyogenes during survival in stationary phase. J Bacteriol. 191(20):6242–6252. doi:10.1128/JB.00440-09.
- Yin W, et al. 2019. Biofilms: the microbial "protective clothing" in extreme environments. Int J Mol Sci. 20(14):3423.
- Zhang Y, Okada R, Isaka M, Tatsuno I, Isobe K-I, Hasegawa T. 2015. Analysis of the roles of NrdR and DnaB from Streptococcus pyogenes in response to host defense. APMIS. 123(3):252–259. doi:10.1111/apm.12340.
- Zhu L, Olsen RJ, Horstmann N, Shelburne SA, Fan J, Hu Y, Musser JM. 2016. Intergenic variable-number tandem-repeat polymorphism upstream of rocA alters toxin production and enhances virulence in Streptococcus pyogenes. Infect Immun. 84(7):2086–2093. doi:10.1128/IAI.00258-16.
- Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, Gottfredsson M, Porter AR, DeLeo FR, Musser JM, et al. 2015. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest. 125(9):3545–3559. doi:10.1172/JCI82478.
- Zutkis AA, Anbalagan S, Chaussee MS, Dmitriev AV. 2014. Inactivation of the Rgg2 transcriptional regulator ablates the virulence of Streptococcus pyogenes. PLoS One. 9(12):e114784. doi:10.1371/journal.pone.0114784.