Abstract
In early-life, the gut microbiota is highly modifiable, being modulated by external factors such as maternal microbiota, mode of delivery, and feeding strategies. The composition of the child’s gut microbiota will deeply impact the development and maturation of its immune system, with consequences for future health. As one of the main sources of microorganisms to the child, the mother represents a crucial factor in the establishment of early-life microbiota, impacting the infant’s wellbeing. Recent studies have proposed that dysbiotic maternal gut microbiota could be transmitted to the offspring, influencing the development of its immunity, and leading to the development of diseases such as obesity. This paper aims to review recent findings in gut microbiota and immune system interaction in early-life, highlighting the benefits of a balanced gut microbiota in the regulation of the immune system.
1. Gut microbiota and early-life development
Humans are heavily colonized by different microbial communities, which constitute the human microbiota. These collections of microorganisms can be encountered in different locations of the human body, as skin, nasal cavity, urogenital tract, oral cavity, and gut, where microbial density and diversity are higher (Amabebe et al. Citation2020). The gut microbiota is composed of commensal bacteria, fungi, viruses, and protozoa, which establish a symbiotic relationship with the host, as well as opportunistic microorganisms that may become pathogenic if a disruption in the intestinal microbial community occurs (Milani et al. Citation2017). The intestinal microbiome is dynamic, suffering alterations due to several host-related factors, such as age, health status, diet, antibiotics intake, and probiotics/prebiotics consumption (Singh et al. Citation2021).
The gut microbiota exerts a crucial role in the maintenance of homeostasis and the overall body balance, being involved in numerous physiological processes, such as 1) breakdown of unabsorbed carbohydrates from digestion; 2) synthesis of beneficial microbial metabolites and vitamins as short-chain fatty acids (SCFA) and vitamin K; 3) mood regulation through the gut-brain-axis, by the production of tryptophan, a serotonin precursor, and other mechanisms; 4) protection against external pathogens by the competition with adhesion sites and/or release of antimicrobials; and 5) immunoregulation, meaning modulation of the immune response to internal or external stimuli (Singh et al. Citation2021; Ruan et al. Citation2020). In fact, the stimulation and regulation of the immune system are two of the most important functions executed by the gut microbiota. The gut microbiota and the immune system are closely related, and any disturbance to the delicate balance of intestinal microbial communities may lead to the development of diseases such as allergies, metabolic and gastrointestinal (GI) disorders like inflammatory bowel disease (Singh et al. Citation2021). This intimate link between gut microbes and immunity begins early in life, with the initial colonization of the GI tract of the infant. It was stated that the colonization of the GI tract starts at birth, during the passage through the birth canal, and further continues after birth according to the “sterile womb” theory (Escherich Citation1988; de Goffau et al. Citation2019). However, this topic remains controversial, since some studies have detected microbial particles, through 16S rRNA sequencing, in the umbilical cord blood, placenta, and meconium from healthy pregnancies (without reporting any infections), suggesting that microbial acquisition rather begins in the utero (Senn et al. Citation2020; Payne and Bayatibojakhi Citation2014; Aagaard et al. Citation2014; Rautava et al. Citation2012). Nevertheless, both theories (the “sterile womb” theory and the theory that suggests in-utero colonization) highlight the important role the maternal microbiome plays as the main source of microorganisms for the offspring in early-life, influencing the initial microbial colonization patterns, through the vertical transmission of microbiota.
At birth, the gut of the infant is rapidly colonized by a wide array of microbes, mainly by facultative anaerobes, such as Staphylococcus spp., Streptococcus spp., Enterobacter spp., and other members of the Enterobacteriaceae family (Lin et al. Citation2022; Martin et al. Citation2016). Although these microorganisms may perform aerobic and anaerobic respiration as well fermentation, normally they prefer aerobic respiration when oxygen is available due to its high energetic gain. Therefore, their presence leads to a gradual depletion of the intestinal oxygen and reducing intestinal oxidation-reduction potential within 48 h of birth, which facilitates the colonization of strict anaerobic bacteria such as Bifidobacterium, Clostridia, and Bacteroides (Lin et al. Citation2022). Moreover, it is believed that microbial colonization patterns are heavily influenced by the mode of delivery. For instance, vaginally delivered infants will be exposed to the vaginal and intestinal microbiota of the mother, essentially composed of Lactobacillus and Prevotella. Infants born through this route normally exhibit a more diverse gut microbiota (Milani et al. Citation2017; Martin et al. Citation2016). Concerning children born by cesarean, the microorganisms from the skin of the mother and hospital environment promote the initial exposure, presenting different colonization patterns in comparison to vaginally delivered infants. In this case, the gut microbiota of the infant will be colonized by Staphylococcus and Clostridium, and it is characterized by low microbial diversity and richness (Milani et al. Citation2017; Martin et al. Citation2016). Furthermore, cesarean-delivered infants experienced a delay in colonization by Bifidobacterium (Isolauri Citation2012).
Due to its plastic and dynamic characteristics, early-life microbiota can be altered or modulated by external factors, namely early-life diet (breastfeeding or formula) (Chichlowski et al. Citation2023; Pärnänen et al. Citation2022), gestational age at birth (Sim et al. Citation2023), and maternal health or habits (Jones et al. Citation2024; Ren et al. Citation2023).
Early-life gut microbiota can differ between term and preterm infants. Compared to term infants, preterm newborns exhibit lower microbial diversity, and experience a delay in the establishment of a healthy microbiota, due to the overgrowth of certain bacterial species, such as Enterococcus (commonly associated with nosocomial infections, and consistent with a hospital stay) (Korpela et al. Citation2018; Rougé et al. Citation2010). It is also known that preterm babies are at higher risk of developing complications, often requiring invasive medical interventions, antibiotic administration, and a stay at the Neonatal Intensive Care Unit (NICU) (Henderickx et al. Citation2019). A study by Chi et al. (Citation2019) found that the gut microbiota of preterm infants was enriched in Klebsiella, a microorganism often implicated in hospital infections, that may be transmitted through medical procedures [22]. Following the line of reasoning that implicates early-life clinical care and hospitalization in preterm intestinal microbiota changes, Yap and colleagues evaluated the impact of neonatal care practices and NICU admission in the microbiota of preterm and term infants, from day one (after birth) to 12 months postpartum. The authors observed significative differences between term and preterm infants, in β-diversity, revealing a clear separation between the two groups, particularly at 6 and 12 months. In terms of taxa, Bacillota (former Firmicutes) and Bacteroidota (former Bacteroidetes) were the taxa that distinguish between term and preterm infants. Furthermore, in the first two weeks of life, the levels of Klebsiella were significantly higher in preterm infants, while the levels of Bacteroides fragilis were lower (but gradually increasing over time). Lastly, when accounting for clinical parameters (such as infections, NICU stay, clinical practices, and feeding regimens), the authors conclude that weight at birth seems the best explanatory variable at birth and month 12, while gestational age seemed to have more impact in the composition of preterm infant microbiota at 12 months; the insertion of a PICC line (peripherally inserted central catheter) for total parenteral nutrition administration and isolation of antibiotic-resistant bacteria, were the best explanatory variables for microbial changes at 6 and 12 months – both clinical practices have been implicated in the disruption of gut microbiota establishment (Yap et al. Citation2021). The evidence presented in the mentioned studies indicate that preterm infants might exhibit a gut microbiota dominated by potentially pathogenic microbes which, combined with invasive treatments and a compromised immune system (discussed later in chapter 2), makes them more susceptible to infections.
Maternal habits during pregnancy, namely the use of antibiotics or probiotics, can also impact the intestinal microbiota of infants. A study in an animal model demonstrated that antibiotic treatment during gestation leads to intestinal microbiota disruption (imbalance between bacterial phyla) and intestinal damage in neonatal mice (Chen et al. Citation2021). Recent studies in human babies revealed similar conclusions. Turta and colleagues demonstrated that intrapartum administration of antibiotics resulted in short-term disruption of infant gut microbiota, with a reduction in microbial richness and an increase in the relative abundances of Clostridiaceae and Erysipelotrichaeceae (Turta et al. Citation2022). In contrast, maternal probiotic treatment seems to have a restorative effect on early-life gut microbiota. Korpela and colleagues (Korpela et al. Citation2018) evaluated the impact of probiotics, when taken during gestation and after birth, on infant gut microbiota, and found that the treatment increased levels of Bifidobacteria and decreased levels of potential pathogens (Clostridia and Pseudomonadota), although this modulation of the gut microbiota was also heavily dependent on the infant’s diet.
Early-life feeding strategies may impact on the infant gut microbial colonization (Robertson et al. Citation2019). As such, an exclusive breastfeeding strategy constitutes an important source of lactic acid-producing bacteria and human milk oligosaccharides (HMO), creating an environment beneficial for the proliferation of Lactobacillus and Bifidobacterium, known probiotic genera (Ventura et al. Citation2019). In comparison, formula-fed infants exhibited a more diverse microbiota (higher number of bacterial genera) than their breastfed counterparts, possibly because formula milk contains a different array of nutrients that modulate the gut microbiota (Milani et al. Citation2017). To demonstrate the impact of an early-life diet on the gut microbiota, Brink et al. (Brink et al. Citation2020) compared fecal microbiota and metabolites in breastfed and formula-fed infants, with dairy-based and soy formula milk, during the first year of life. Soy-based infant formula differs from the common dairy-based formula, due to the presence of soy protein isolates instead of bovine protein isolates (Byrne et al. Citation2021). The different diet regimens lead to differences in overall microbial diversity, with breastfed infants displaying lower diversity and higher levels of Bifidobacterium. In a cross-sectional study, Ma et al. compared the microbial composition and diversity of the gut microbiota of infants fed with breastmilk or formula. It was found that despite Bifidobacterium being the most abundant genera in both groups, the relative abundance of potentially pathogenic bacteria (such as Streptococcus, Clostridium, and Enterococcus) was higher in formula-fed babies (Ma et al. Citation2020), probably due to the low abundance of protective bacteria. To further highlight the beneficial effects of breastfeeding, Liu et al. evaluated the restorative effects of breastmilk in cesarean-delivered infants. The study has shown that infants born by C-section and exclusively breastfeed, not only possess a gut microbiota that closely resembles one of the vaginally delivered infants, but also that breastmilk was able to restore the presence of beneficial bacteria (such as Faecalibacterium) and to reduce the abundance of bacterial genera typically associated with this mode of delivery (such as Enterococcus and Veillonella) (Liu et al. Citation2019). A randomized clinical trial by Embleton et al. evaluated different feeding regimens in the microbial composition of 120 preterm infants, to better understand if an exclusive human milk diet could impact microbial and clinical parameters in preterm newborns (particularly, when a mother’s own breastmilk experiences a shortfall in supply). The participants were divided into two groups: intervention, characterized by exclusive human milk diet, in which infants were fed maternal breastmilk or pasteurized human milk, in case of shortfall of breastmilk supply; and control, in which infants were fed maternal breastmilk or artificial bovine formula, in case of shortfall of breastmilk supply. The study groups received milk fortifiers, either pasteurized human milk-derived or bovine-derived fortifiers, depending if the infant belonged to the intervention or control group. Stool samples were collected and analyzed at five timepoints: (A) baseline (following enrollment), (B) day of life 10, (C) “full feeds,” (D) day of life 21–28, and (E) final sample collected before study completion (34 weeks postpartum). No significant differences among groups and throughout time were observed, and the type of feeding did not impact clinical outcomes (namely, the development of necrotizing enterocolitis and other neonatal pathologies). The only variables that did influence the bacterial profiles, at more than one timepoint, were the place of NICU stay and the intake of probiotics. In terms of microbial abundance, it was shown that infants in NICU using probiotics, displayed a higher relative abundance of Bifidobacterium, and it positively correlated with the number of days of mother’s own milk consumption. In the intervention branch, preterm infants showed a reduced abundance of Lactobacillus (Embleton et al. Citation2023). The results stated in the study indicate that, while milk from human donors can be useful for infants when maternal breastmilk supply is running low, it cannot substitute it.
After 6 months, with the introduction of solid foods (weaning period), there is a significant increase in the α-diversity, a diversity parameter used to evaluate the richness and evenness of microbial species of a given sample, in a certain environment (Walters and Martiny Citation2020). The gut microbiota shifts its composition from Pseudomonadota (previously Proteobacteria) and Actinomycetota (previously Actinobacteria) to Bacillota (previously Firmicutes) and Bacteroidota (previously Bacteroidetes) (Milani et al. Citation2017). Also, in the transition from an exclusively-breastmilk regimen to solid foods, a significant decrease in bifidobacteria was reported (Zhuang et al. Citation2019). During this period, the child’s gut microbiota changes, leading to a more adult-like composition, reaching a stable composition at the age of 3. The adult microbiota is dominated by bacterial strains capable of producing short-chain fatty acids (SCFA), as well as degrading glycans and complex carbohydrates. Hence, proper nutrition during the weaning period is crucial for the establishment and maintenance of mature microbiota. Malnourishment and/or a high-fat diet, prevalent in Western societies, may lead to gut dysbiosis and growth impairment (Singh et al. Citation2020).
Other factors that may influence the composition and colonization patterns of infant gut microbiota include geographical location (closely related to cultural and dietary practices), family lifestyle (close relatives, siblings, and pets), and host genetics (Zhuang et al. Citation2019). The contribution of the host genetics to the composition of the gut microbiota remains unclear, as most conclusions are drawn from studies on twins. While some studies demonstrate that monozygotic twins exhibit a very similar microbiota, other studies debunked those theories, demonstrating no significant differences were found between identical twins (Rodríguez et al. Citation2015). Therefore, more studies are necessary.
Interaction with household members such as siblings and pets can also modulate the gut microbiota of the child. Studies have found that living with older siblings was associated with increased microbial diversity and richness and that the number of siblings was positively correlated with microbial diversity (Christensen et al. Citation2022; Laursen et al. Citation2015). Furthermore, the existence of siblings may offer protection against allergies (Christensen et al. Citation2022). Similarly, living with pets is related to increased microbial diversity and richness in the intestinal microbiota of infants, with an increased abundance of certain bacterial species, such as Ruminococcus and Oscilospirra (Zoratti et al. Citation2020; Tun et al. Citation2017). Furthermore, it was found that prenatal exposure to domestic pets might be associated with a decreased streptococcal colonization and, thus, may lead to decreased risk of developing metabolic diseases and atopy (Tun et al. Citation2017). However, additional studies are necessary to further comprehend the link between pet-infant gut microbiota and its impact on the child’s health.
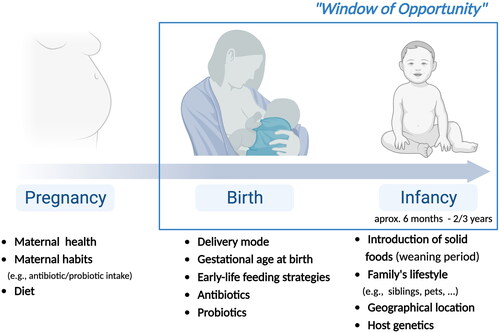
It is postulated that, after the first 1000 days of life, the microbiome is already established and maintained by the host. During this period, the microbiota and its metabolites modulate the development of the infant’s immune system, offering the antigenic stimulus necessary for its maturation, through the establishment of immune tolerance, that differentiates commensal microbiota from pathogenic microorganisms (Zhuang et al. Citation2019). Of particular interest are the emerging evidence on the cellular mechanisms that trigger trained immunity or immune tolerance. Specifically, trained immunity is primed by specific Pathogen-associated molecular patterns (PAMPs) and Damage-associated molecular patterns (DAMPs), whereas immune tolerance is promoted by Gram-negative endotoxin, such as Lipopolysaccharide (LPS) (reviewed by (Lajqi et al. Citation2023)). Therefore, throughout the first days of life, an important “window of opportunity” is established (), in which the interactions between the gut microbiota and the host are crucial for the development of a balanced immune system (Zhuang et al. Citation2019).
Figure 1. Factors that influence microbiota composition in early-life, from pregnancy to birth and infancy (created with BioRender).
In summary, the infant gut microbiota is dominated by specific bacterial genera, depending on the development stage. After birth, the early-life gut microbiota is mainly composed of Bifidobacterium and Lactobacillus, due to maternal transmission of microbiota and breastfeeding (as these bacterial genera thrive in the presence of human breastmilk oligosaccharides (HMO)). Other bacterial genera that may be present in infant gut microbiota include Clostridium (despite being a potential pathogenic organism, it can remain asymptomatic in newborns), Bacteroides, Enterobacteriaceae, Streptococcus, and other genera found in lower relative abundances, such as Akkermansia (Yao et al. Citation2021; Moore and Townsend Citation2019).
2. Gut microbiota and immune system Interactions in early-life
Under normal conditions, the dynamic interactions between the gut microbiota and the host’s immune system aid in the maintenance of the overall body balance. Along with intestinal and immune cells, the gut microbiota constitutes another barrier of protection, competing with pathogens for nutrients and adhesion sites in the intestinal mucosa and preventing the colonization and replication of external pathogens (Yoo et al. Citation2020). Moreover, the gut microbiota can regulate both local and systemic immune responses either directly, by interacting with adaptive and innate immune cells, or indirectly through the production of microbial metabolites (Yoo et al. Citation2020). As the infant’s gut is colonized by different microbial populations, it triggers the immature immune system and induces tolerance toward this new commensal microbiota.
2.1. Early-Life immune system development
The immune system, alongside other protective barriers such as the skin, provides the body protection against disease and has an innate and an acquired component.
The innate immune response is nonspecific, immediate, with no immunological memory, and is mainly mediated by macrophages, neutrophils, and dendritic cells (DCs), known as professional phagocytes, and innate lymphoid cells (ILCs), mast cells, eosinophils, basophils, and natural killer cells. In turn, the acquired immune response is pathogen and antigen-specific, has a cell-mediated and a humoral component, and triggers immunological memory. The adaptive immune system is composed of T cells and B cells. Regarding T cells, there are two main subsets: CD4+ T helper (Th) cells and CD8+ cytotoxic T cells. CD4+ T cells can be subdivided into Th1, Th2, Th9, Th17, Th22, T regulatory cells (Tregs), and follicular helper (Tfh) cells. All these subsets have distinct tasks: Th1 produce cytokines such as IL-2 (interleukin 2) and IFNγ (interferon-gamma), responding promptly to virus and bacteria; Th2 direct their response to parasites, through the production of IL-4 and IL-10; Th9 are central to control helminth infection and anti-tumor immunity through secretion of IL-9; Th17 have a crucial role in the response to extracellular bacteria and fungi; Th22 support mucosal immunity to microbial infection through the secretion of IL-22; Tregs are negative regulators of the immune response and protect against auto-immunity; and finally, the main role of Tfh cells is to regulate B cell immunity in the germinal centers of the lymph nodes (Wilson and Messaoudi Citation2015). The main function of CD8+ T cells is clearing intracellular pathogens and tumors (Kurachi Citation2019; Wang et al. Citation2019). This T cell population, termed as cytotoxic, is divided into three major subpopulations: Tc1, with central cytotoxic activity through the secretion of granzyme and perforin, tumor necrosis factor-alpha (TNF-alpha), and IFN-gamma; Tc17 involved in mucosal immunity and production of IL-17 and defensins; and the immune suppressor Tc2 cells that produce IL-4, IL-5, and IL-13 ((Annunziato et al. Citation2015; Mittrücker et al. Citation2014)). T cells can be activated and differentiated after contact with antigen-presenting cells (APCs), such as macrophages or DCs, by the presentation of pathogen-derived peptides through major histocompatibility complex class II (MHC II), in the case of CD4+ Tcells, or by the presentation of cellular antigens through MHC I, in the case of CD8+ T cells (Yao et al. Citation2021). B cells are responsible for antibody secretion, which then coat pathogens, targeting them for opsonization, activation of the complement system, and neutralization (Gaudino and Kumar Citation2019).
The immune system development starts in utero, as early as at the 8 weeks of development, when the thymus is seeding by hematopoietic cells, and at 12 weeks of gestation the lymph nodes of the fetus are already evident. T-cell development in the thymus begins around 14 to 17 weeks of gestation and, around this time, these cells are already detected at the lymph nodes as well. On the other hand, B cells are already present in the primary follicles of lymph nodes by the 17 weeks of gestation. Conversely, DCs and circulating monocytes are only detected in the second trimester of pregnancy, increasing until the end of pregnancy. As for neutrophils, these are not present until the third trimester of pregnancy but are the predominant population of cells at birth (Wilson and Messaoudi Citation2015). Although throughout this period, the exposure of the fetus to live bacteria remains controversial, it is known that the presentation to diverse microbial metabolites and fragments may happens via the placenta (Jain Citation2020). Gut commensal-specific IgG antibodies may be transferred to the fetus, regulating its mucosal CD4+ T-cell response to microbial antigens at birth. Interestingly, in a mouse model, maternal antibodies that only were present during pregnancy improved the transmission of microbial molecules across the placenta, which primed their pup’s immune system and suppressed allergic airway diseases by increasing the number and functionality of Treg cells later in life (Thorburn et al. Citation2015).
Still in utero, the fetal immune system has a Th2 phenotype to prevent alloimmune responses against the mother (Abenavoli et al. Citation2019). This phenotype remains after delivery, with the infant’s immune system still biased toward a Th2 phenotype, presenting low IL-2 and IFNγ (Wilson and Messaoudi Citation2015). The low production of such cytokines can be due to functional differences in DCs which may prevent the differentiation of naïve CD4+ T cells toward Th1 (Wilson and Messaoudi Citation2015). However, after multiple pathogenic encounters and in a time- and age-dependent manner, the infant switches toward a Th1 polarization, promoting a pro-inflammatory profile, with negative consequences in the child’s long-term health (Abenavoli et al. Citation2019).
During delivery, infants receive “a bacterial boost” mainly from the mother, exposing the immature immune system of the newborns to a significant microbial load (Abenavoli et al. Citation2019). After delivery, the immune system of the newborn is still functionally immature and is markedly modulated to avoid excessive inflammatory responses. For this reason, it is observed a dampening of the immune system mechanisms, characterized by a decrease in the complement function, decrease in neutrophils amount and functions, decrease in the production of pro-inflammatory cytokines by APCs (IL-1β, tumor necrosis factor (TNF)-α, and IL-12p70), and an increased expression of inhibitory receptors by NK cells, such as NKG2A/CD94 (Esteve-Solé et al. Citation2018; Wang et al. Citation2007). Simultaneously, CD8 T cells present low activity, and monocytes and macrophages still show immaturity (Méndez et al. Citation2021).
Likewise, B cells are mostly naïve at birth and their ability to respond is limited possibly due to their immaturity, poor B cell repertoire, or reduced strength of B cell receptor (BCR) signaling (Wilson and Messaoudi Citation2015; Basha et al. Citation2014). Additionally, neonates have underdeveloped germinal centers in lymph nodes and spleen and low expression of BCRs, resulting in low levels of primary IgG responses to infections and vaccines (Basha et al. Citation2014). Therefore, neonates exhibit an increased susceptibility to infections because of the immaturity of their lymphocytes, low numbers of effector-memory T cells, low Th1 cytokine secretion, and reduced strength of BCR signaling.
In this context it is of note that preterm infants, compared with term infants, have an underdeveloped immune system, characterized by diminished innate and adaptive immunity. They have a smaller pool of monocytes and neutrophils, and the compromised capacity of these cells to eliminate pathogens, coupled with decreased cytokine production, restricts T cell activation and undermines the ability to combat bacteria and detect viruses within cells (Melville and Moss Citation2013). In normal conditions, the adaptive immune system is only completely developed in the early childhood, meaning that the neonates strongly rely on innate immune response, which in the case of preterm infants is a critical issue, since they have an immature innate immune system (Melville and Moss Citation2013). Moreover, antigen-specific IgG are abundantly transferred across the placenta from the maternal circulation, particularly after the 32nd week of gestation, and the transfer increases with fetal age, which can result, in preterm infants in lack of pathogens opsonization and deficiencies in phagocytosis(van den Berg et al. Citation2011).
Since the newborn adaptive immune system is exclusively constituted by naïve T and B cells, during the first 6 months of life, maternal antibodies acquired in utero and breastmilk provide additional protection against pathogens in the GI and respiratory tract (Wilson and Messaoudi Citation2015). Indeed, the feeding strategy may also impact early-life immunity by modulating the production of microbial metabolites, with breastfed infants displaying higher concentrations of fecal butyric acid (derived from the metabolism of SCFA and able to increase regulatory T cell differentiation), d-sphingosine (involved in immune cell trafficking) and betaine (inhibits inflammatory responses) (Brink et al. Citation2020). Furthermore, breastmilk also provides IgA, the main immunoglobulin in mucosal protection, enabling a tolerant and balanced immune system (Milani et al. Citation2017). The secretory IgA (sIgA) present in the breastmilk is produced by plasma cells in the mammary gland but originally from the gut of the mother. Therefore, the specificity of the sIgA is modulated by maternal exposure to her enteric bacteria, both commensals, and pathogens, therefore protecting from the expansion of the latter while the child’s immune system is maturing (van den Elsen et al. Citation2019). Furthermore, sIgA also seems to shape the gut microbiota of the child with a long-lasting effect, which is fundamental to prevent excessive expansion of pro-inflammatory microbial taxa, and in the modulation of microbial gene expression and metabolic function (van den Elsen et al. Citation2019).
The number of immune cells varies in different age groups (Méndez et al. Citation2021; Valiathan et al. Citation2016). Lymphocytes, including CD4+ T cells and B cells, appear to decrease significantly with age, whereas neutrophils and CD8+ T cells increase in adulthood, and NK cells increase in adolescence (Méndez et al. Citation2021; Valiathan et al. Citation2016). Throughout infancy and early childhood, the Th phenotype changes to a Th1 phenotype, potentiating macrophage activation and cellular immunity (Wilson and Messaoudi Citation2015). At the age of 2, DCs start producing the cytokine IL-12p70, which then stimulates the Th1 response. Furthermore, with further antigen exposure, T cell response increases, and cytotoxic and Th cells develop memory toward the antigen (Méndez et al. Citation2021).
Innate lymphoid cells (ILCs) are another component of the innate immune system, and that has an important role in neonatal immunity. In adulthood, ILCs originate from a common lymphoid precursor (CLP) located in the bone marrow, which originates a common helper-ILC precursor (CHILP), eventually differentiating in different subsets, depending on their cytokine expression profile (namely, ILCs group 1, group 2, and group 3) (Ganal-Vonarburg and Duerr Citation2020). In early-life, ILCs derive from a progenitor located in the fetal liver, but have been found in other sites of the fetal environment, including the secondary lymphoid organs (such as the mesenteric lymph nodes and Peyer’s patches) and the intestine. Despite the gaps in the current literature, studies in rodent models claim that a different subset of ILCs, the lymphoid tissue inducer cells (LTi), have a central role in the proper development of SLOs, as reviewed + (Mendes et al. Citation2020). Moreover, it is suggested that ILCs can confer protection to the newborn, as it transitions from the sterile womb to a microbially-rich environment, and establishes the tolerance system which distinguishes between commensal and harmful microbes (Yu et al. Citation2018).
In sum, the first 1000 days of life are fundamental not only to the microbiome establishment, but also to the concomitant development and maturation of the immune system and, consequently, to the interplay between both. Hence, this period is considered to represent a microbiological and immune ‘window of opportunity’ during which any event will have a pivotal impact on the metabolic, immunological, and microbiological priming, which may later affect human health (Selma-Royo et al. Citation2019).
2.2. Gut microbiota and immune system Interactions
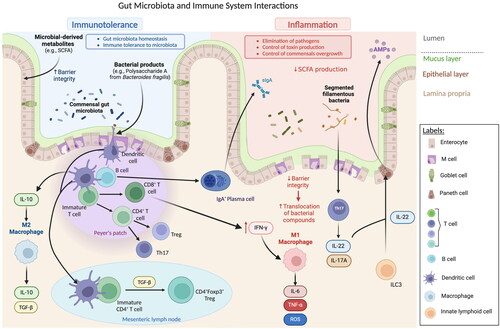
The complex balance between host immunity and gut microbiota is established when the commensal microorganisms interact with intestinal epithelial cells (IEC). The immune response begins with the recognition of microbe-associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs), located on IECs or DCs, which can then recruit activated B and T cells. PRRs, which can include Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors, when activated by MAMPs, induce the production of antimicrobial peptides, such as regenerating islet-derived protein III-gamma (RegIIIγ), and immunological mediators such as IL-18, IL-33, IL-25, and tumor growth factor-β (TGF-β) which, consequently, promotes the development tolerogenic macrophages (producers of high levels of IL-10) and DCs, which stimulate the differentiation of Tregs (Maranduba et al., Citation2015; Thaiss et al. Citation2016). In the specific case of TLRs, they can be activated by a wide variety of microbial antigens (including LPS, peptidoglycan, and flagellin) and trigger a signaling cascade that results in the activation of the NF-kB transcription factor, which regulates the expression of cytokines, chemokines, and of other immunological mediators. A known example of the role of TLRs in gut eubiosis (balanced gut microbiota) is the model of polysaccharide A (PSA) of Bacteroides fragilis. PSA can be detected by TLR2/TLR1 heterodimer in collaboration with Dectin (a C-type lectin PRR), promoting the activation of anti-inflammatory genes. Furthermore, Dectin-1 can control gut immunity by altering the microbiota pattern, leading to Treg differentiation (Al-Rashidi, Citation2022). Moreover, PRRs can also contribute to the elimination of pathogenic microorganisms, through the activation of the inflammasome-forming NLR family CARD-domain containing protein 4 (NLRC4) which triggers the detachment of the infected epithelial cells when detected as infected by a pathogen (Thaiss et al. Citation2016).
Considering the mechanism of microbiota recognition and the importance of the early-life microbiota in immune system priming, it is important to highlight a recent study by Wampach et al. in which it assessed the immunostimulatory potential of maternal microbiota transferred to infants born through vaginal or cesarean delivery. This study reported that fecal LPS isolated 3 days after vaginally-delivered neonates induced higher levels of cytokines in monocyte-derived DCs, compared with fecal LPS from cesarean-delivered neonates (Wampach et al. Citation2018). Since LPS is recognized by TLR-4 on the membranes of intestinal epithelial cells and it stimulates components of the immune system, these results suggest that disturbances in the initial microbiota inherited by the mode of delivery may impact the immune system stimulation and, thus, the immune response of the infant (Jašarević and Bale, Citation2019). Moreover, a recent study in mice observed that vaginally-delivered offspring acquired immune tolerance via spontaneous activation of the intestinal epithelial cells and acquired resistance to LPS shortly after delivery, whereas C-section-born pups and TLR4-deficient mice did not (Lotz et al., Citation2006).
2.2.1. Gut microbiota and innate immune system Interactions
Within the gut epithelium, specialized cell types are involved in immune recognition. At the gut-associated lymphoid tissue (GALT), Peyer’s patches, isolated lymphoid follicles (ILFs), and epithelial M cells are part of the local immune system, sampling antigens from the lumen and presenting them to APCs located under the epithelium. Enteroendocrine cells, by their side, act in the local immune system through the production of glucagon-like peptide 2 (GLP-2), involved in the regulation of the innate immune response, by controlling the expression of antimicrobial peptides (AMPs) (Maranduba et al., Citation2015). Additionally, in the lamina propria, the commensal microbiota can also stimulate and interact with innate lymphoid cells (ILC), tissue resident cells particularly abundant in the intestinal mucosa. Despite some similarities with Th cells, ILCs respond faster to pathogens, by presenting antigens to Th cells and promoting their differentiation and activation, and acting as essential players in the maintenance of intestinal barrier integrity. Upon damage to the epithelium, ILCs are activated to restore barrier function (Zheng and Zhu Citation2022). The commensal microbiota induces the secretion of intestinal cytokines, and in turn, ILCs can react to the gut microbiota by altering their structure, with consequences on gut immunity (for example, promoting the production of antimicrobial peptides by IECs) (Han et al., Citation2019).
Other immune cells pivotal to the initiation of the primary immune response are intestinal DCs, by limiting the reactivity toward the commensal gut microbiota and by recognizing potential pathogens (D’Amelio and Sassi, Citation2018). Intestinal DCs, typically found in the GALT, are continuously exposed to luminal antigens and thus, in order to distinguish between innocuous microbiota and pathogenic microbes, they sample the luminal content (Stagg Citation2018; Owen and Mohamadzadeh Citation2013). These cells have distinct ways of antigen recognition and capture: they can extend dendrites across the epithelial layer and capture translocated IgA immune complexes, or sample the luminal space through M cells, which internalize microorganisms and deliver to DCs (Stagg Citation2018; Swiatczak and Rescigno Citation2012). Activation of gut DCs is facilitated by PRRs, such as TLRs. Concerning DC interaction with gut microbiota, the typical response is tolerance to prevent inflammation, a mechanism known as oral tolerance, a state of non-response toward antigens from food (Sun et al. Citation2020). The main location for this process is within the mesenteric lymph nodes (MLNs), in which DCs are strong promotors of tolerance. Additionally, intestinal DCs can promote IgA class switching in B cells, further enhancing the maintenance of gut homeostasis (Tezuka and Ohteki Citation2019). For instance, bacterial products, and certain species of probiotics have been shown to promote tolerance in DCs, simply by attaching to their surface. A few examples include the interaction between intestinal DCs and Lactobacillus acidophilus NCFM (a food supplement), in which the bacteria was able to promote the secretion of IL-10, an anti-inflammatory cytokine (Swiatczak and Rescigno Citation2012). Besides live bacteria, bacterial products can also induce a tolerogenic profile in intestinal DCs. Wang et al. (Citation2006) showed that, when exposed to the bacterial product PSA, derived from Bacteroides fragilis, intestinal DCs promote inducible nitric oxide synthase (iNOS), to produce nitric oxide, which appears to exert an impact in the maintenance of intestinal homeostasis (Tezuka and Ohteki Citation2019; Wang et al. Citation2006).
2.2.2. Gut microbiota and adaptive immune system Interactions
Bacterial-loaded DCs can also induce the differentiation of B cells into IgA+ plasma cells, leading to IgA secretion, IgA coating, and opsonization of the pathogen for elimination, which can occur through T-cell-dependent or independent mechanisms (Tezuka and Ohteki Citation2019). This antibody production, affinity maturation, and class switch recombination require complex interaction between DCs and B cells in the Peyer patches, dependent on the secretion of TGF-β by ILCs. Still regarding the interaction between B cells and the gut microbiota, and as previously mentioned, the low production of sIgA in newborns is compensated by the presence of these antibodies in the mother’s breastmilk. As so, breastmilk IgA plays an important role in modulating the gut microbiota in early-life by driving microbial colonization and immune system maturation and priming (Thorburn et al. Citation2015). As the neonate gut begins to be progressively colonized by different species, the immune system responds by producing endogenous sIgA, which will then modulate the composition of the microbiota (Thorburn et al. Citation2015). The mechanisms behind the control and proliferation of commensal bacteria are still to be unveiled, but it is hypothesized that it might be related to the ability of sIgA to bind to bacteria, promoting changes in the regulation of gene expression which will then influence their metabolic processes and the biogeography of the gut (Kato et al., Citation2014).
Regarding T cells and microbiota, the most studied CD4+ T cell subtypes are Th17 and Treg. In the case of Th17, in response to stimuli (such as segmented filamentous bacteria – SFB), naïve CD4+ T cells can migrate to the intestinal lamina propria and differentiate into IL-17A-producing Th17 cells. Consequently, the cytokines produced by these cells (IL-17A, IL-17F, IL-22) stimulate IECs to produce AMPs, helping to preserve a non-inflammatory state of the intestinal barrier (Wang et al. Citation2019; Ivanov et al., Citation2009; Weaver et al. Citation2013). On the other hand, Treg cells located in the intestinal lamina propria have a fundamental role regarding tolerance toward commensal microbiota. For instance, RORγt+ Tregs population located in the colon seems to be expressed in a microbiota-dependent manner. In germ-free mice, this population seems to decrease, and, during weaning, the generation of RORγt+ Tregs is associated with a decreased susceptibility to allergic inflammation later in life. Moreover, the induction of peripheral Treg cells can suppress abnormal inflammatory responses (Wiertsema et al. Citation2021). Overall, intestinal homeostasis is maintained by a balance between the effector T cells and Treg cells, which mediate immune responses and confine excessive immune activation, respectively (Wang et al. Citation2019). Likewise, some probiotic Lactobacillus strains improve intestinal inflammation by modulating the ratio between Tregs and Th17 cells (Wang et al. Citation2017).
The production and release of gut microbial metabolites, such as bile acids, branched-chain amino acids, and trimethylamine N-oxide, can also greatly impact the host immune response, influencing inflammatory signaling and interactions with immune cells (Agus et al., Citation2021). Certain gut bacteria, such as Faecalibacterium prausnitzii and Roseburia intestinalis (known probiotics), can generate microbial metabolites through the anaerobic fermentation of complex carbohydrates (dietary fibers and resistant starch), namely SCFA (Silva et al. Citation2020). Shortly after their production, SCFA are absorbed by colonocytes, and then transferred to the liver, where they are used as an energy supply for hepatocytes, with the exception of acetate, which is oxidized. A small fraction of SCFA can also be carried through the systemic circulation to other tissues (Silva et al. Citation2020). Being the most abundant microbial-derived metabolites, SCFA influences gut health through several mechanisms, specifically, maintenance of gut barrier integrity, mucus production, and protection against inflammation and pathogens. These mechanisms occur due to signaling triggered by SCFA, which can bind to G protein-coupled receptors (GPCRs), expressed in several cell types, or through the inhibition of histone deacetylases (HDACs), a mechanism often linked to immune tolerance and anti-inflammatory phenotype, aiding in the maintenance of immune homeostasis (Yoo et al. Citation2020; Silva et al. Citation2020). Nevertheless, the activation of these receptors will result in different outcomes, depending on the type of cells involved. For example, SCFA, particularly butyrate, influences the systemic immune response through the control of the differentiation process of T cells, thus, influencing the adaptive immune response. These microbial metabolites can impact the differentiation of T cells either in Th1, Th2, and Th17 cells, or into Tregs. Moreover, SCFA can also reduce inflammation, regulating the expression of pro-inflammatory cytokines (such as IL-6, IL-12, and TNF-α) by activating macrophages and DCs (Yoo et al. Citation2020). SCFA can also lead to the induction of regulatory B cells, inhibiting the generation of Th17 cells, which may play a role in preventing autoimmune disorders or gastrointestinal diseases such as inflammatory bowel disease (IBD) (Wiertsema et al. Citation2021). Furthermore, the commensal bacteria can impact the innate immune response, for example, through lymphoid stimulation in the spleen, activation of macrophages, and stimulation of the maturation process of NK cells (Wiertsema et al. Citation2021).
Of note are the observations from germ-free mice demonstrating that gut microbiota colonization in early-life is essential for optimal immune development, as these animals presented underdeveloped mucosal immunity, reduced number of immune cells as IgA-producing plasma cells, CD4+ and CD8+ T-cells, and reduced capacity of resisting to pathogenic bacteria (Sommer and Bäckhed Citation2013). Therefore, early-life gut colonization is a sequential process that influences the maturation and development of the child’s immunity, such as through the production of SCFA (Silva et al. Citation2020; Yang et al. Citation2020) or the control of proliferation and differentiation of T and B cells (Tanaka and Nakayama Citation2017).
Overall, the interactions between gut epithelial and immune cells contribute to the tolerogenic state of the intestinal environment (Wang et al. Citation2019). Under physiological conditions, the gut microbiota contributes to intestinal tolerance via sIgA production and Treg differentiation, and tolerogenic DCs and macrophages contribute further to this, via IL-10 secretion. Gut microbiota, SCFAs, and IL-21 secreted from Tfh cells contribute to the secretion of specific sIgA, which will then opsonize the gut commensal bacteria to control their proliferation and toxin production. Commensal bacteria components induce further Treg and B cell production and promote IFNγ secretion from CD8+ cells. On the other hand, sIgA and tolerogenic DCs negatively regulate the stimulation of Th17 cells, inhibiting a strong pro-inflammatory response against commensal bacteria. Tregs also downregulate DCs and Th17 cells by producing TGF-β (Wang et al. Citation2019). In this scenario, the intestinal balance is well maintained ().
3. Early-Life obesity-associated gut dysbiosis and alterations in the immune system
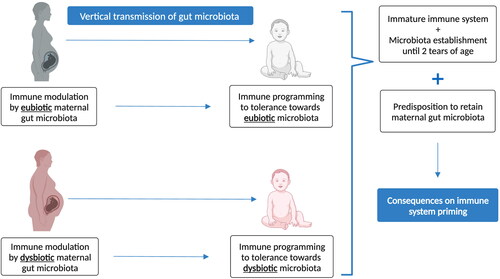
During early-life, the interplay between the infant’s intestinal microbiota and the immune system significantly influences each other development: the colonization of commensal microbes affects the maturation of lymphoid tissues and lymphocytes, while the host’s immune response influences the selection and maintenance of commensal microbiota, fostering a stable equilibrium and ecological balance among microbial communities (Hong and Medzhitov, Citation2023). For example, loss of microbial diversity leads to increased serum IgE production in mice which could be rescued by neonatal but not adult microbial colonization (Cahenzli et al., Citation2013). At the same time, infant’s intestinal microbiota is highly plastic and shaped by several external factors, including the maternal microbiota. During this period of development, the mother plays a crucial role in the establishment of early-life gut microbiota, since mothers vertically transmit their microbiome and their metabolites to the offspring maybe in utero, but certainly during delivery and breastfeeding (Soderborg et al. Citation2016). Therefore, the mother impacts the composition of the infant’s gut microbiota, as well as its immunity. The composition of the maternal microbiota can vary depending on different factors such as diet, exposure to antibiotics, systemic health status, and diseases such as obesity, which in turn, affect the infant’s gut microbiota (Neff et al. Citation2018; Arrieta et al., Citation2015).
Obesity has become a worldwide problem, with over 500 million people suffering from this condition according to the World Health Organization. Obesity is often associated with other comorbidities, such as type 2 diabetes and cardiovascular disease (Milano et al. Citation2022). High-calorie diets and sedentarism might have contributed to the steady rise of these numbers. Still, obesity is a complex disease, influenced not only by the environment, but also by the genetic makeup of the host (genetic polymorphisms) and, as unveiled more recently, by the gut microbiota (Milano et al. Citation2022). Altered gut microbiota can contribute to obesity by increasing energy intake, inflammation, and alterations in metabolic pathways (Kincaid et al., Citation2020). It is estimated that maternal obesity affects two out of five pregnancies, representing a serious health issue since maternal obesity can influence the health of the infant, which is already vulnerable to external threats in early-life (Di Gesù et al., Citation2021).
3.1. Obesity and gut microbiota
The link between obesity and gut microbiota has been studied in animal models and human patients. Menni et al. observed that patients with a more diverse gut microbiota were less prone to develop long-term weight gain, suggesting a preventive effect of the gut microbiota (Menni et al. Citation2017). Other studies focused on the differences observed in the Bacillota/Bacteroidota ratio between obese and lean individuals (Kasai et al., Citation2015). However, there is some controversy in using the Bacillota/Bacteroidota ratio as a marker for obesity since discrepancies were reported in other studies in which obese patients showed decreased Bacillota/Bacteroidota ratio, as reviewed (Magne et al., Citation2020). Another factor that contributes to these inconsistencies is related to the metabolic endotoxemia hypothesis, which proposes that increased adiposity and development of low-grade inflammation could be associated with the dissemination of LPS – the theory is at odds with decreased levels of Bacteroidota observed in obese patients since this phylum is mainly composed of Gram-negative bacteria (rich in LPS) (Magne et al., Citation2020). These divergences could also be related to the experimental procedure applied in different studies, such as the number of participants and the methods used, or even the heterogeneity between participants (Magne et al., Citation2020).
Additionally, scientific evidence suggests that another putative mechanism for this causal relationship between the gut microbiota and obesity may be a higher energy extraction from the diet by the gut microorganisms. The gut microbiota metabolizes certain compounds from the diet that would otherwise not be digested, such as some carbohydrates and fibers. These are then converted by the intestinal microbiota into SCFA, which, in turn, correspond to 10% of the human daily energy intake and are essential for colon and liver cells. It has been found that when lean and obese adults have a diet with excessive calorie content, lean subjects lose more energy through stool than obese subjects (Jumpertz et al., Citation2011). But a core theme in the link between obesity-microbiota is that diet has a deep impact on the composition of the intestinal microbiota and, consequently, on the host health. A Westernized diet, rich in saturated fats and poor in fiber, can cause a shift in the individuals gut microbiota, leading to a reduction in the abundance of beneficial microorganisms (dysbiosis) and increasing inflammation, a profile often associated with the development of metabolic disorders such as obesity (Kim et al., Citation2019).
The bidirectional connection between gut microbiota and the central nervous system, known as the microbiota-gut-brain-axis (MGBA), has been extensively studied in recent years as a novel route to control obesity (as reviewed by (Longo et al., Citation2023)). A few studies have highlighted the key influence of gut microbes, and its metabolites, on the vagal nerve, particularly in the regulation of eating behaviors and development of obesity (Asadi et al., Citation2022; Sen et al. Citation2017). The MGBA has been explored in the context of early-life and brain development, since this process occurs in parallel with the maturation of the gut microbiota, and it is influenced by environmental factors, including diet, lifestyle and, maternal health (Cerdó et al., Citation2023). Obesity-induced maternal gut-dysbiosis has been associated with behavioral complications in children, with recent studies indicating that maternal cardiometabolic disease and gut dysbiosis, were linked to negative psychiatric outcomes in children (such as behavioral issues and attention deficits), and a disturbed infant gut microbiota composition, with increased prevalence of bacterial species positively correlated with psychological conditions in the offspring (Nieto-Ruiz et al. Citation2023; Liu et al., Citation2021). Although the maternal gut microbiota has been proposed as a possible mediator between maternal obesity and infant brain development, a definite mechanism is yet to be determined. Research suggests that an disturbed transfer of metabolites (including microbial SCFA) and nutrients from mother to fetus during gestation, and a chronic inflammation state (in part, due to increased levels of LPS caused by the dysbiotic gut microbiota), could negatively impact fetal brain development, eventually leading to impairment of behavioral and cognitive pathways (Di Gesù et al., Citation2021; Nieto-Ruiz et al. Citation2023; Sajdel-Sulkowska Citation2023)).
3.2. Maternal role on offspring obesity
The influence of maternal obesity in the child’s future health may start even before delivery, as dietary, metabolic, and microbial changes during pregnancy may already predispose metabolic changes in the offspring (Rubini et al. Citation2022). Maternal obesity is associated with irregular feto-placental function and an increased body fat percentage and insulin resistance during gestation (Beckers et al., Citation2024). Regarding maternal diet, it was also observed that a high-fat diet during pregnancy is linked to gut dysbiosis in primate infants, characterized by reduced diversity at 1 year of age even after switching to a healthy diet at the time of weaning (Ma et al., Citation2014). Kimura et al. found that pups born to germ-free mothers (who consumed a high-fat diet) and/or mothers fed a low-fiber diet, exhibited a pronounced obesity phenotype, characterized by an increase in body weight and food consumption (Kimura et al., Citation2020). Furthermore, infants born to mothers with high-fat intake during pregnancy were found to have lower levels of Bacteroides, which persisted from birth until 6 weeks of age (Chu et al., Citation2016).
Another study by García-Mantrana and colleagues centered around maternal BMI and diet, and its impact on the offspring anthropometric measures, at 1 month and 18 months after birth. Firstly, the study describes that maternal microbiota could be divided into two clusters: Cluster I, dominated by Prevotella, and Cluster II with a prevalence of Ruminococcus. Moreover, maternal microbiota composition was diet-dependent: for instance, increased consumption of fibers, omega-3, and polyphenols (associated with a vegetarian diet) was linked to an increased abundance of Christensellaceae, Dehalobacterium, and Eubacterium, and low levels of Dialister e Campylobacter. The second part of the study stated that maternal clusters were associated with neonatal gut microbiota and growth measures (namely BMI and Weight for Length - WFL), but dependent on the mode of delivery. Thus, infants from Cluster I and born by c-section presented higher z-scores at 18 months, and a higher risk of weight gain; infants from Cluster II (who also showed the highest levels of exclusive breastfeeding), showed lower BMI and WFL at the two timepoints. The authors concluded that infant anthropometric data (particularly at 18 months) was strongly influenced by maternal diet, maternal microbiota clusters, and mode of delivery (García-Mantrana et al., Citation2020). Recently, Dreisbach and team evaluated the link of pre-pregnancy BMI and maternal gut microbiota, with weight at birth. Following the analysis of 102 fecal samples of pregnant women in the second trimester, and after adjustment for confounding factors (antibiotic use and total weight gain during gestation), the researchers found that pre-pregnancy BMI and the component PC3 of the maternal microbiota (when controlled for the glucose levels) were predictive variables of neonatal weight. Within the microbial consortium that comprised the PC3 component, the probiotic Lactobacillus kefir, and respective virus Lactobacillus phage 3 SAC12, were the highest contributors to changes in neonatal weight (adjusted to gestational age). The researchers also observed several microbial taxa, commonly isolated from freshwater sources, represented in the PC3 component, but there is insufficient data exploring the impact of water quality and microbiota changes in the early-life (Dreisbach et al., Citation2023).
Moreover, the researchers verified that SCFA produced by the maternal microbiota can confer resistance to obesity to the offspring, by interacting with GPR41 and GPR43 receptors (G-protein coupled receptors), during fetal development. The activation of these receptors during embryonic maturation aids in the maintenance of energy homeostasis by promoting the differentiation of sympathetic neuronal, enteroendocrine, and pancreatic cells (Kimura et al., Citation2020). More recently, in a randomized controlled trial, Sugino and colleagues found that women with gestational diabetes mellitus that consumed a diet rich in complex carbohydrates with reduced fat had an advantageous impact on their microbiome, with increase in the probiotic family Bifidobacteriaceae, specifically B. adolescentis, enhancing the diversity in the infant gut microbiome, during the first four months of life, while mitigating the presence of opportunistic pathogens that could influence obesity and immune system development (Sugino et al. Citation2022). Thus, the effect of maternal diet exposure, metabolism, and microbiota during gestation may modulate the composition of the microbial community of the child, with long-lasting effects.
The transmission of microbiota between mother and child with the potential to promote obesity has been suggested as a possible route of intergenerational transmission of obesity, with maternal weight being the factor unrelated to the child that most influences the development of obesity in childhood and throughout life (Skrypnik et al. Citation2019). Stanislawski et al. followed up a cohort of 165 children 4, 10, 30, 120, 365, and 730 days after delivery and at 12 years of age. The authors observed that, in early-life, taxa within the gut microbiota that best predicted later childhood body mass index (BMI), substantially overlapped with the maternal taxa most strongly associated with overweight and obesity, namely F. prausnitzii and Ruminococcacae. In other words, certain bacterial taxa associated with the BMI of children later in life might derive from bacterial taxa found in the mother’s gut microbiota, and that is often associated with obesity. The results show an association between the infant gut microbiota and later BMI, and they offer preliminary evidence that the infant gut microbiota, particularly at 2 years of age, may have the potential to help identify children at risk for obesity (Stanislawski et al. Citation2018). Singh et al. considered birth mode and pre-pregnancy BMI when analyzing the gut microbiome composition from fecal samples from 6-week-old infants. In the vaginal-delivery group, maternal overweight or obesity was associated with higher infant gut microbiome diversity and higher relative abundance of 15 operational taxonomic units (OTUs), including the overrepresentation of Bacteroides fragilis, Escherichia coli, Veillonella dispar, and OTUs in the genera Staphylococcus and Enterococcus (Singh et al. Citation2020). Another recent study reported that high maternal BMI is associated with a lower abundance of butyrate-producing bacteria (such as Ruminococcus, Turicibacter, and Roseburia) in their children 1 month after delivery and that higher microbial diversity at this stage may predict higher adiposity later in life (Gilley et al., Citation2022). Additionally, at 6 months, infants from obese mothers had a lower abundance of bacteria from the family Lachnospiraceae, and, at 12 months, these infants had a lower abundance of Desulfovibrionaceae, Porphyromonadaceae, and an increased abundance of Enterobacteriaceae (Gilley et al., Citation2022).
Since early-life represents a critical window for immune stimulation, maternal obesity may reprogram the neonatal immune system (Neff et al. Citation2018). For instance, a study that assessed umbilical cord blood samples from babies born from lean mothers versus babies of obese mothers reported that the latter had fewer eosinophils and CD4+ T helper cells, reduced monocyte and DCs responses to TLR ligands, and increased levels of IFN-α2 and IL-6 (Wilson et al. Citation2015). More recently, a study comparing cord blood leukocytes from pregnancies with and without obesity demonstrated that pregnancy obesity is linked to a unique pattern of gene expression and DNA methylation in cord blood monocytes, in leptin-related genes, suggesting that this feature serves as a distinguishing characteristic of neonatal monocytes born to mothers with obesity (Krause et al., Citation2023). Moreover, similar transcriptional characteristics were reproduced in neonatal monocytes from the cord blood of healthy women subjected to pro-inflammatory stimuli. As such, this study indicates that the leptin pathway influences the inflammatory response in neonates and contributes to the programming of fetal and neonatal monocytes, a process influenced by pre-pregnancy or gestational obesity (Krause et al., Citation2023). In the same line, other authors showed that the maternal triglycerides levels are associated with, cord blood leukocytes epigenetic differences in immune and lipid metabolism-associated genes, which in turn was associated with child adiposity at 4–6 months and 4–6 years (Waldrop et al. Citation2024), highlighting the role of the epigenetic mechanism as a link between healthy conditions of the mother and the offspring.
The importance of maternal-associated mechanism of epigenetic regulation in the child is also emphasized by two studies with non-human primates models subject to western-style diet, associated with obesity (Nash et al. Citation2023; Sureshchandra et al. Citation2023). However, while in one study a mother western-style diet was associated with increased chromatin accessibility in inflammatory genes of juvenile’s hematopoietic stem and progenitor cells and bone marrow-derived macrophages (Nash et al. Citation2023), the other described that pregravid obesity is associated with reduced chromatin accessibility and lack of chromatin remodeling in umbilical cord monocytes, which resulted in attenuated response to bacterial and virus pathogens stimulation (Sureshchandra et al. Citation2023). The blunt reaction was not only observed in umbilical cord monocytes but also in peripheral blood monocytes and tissue-resident macrophages obtained from animals born from obese mothers, fed with a high-fat diet (Sureshchandra et al. Citation2023).
Another study in a human cohort verified that maternal obesity was associated with a higher level of plasma superoxide-dismutase activity, IL-6, and IL-7 in neonates (Hernández-Trejo et al., Citation2017). A study by Enninga et al. (Citation2021) reported that newborns from women with high BMI presented significantly increased levels of CD4+ T cells and decreased myeloid cell populations, as well as increased concentrations of IL-12p40 and macrophage-derived chemokine (Enninga et al., Citation2021). However, it remains to be clarified if these changes in immune and inflammatory profiles persist during child development. Interestingly, with the information that maternal obesity correlates with increased susceptibility to obesity and nonalcoholic fatty liver disease (NAFLD) in offspring, Soderborg and colleagues explored the effect of colonization of germ-free mice with stool microbes from 2-week-old infants form obese or lean mothers, (born vaginally, exclusively breastfed, and without exposure to antibiotics after delivery) (Soderborg et al. Citation2018). Colonization resulted, in the first animal group, in increased intestinal permeability, reduced macrophage phagocytosis, and decreased cytokine production suggestive of impaired macrophage function. Moreover, feeding the animals with a western-style diet resulted in excess weight and accelerated NAFLD in mice colonized with the microbiota from infants from obese mothers first (Soderborg et al. Citation2018). These results highlighted that the early gut dysbiosis have a clear impact in metabolism and inflammation, and may impact the proper myeloid development ().
Figure 3. Impact of maternal cardiometabolic disease and gut dysbiosis on early-life immunity (created with BioRender).
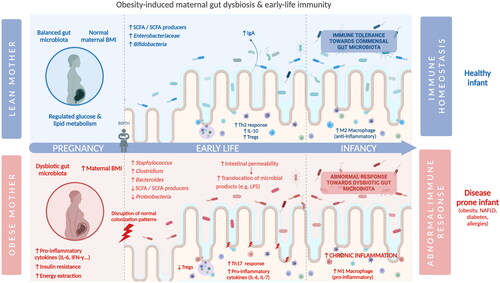
The importance of the initial seeding of early-life gut microbiota with maternal microbes in the development of the immune system of the child may be the reason why maternally transmitted strains are more likely to adapt and persist in the gut of the infant than non-maternally acquired strains (Azevedo et al., Citation2020; McDonald and McCoy Citation2019; Tamburini et al. Citation2016) (). In the case of obese mothers, it is possible to hypothesize that, if the maternal microbiota is vertically transmitted to the child and if this microbiota is already dysbiotic, combined with the fact that the immune system of the infant is influenced by the mother in early-life (e.g. in utero transmission of immune cells, immunoglobulins, and cytokines stimulated with maternal gut microbiota, IgA transmitted via breastfeeding stimulated with the maternal gut microbiota), this may predispose the colonization and selection of specific obesity-associated microorganisms. This microbiota would remain in the gut of the child and would perpetuate an obesogenic-microbiome-associated phenotype in early-life. The child may therefore be “primed” to be colonized by a dysbiotic maternally-transmitted gut microbiota and impairing the immune tolerance profile and overall obesogenic phenotype of the child.
3.3. The impact of other early-life factors on obesity
Other studies, while evaluating the potential role of maternal gut microbiota on the offspring gut microbiota, also reflected on the impact of additional factors, such as early-life nutrition and maternal diet, and how they correlated with early-life risk of developing obesity. In fact, as described in the first part of the review, the early-life gut microbiome is influenced by the birth mode, antibiotics, diet, and breast feeding. As such, studies linking obesity and gut microbiota in early-life that do not account for those variables may bias the conclusions obtained, and caution should be taken when analyzing the results. In humans, Tun et al. (Tun et al. Citation2018) also verified that, in a cohort of 935 mother-child pairs (aged 1 and 3 years), birth mode and gut microbiota of the infant (Bacillota richness, especially of the Lachnospiraceae family) mediated the association between maternal pre-pregnancy overweight and childhood overweight at ages 1 and 3 years, with children born via cesarean to overweight mothers being more at risk of developing childhood obesity, compared to infants born to healthy mother and delivered vaginally. However, the genera of Lachnospiraceae family differed between infants delivered vaginally and those delivered via cesarean birth. Haddad and colleagues focused on the impact of breastmilk exposure and maternal pre-pregnancy BMI in early-life gut microbiota and infant growth measures (BAZ – BMI-for-age z-score; LAZ – Length-for-age z-score; LWZ – Length-for-weight z-score; and WAZ – Weight-for-age z-score) in 33 mother-baby dyads, at 6 and 12 months postpartum. The authors observed that, although maternal BMI was positively correlated with infant anthropometric measures, it was not associated with gut microbiota composition. The differences in breastmilk exposure were the key drivers in microbial composition differences, with infants who did not receive breastmilk at or after the 6-month timepoint, displaying a profoundly different gut microbiota from infants who continued breastfeeding. Nonetheless, results indicated that increased exposure to maternal breastmilk resulted in decreased microbial richness, possibly due to the selective enrichment of bacterial species associated with HMO degradation. The authors suggest that other external factors, besides maternal BMI, such as diet, host genetics, and lifestyle may also impact early-life microbiota composition and, consequently, the child predisposition to develop diseases (Haddad et al., Citation2021).
The gut microbiota, can also be impacted by host-related factors, such as geographical location and ethnicity. Despite the fact that variations in gut microbiota composition across distinct ethnicities remains poorly explored, a few studies have started to uncover these differences. Concerning early-life, a recent study by Balakrishnan et al. assessed ethnic variability in the gut and oral microbiome of African American and European American children, and its correlation with childhood obesity (Balakrishnan et al., Citation2021). The authors observed significant differences in gut microbiota diversity (alpha and beta diversity) between both groups, and a prevalence of specific taxa depending on ethnicity (for instance, African American children displayed higher levels of Ruminococcaceae and Oxalobacter, which have been associated with obesity) (Balakrishnan et al., Citation2021). Other studies have found that these ethnicity-dependent differences in gut microbiota composition could be observed in infants as young as 3 months old, before the introduction of more complex diets, and that can persist throughout childhood (Mallott et al., Citation2023; Xu et al. Citation2020).
Important to note that the gut microbiome is not the only being associated with obesity. Many studies are now also exploring the role of oral microbiome in obesity. The oral microbiota harbors a diverse community of microbes that sustain the homeostasis of the oral cavity (Schamarek et al. Citation2023). The oral-gut axis has been explored not only due to its bidirectional link (since these niches share some taxa), but also for its impact in the development of metabolic pathologies, as recently reviewed (Yamazaki and Kamada Citation2024 and Schamarek et al. Citation2023)). In early-life, recent research have started to unravel the role of gut and oral in childhood obesity (Wang et al. Citation2023). Ma and colleagues characterized the oral and gut microbiota of a cohort of children, aged 3 to 5 years old, with and without obesity (Ma et al., Citation2023). The results indicated a significant difference in alpha-diversity between the two groups, with obese children displaying the highest diversity in the oral microbiota, and the lowest in the gut (Ma et al., Citation2023). Furthermore, oral samples of obese children were enriched in Filifactor and Butyrivibrio, while fecal samples presented higher abundance of Faecalibacterium, Tyzella and Klebsiella, considered biomarkers for obesity (Ma et al., Citation2023). Coker and co-authors assessed the relationship of oral microbiota composition and weigh gain and found that. beta-diversity was inversely correlated with infant BMI and z-scores (Coker et al., Citation2022). Concerning oral microbiota composition, the authors found certain taxa, namely Streptococcus mitis and Corynebacterium matruchotii, to be associated with weight gain (Coker et al., Citation2022). Despite these recent findings, both articles highlighted the need for further research, as the impact of oral microbiota (and its link to gut microbiota) in the development of childhood obesity remains poorly studied. It is also relevant to emphasize the potential role of the maternal oral microbiota, in the establishment of the offspring oral microbiota and health outcomes (Saadaoui et al. Citation2021).
4. Conclusion
The infant gut microbiota has a profound impact on the development and maturation of early-life immunity, by establishing a symbiotic link between the commensal microorganisms of the child and its immune system. In homeostasis, there is a tolerance system that strictly regulates and prevents an overreactive immune response toward the gut microbiota, allowing the proper development of the immune system of the child. However, disruptions to the gut microbiota during this period have been linked to the development of diseases, such as obesity. Recent studies have highlighted the importance of the mother as the main source of microorganisms to the offspring, particularly in the transmission of dysbiotic gut microbiota and its potential impact on future health. Research conducted to this point seems to support the proposed hypothesis that obesogenic maternal microbiota could lead to the development of obesity in children, namely through the transfer of bacterial species associated with higher BMI that persist and modulate the immune system of the infant, generating an abnormal immune response. Nevertheless, further studies (in comprehensive cohorts of mothers and infants) are necessary to understand the mechanisms associated with the interaction of dysbiotic gut microbiota and childhood obesity, as well as the mechanism that explains the reason why maternal strains are more likely to adapt and remain in the child’s gut.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. 2014. The placenta harbors a unique microbiome. Sci Transl Med. 6(237):237ra65. doi:10.1126/scitranslmed.3008599.
- Abenavoli L, Scarpellini E, Colica C, Boccuto L, Salehi B, Sharifi-Rad J, Aiello V, Romano B, De Lorenzo A, Izzo AA, et al. 2019. Gut microbiota and obesity: a role for probiotics. Nutrients. 11(11):2690. doi:10.3390/nu11112690.
- Agus A, Clément K, Sokol H. 2021. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 70(6):1174–1182. doi:10.1136/gutjnl-2020-323071.
- Al-Rashidi HE. 2022. Gut microbiota and immunity relevance in eubiosis and dysbiosis. Saudi J Biol Sci. 29(3):1628–1643. doi:10.1016/j.sjbs.2021.10.068.
- Amabebe E, Robert FO, Agbalalah T, Orubu ESF. 2020. Microbial dysbiosis-induced obesity: role of gut microbiota in homoeostasis of energy metabolism. Br J Nutr. 123(10):1127–1137. doi:10.1017/S0007114520000380.
- Annunziato F, Romagnani C, Romagnani S. 2015. The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol. 135(3):626–635. doi:10.1016/j.jaci.2014.11.001.
- Arrieta M-C, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 7(307):307ra152. doi:10.1126/scitranslmed.aab2271.
- Asadi A, Shadab Mehr N, Mohamadi MH, Shokri F, Heidary M, Sadeghifard N, et al. 2022. Obesity and gut-microbiota-brain axis: A narrative review. J Clin Lab Anal. 36(5):e24420.
- Azevedo MJ, Pereira ML, Araujo R, Ramalho C, Zaura E, Sampaio-Maia B. 2020. Influence of delivery and feeding mode in oral fungi colonization - a systematic review. Microb Cell. 7(2):36–45. doi:10.15698/mic2020.02.706.
- Balakrishnan B, Selvaraju V, Chen J, Ayine P, Yang L, Ramesh Babu J, Geetha T, Taneja V. 2021. Ethnic variability associating gut and oral microbiome with obesity in children. Gut Microbes. 13(1):1–15. doi:10.1080/19490976.2021.1882926.
- Basha S, Surendran N, Pichichero M. 2014. Immune responses in neonates. Expert Rev Clin Immunol. 10(9):1171–1184. doi:10.1586/1744666X.2014.942288.
- Beckers KF, Flanagan JP, Sones JL. 2024. Microbiome and pregnancy: focus on microbial dysbiosis coupled with maternal obesity. Int J Obes. 48(4):439–448. doi:10.1038/s41366-023-01438-7.
- Brink LR, Mercer KE, Piccolo BD, Chintapalli SV, Elolimy A, Bowlin AK, Matazel KS, Pack L, Adams SH, Shankar K, et al. 2020. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am J Clin Nutr. 111(6):1190–1202. doi:10.1093/ajcn/nqaa076.
- Byrne ME, O’Mahony JA, O’Callaghan TF. 2021. Compositional and functional considerations for bovine-, caprine-and plant-based infant formulas. Dairy. 2(4):695–715. doi:10.3390/dairy2040054.
- Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. 2013. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 14(5):559–570. doi:10.1016/j.chom.2013.10.004.
- Cerdó T, Nieto-Ruíz A, García-Santos JA, Rodríguez-Pöhnlein A, García-Ricobaraza M, Suárez A, Bermúdez MG, Campoy C. 2023. Current knowledge about the impact of maternal and infant nutrition on the development of the microbiota–gut–brain axis. Annu Rev Nutr. 43(1):251–278. doi:10.1146/annurev-nutr-061021-025355.
- Chen C-M, Chou H-C, Yang Y-C. 2021. Maternal antibiotic treatment disrupts the intestinal microbiota and intestinal development in neonatal mice. Front Microbiol. 12:684233. doi:10.3389/fmicb.2021.684233.
- Chi C, Xue Y, Lv N, Hao Y, Liu R, Wang Y, Ding X, Zeng H, Li G, Shen Q, et al. 2019. Longitudinal gut bacterial colonization and its influencing factors of low birth weight infants during the first 3 months of life. Front Microbiol. 10:1105. doi:10.3389/fmicb.2019.01105.
- Chichlowski M, van Diepen JA, Prodan A, Olga L, Ong KK, Kortman GAM, Dunger DB, Gross G. 2023. Early development of infant gut microbiota in relation to breastfeeding and human milk oligosaccharides. Front Nutr. 10:1003032. doi:10.3389/fnut.2023.1003032.
- Christensen ED, Hjelmsø MH, Thorsen J, Shah S, Redgwell T, Poulsen CE, Trivedi U, Russel J, Gupta S, Chawes BL, et al. 2022. The developing airway and gut microbiota in early life is influenced by age of older siblings. Microbiome. 10(1):106. doi:10.1186/s40168-022-01305-z.
- Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. 2016. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 8(1):77. doi:10.1186/s13073-016-0330-z.
- Coker MO, Lebeaux RM, Hoen AG, Moroishi Y, Gilbert-Diamond D, Dade EF, Palys TJ, Madan JC, Karagas MR. 2022. Metagenomic analysis reveals associations between salivary microbiota and body composition in early childhood. Sci Rep. 12(1):13075. doi:10.1038/s41598-022-14668-y.
- D’Amelio P, Sassi F. 2018. Gut microbiota, immune system, and bone. Calcif Tissue Int. 102(4):415–425. doi:10.1007/s00223-017-0331-y.
- de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, Parkhill J, Charnock-Jones DS, Smith GCS. 2019. Human placenta has no microbiome but can contain potential pathogens. Nature. 572(7769):329–334. doi:10.1038/s41586-019-1451-5.
- Di Gesù CM, Matz LM, Buffington SA. 2021. Diet-induced dysbiosis of the maternal gut microbiome in early life programming of neurodevelopmental disorders. Neurosci Res. 168:3–19. doi:10.1016/j.neures.2021.05.003.
- Dreisbach C, Prescott S, Siega-Riz AM, McCulloch J, Habermeyer L, Dudley D, Trinchieri G, Kelsey C, Alhusen J. 2023. Composition of the maternal gastrointestinal microbiome as a predictor of neonatal birth weight. Pediatr Res. 94(3):1158–1165. doi:10.1038/s41390-023-02584-4.
- Embleton ND, Sproat T, Uthaya S, Young GR, Garg S, Vasu V, Masi AC, Beck L, Modi N, Stewart CJ, et al. 2023. Effect of an exclusive human milk diet on the gut microbiome in preterm infants: a randomized clinical trial. JAMA Netw Open. 6(3):e231165. doi:10.1001/jamanetworkopen.2023.1165.
- Enninga EAL, Jang JS, Hur B, Johnson EL, Wick MJ, Sung J, et al. 2021. Maternal obesity is associated with phenotypic alterations in fetal immune cells by single-cell mass cytometry. Am J Reprod Immunol. 85(3):e13358.
- Escherich T. 1988. The intestinal bacteria of the neonate and breast-fed infant. 1884. Rev Infect Dis. 10(6):1220–1225. doi:10.1093/clinids/10.6.1220.
- Esteve-Solé A, Luo Y, Vlagea A, Deyà-Martínez Á, Yagüe J, Plaza-Martín AM, et al. 2018. B regulatory cells: players in pregnancy and early life. Int J Mol Sci. 19(7):2099.
- Ganal-Vonarburg SC, Duerr CU. 2020. The interaction of intestinal microbiota and innate lymphoid cells in health and disease throughout life. Immunology. 159(1):39–51. doi:10.1111/imm.13138.
- García-Mantrana I, Selma-Royo M, González S, Parra-Llorca A, Martínez-Costa C, Collado MC. 2020. Distinct maternal microbiota clusters are associated with diet during pregnancy: impact on neonatal microbiota and infant growth during the first 18 months of life. Gut Microbes. 11(4):962–978. doi:10.1080/19490976.2020.1730294.
- Gaudino SJ, Kumar P. 2019. Cross-talk between antigen presenting cells and T cells impacts intestinal homeostasis, bacterial infections, and tumorigenesis. Front Immunol. 10:360. doi:10.3389/fimmu.2019.00360.
- Gilley SP, Ruebel ML, Sims C, Zhong Y, Turner D, Lan RS, et al. 2022. Associations between maternal obesity and offspring gut microbiome in the first year of life. Pediatr Obes. 17(9):e12921.
- Gilley SP, Ruebel ML, Sims C, Zhong Y, Turner D, Lan RS, Pack LM, Piccolo BD, Chintapalli SV, Abraham A, et al. 2022. Associations between maternal obesity and offspring gut microbiome in the first year of life. Pediatric Obesity. 17(9):e12921. doi:10.1111/ijpo.12921.
- Haddad EN, Sugino KY, Kerver JM, Paneth N, Comstock SS. 2021. The infant gut microbiota at 12 months of age is associated with human milk exposure but not with maternal pre-pregnancy body mass index or infant BMI-for-age z-scores. Curr Res Physiol. 4:94–102. doi:10.1016/j.crphys.2021.03.004.
- Han L, Wang X-m, Di S, Gao Z-z, Li Q-w, Wu H-r, Wang Q, Zhao L-h, Tong X-l 2019. Innate lymphoid cells: a link between the nervous system and microbiota in intestinal networks. Mediators Inflamm. 2019:1–11. doi:10.1155/2019/1978094.
- Henderickx JGE, Zwittink RD, van Lingen RA, Knol J, Belzer C. 2019. The preterm gut microbiota: an inconspicuous challenge in nutritional neonatal care. Front Cell Infect Microbiol. 9:85. doi:10.3389/fcimb.2019.00085.
- Hernández-Trejo M, Montoya-Estrada A, Torres-Ramos Y, Espejel-Núñez A, Guzmán-Grenfell A, Morales-Hernández R, Tolentino-Dolores M, Laresgoiti-Servitje E. 2017. Oxidative stress biomarkers and their relationship with cytokine concentrations in overweight/obese pregnant women and their neonates. BMC Immunol. 18(1):3. doi:10.1186/s12865-016-0184-6.
- Hong JY, Medzhitov R. 2023. On developmental programming of the immune system. Trends Immunol. 44(11):877–889. doi:10.1016/j.it.2023.09.004.
- Isolauri E. 2012. Development of healthy gut microbiota early in life. J Paediatrics Child Health. 48(s3):1–6. doi:10.1111/j.1440-1754.2012.02489.x.
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139(3):485–498. doi:10.1016/j.cell.2009.09.033.
- Jain N. 2020. The early life education of the immune system: moms, microbes and (missed) opportunities. Gut Microbes. 12(1):1824564. doi:10.1080/19490976.2020.1824564.
- Jašarević E, Bale TL. 2019. Prenatal and postnatal contributions of the maternal microbiome on offspring programming. Front Neuroendocrinol. 55:100797. doi:10.1016/j.yfrne.2019.100797.
- Jones JM, Reinke SN, Mousavi-Derazmahalleh M, Garssen J, Jenmalm MC, Srinivasjois R, Silva D, Keelan J, Prescott SL, Palmer DJ, et al. 2024. Maternal prebiotic supplementation during pregnancy and lactation modifies the microbiome and short chain fatty acid profile of both mother and infant. Clin Nutr. 43(4):969–980. doi:10.1016/j.clnu.2024.02.030.
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. 2011. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 94(1):58–65. doi:10.3945/ajcn.110.010132.
- Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y, et al. 2015. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 15(1):100. doi:10.1186/s12876-015-0330-2.
- Kato LM, Kawamoto S, Maruya M, Fagarasan S. 2014. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev. 260(1):67–75. doi:10.1111/imr.12185.
- Kim B, Choi HN, Yim JE. 2019. Effect of diet on the gut microbiota associated with obesity. JOMES. 28(4):216–224. doi:10.7570/jomes.2019.28.4.216.
- Kimura I, Miyamoto J, Ohue-Kitano R, Watanabe K, Yamada T, Onuki M, Aoki R, Isobe Y, Kashihara D, Inoue D, et al. 2020. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. 367(6481), eaaw8429. doi:10.1126/science.aaw8429.
- Kincaid HJ, Nagpal R, Yadav H. 2020. Microbiome-immune-metabolic axis in the epidemic of childhood obesity: evidence and opportunities. Obes Rev. 21(2):e12963.
- Korpela K, Blakstad EW, Moltu SJ, Strømmen K, Nakstad B, Rønnestad AE, Brække K, Iversen PO, Drevon CA, de Vos W, et al. 2018. Intestinal microbiota development and gestational age in preterm neonates. Sci Rep. 8(1):2453. doi:10.1038/s41598-018-20827-x.
- Korpela K, Salonen A, Vepsäläinen O, Suomalainen M, Kolmeder C, Varjosalo M, Miettinen S, Kukkonen K, Savilahti E, Kuitunen M, et al. 2018. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome. 6(1):182. doi:10.1186/s40168-018-0567-4.
- Krause BJ, Vega-Tapia FA, Soto-Carrasco G, Lefever I, Letelier C, Saez CG, Castro-Rodriguez JA. 2023. Maternal obesity and high leptin levels prime pro-inflammatory pathways in human cord blood leukocytes. Placenta. 142:75–84. doi:10.1016/j.placenta.2023.08.069.
- Kurachi M. 2019. CD8+ T cell exhaustion. Semin Immunopathol. 41(3):327–337. doi:10.1007/s00281-019-00744-5.
- Lajqi T, Köstlin-Gille N, Bauer R, Zarogiannis SG, Lajqi E, Ajeti V, Dietz S, Kranig SA, Rühle J, Demaj A, et al. 2023. Training vs. Tolerance: the Yin/Yang of the Innate Immune System. Biomedicines. 11(3):766. doi:10.3390/biomedicines11030766.
- Laursen MF, Zachariassen G, Bahl MI, Bergström A, Høst A, Michaelsen KF, Licht TR. 2015. Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol. 15(1):154. doi:10.1186/s12866-015-0477-6.
- Lin C, Lin Y, Zhang H, Wang G, Zhao J, Zhang H, Chen W. 2022. Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life. Nutrients. 14(7):1498. doi:10.3390/nu14071498.
- Liu X, Li X, Xia B, Jin X, Zou Q, Zeng Z, Zhao W, Yan S, Li L, Yuan S, et al. 2021. High-fiber diet mitigates maternal obesity-induced cognitive and social dysfunction in the offspring via gut-brain axis. Cell Metab. 33(5):923–938. e6. doi:10.1016/j.cmet.2021.02.002.
- Liu Y, Qin S, Song Y, Feng Y, Lv N, Xue Y, Liu F, Wang S, Zhu B, Ma J, et al. 2019. The perturbation of infant gut microbiota caused by cesarean delivery is partially restored by exclusive breastfeeding. Front Microbiol. 10:598. doi:10.3389/fmicb.2019.00598.
- Longo S, Rizza S, Federici M. 2023. Microbiota-gut-brain axis: relationships among the vagus nerve, gut microbiota, obesity, and diabetes. Acta Diabetol. 60(8):1007–1017. doi:10.1007/s00592-023-02088-x.
- Lotz M, Gütle D, Walther S, Ménard S, Bogdan C, Hornef MW. 2006. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med. 203(4):973–984. doi:10.1084/jem.20050625.
- Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, Zhuo N, Wang H, Wang L, Wu D, et al. 2020. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep. 10(1):15792. doi:10.1038/s41598-020-72635-x.
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, et al. 2014. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. 5(1):3889. doi:10.1038/ncomms4889.
- Ma T, Wu Z, Lin J, Shan C, Abasijiang A, Zhao J. 2023. Characterization of the oral and gut microbiome in children with obesity aged 3 to 5 years. Front Cell Infect Microbiol. 13:1102650. doi:10.3389/fcimb.2023.1102650.
- Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, Balamurugan R. 2020. The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients. 12(5):1474. doi:10.3390/nu12051474.
- Mallott EK, Sitarik AR, Leve LD, Cioffi C, Camargo CA, Hasegawa K, Bordenstein SR. 2023. Human microbiome variation associated with race and ethnicity emerges as early as 3 months of age. PLoS Biol. 21(8):e3002230. doi:10.1371/journal.pbio.3002230.
- Maranduba CMdC, De Castro SBR, Souza GTd, Rossato C, da Guia FC, Valente MAS, Rettore JVP, Maranduba CP, Souza CMd, Carmo AMRd, et al. 2015. Intestinal microbiota as modulators of the immune system and neuroimmune system: impact on the host health and homeostasis. J Immunol Res. 2015:1–14. doi:10.1155/2015/931574.
- Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, et al. 2016. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS ONE. 11(6):e0158498. doi:10.1371/journal.pone.0158498.
- McDonald B, McCoy KD. 2019. Maternal microbiota in pregnancy and early life. Science. 365(6457):984–985. doi:10.1126/science.aay0618.
- Melville JM, Moss TJ. 2013. The immune consequences of preterm birth. Front Neurosci. 7:79. doi:10.3389/fnins.2013.00079.
- Mendes J, Areia AL, Rodrigues-Santos P, Santos-Rosa M, Mota-Pinto A. 2020. Innate lymphoid cells in human pregnancy. Front Immunol. 11:551707. doi:10.3389/fimmu.2020.551707.
- Méndez CS, Bueno SM, Kalergis AM. 2021. Contribution of Gut Microbiota to Immune Tolerance in Infants. J Immunol Res. 2021:1–11. doi:10.1155/2021/7823316.
- Menni C, Jackson MA, Pallister T, Steves CJ, Spector TD, Valdes AM. 2017. Gut microbiome diversity and high-fibre intake are related to lower long-term weight gain. Int J Obes. 41(7):1099–1105.). doi:10.1038/ijo.2017.66.
- Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, et al. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 81(4):e00036-17. doi:10.1128/MMBR.00036-17.
- Milano W, Carizzone F, Foia M, Marchese M, Milano M, Saetta B, Capasso A. 2022. Obesity and its multiple clinical implications between inflammatory states and gut microbiotic alterations. Diseases. 11(1):7. doi:10.3390/diseases11010007.
- Mittrücker HW, Visekruna A, Huber M. 2014. Heterogeneity in the differentiation and function of CD8+ T cells. Arch Immunol Ther Exp. 62(6):449–458. doi:10.1007/s00005-014-0293-y.
- Moore RE, Townsend SD. 2019. Temporal development of the infant gut microbiome. Open Biol. 9(9):190128. doi:10.1098/rsob.190128.
- Nash MJ, Dobrinskikh E, Soderborg TK, Janssen RC, Takahashi DL, Dean TA, Varlamov O, Hennebold JD, Gannon M, Aagaard KM, et al. 2023. Maternal diet alters long-term innate immune cell memory in fetal and juvenile hematopoietic stem and progenitor cells in nonhuman primate offspring. Cell Rep. 42(4):112393. doi:10.1016/j.celrep.2023.112393.
- Neff CP, Krueger O, Xiong K, Arif S, Nusbacher N, Schneider JM, Cunningham AW, Armstrong A, Li S, McCarter MD, et al. 2018. Fecal microbiota composition drives immune activation in HIV-infected individuals. EBioMedicine. 30:192–202. doi:10.1016/j.ebiom.2018.03.024.
- Nieto-Ruiz A, Cerdó T, Jordano B, Torres-Espínola FJ, Escudero-Marín M, García-Ricobaraza M, Bermúdez MG, García-Santos JA, Suárez A, Campoy C, et al. 2023. Maternal weight, gut microbiota, and the association with early childhood behavior: the PREOBE follow-up study. Child Adolesc Psychiatry Ment Health. 17(1):41. doi:10.1186/s13034-023-00589-9.
- Owen JL, Mohamadzadeh M. 2013. Microbial activation of gut dendritic cells and the control of mucosal immunity. J Interferon Cytokine Res. 33(11):619–631. doi:10.1089/jir.2013.0046.
- Pärnänen KMM, Hultman J, Markkanen M, Satokari R, Rautava S, Lamendella R, Wright J, McLimans CJ, Kelleher SL, Virta MP, et al. 2022. Early-life formula feeding is associated with infant gut microbiota alterations and an increased antibiotic resistance load. Am J Clin Nutr. 115(2):407–421. doi:10.1093/ajcn/nqab353.
- Payne MS, Bayatibojakhi S. 2014. Exploring preterm birth as a polymicrobial disease: an overview of the uterine microbiome. Front Immunol. 5:595. doi:10.3389/fimmu.2014.00595.
- Rautava S, Luoto R, Salminen S, Isolauri E. 2012. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol. 9(10):565–576. doi:10.1038/nrgastro.2012.144.
- Ren Y, Zeng Y, Wu Y, Yu J, Zhang Q, Xiao X. 2023. The Role of Gut Microbiota in Gestational Diabetes Mellitus Affecting Intergenerational Glucose Metabolism: possible Mechanisms and Interventions. Nutrients. 15(21):4551. doi:10.3390/nu15214551.
- Robertson RC, Manges AR, Finlay BB, Prendergast AJ. 2019. The human microbiome and child growth - first 1000 days and beyond. Trends Microbiol. 27(2):131–147. doi:10.1016/j.tim.2018.09.008.
- Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, et al. 2015. The composition of the gut microbiota throughout life, with an emphasis on early life. Microbial Ecol Health Dis. 26(0):26050. doi:10.3402/mehd.v26.26050.
- Rougé C, Goldenberg O, Ferraris L, Berger B, Rochat F, Legrand A, Göbel UB, Vodovar M, Voyer M, Rozé J-C, et al. 2010. Investigation of the intestinal microbiota in preterm infants using different methods. Anaerobe. 16(4):362–370. doi:10.1016/j.anaerobe.2010.06.002.
- Ruan W, Engevik MA, Spinler JK, Versalovic J. 2020. Healthy human gastrointestinal microbiome: composition and function after a decade of exploration. Dig Dis Sci. 65(3):695–705. doi:10.1007/s10620-020-06118-4.
- Rubini E, Schenkelaars N, Rousian M, Sinclair KD, Wekema L, Faas MM, Steegers-Theunissen RPM, Schoenmakers S. 2022. Maternal obesity during pregnancy leads to derangements in one-carbon metabolism and the gut microbiota: implications for fetal development and offspring wellbeing. Am J Obstet Gynecol. 227(3):392–400. doi:10.1016/j.ajog.2022.04.013.
- Saadaoui M, Singh P, Al Khodor S. 2021. Oral microbiome and pregnancy: A bidirectional relationship. J Reproduct Immunol. 145:103293. doi:10.1016/j.jri.2021.103293.
- Sajdel-Sulkowska EM. 2023. The impact of maternal gut microbiota during pregnancy on fetal gut–brain axis development and life-long health outcomes. Microorganisms. 11(9):2199. doi:10.3390/microorganisms11092199.
- Schamarek I, Anders L, Chakaroun RM, Kovacs P, Rohde-Zimmermann K. 2023. The role of the oral microbiome in obesity and metabolic disease: potential systemic implications and effects on taste perception. Nutr J. 22(1):28. doi:10.1186/s12937-023-00856-7.
- Selma-Royo M, Tarrazó M, García-Mantrana I, Gómez-Gallego C, Salminen S, Collado MC. 2019. Shaping microbiota during the first 1000 days of life. Adv Exp Med Biol. 1125:3–24.
- Sen T, Cawthon CR, Ihde BT, Hajnal A, DiLorenzo PM, de La Serre CB, Czaja K. 2017. Diet-driven microbiota dysbiosis is associated with vagal remodeling and obesity. Physiology & Behavior. 173:305–317. doi:10.1016/j.physbeh.2017.02.027.
- Senn V, Bassler D, Choudhury R, Scholkmann F, Righini-Grunder F, Vuille-dit-Bille RN, Restin T. 2020. Microbial Colonization From the Fetus to Early Childhood-A Comprehensive Review. Front Cell Infect Microbiol. 10:573735. doi:10.3389/fcimb.2020.573735.
- Silva YP, Bernardi A, Frozza RL. 2020. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol. 11:25. doi:10.3389/fendo.2020.00025.
- Sim K, Powell E, Cornwell E, Simon Kroll J, Shaw AG. 2023. Development of the gut microbiota during early life in premature and term infants. Gut Pathog. 15(1):3. doi:10.1186/s13099-022-00529-6.
- Singh R, Zogg H, Wei L, Bartlett A, Ghoshal UC, Rajender S, Ro S. 2021. Gut microbial dysbiosis in the pathogenesis of gastrointestinal dysmotility and metabolic disorders. J Neurogastroenterol Motil. 27(1):19–34. doi:10.5056/jnm20149.
- Singh SB, Madan J, Coker M, Hoen A, Baker ER, Karagas MR, Mueller NT. 2020. Does birth mode modify associations of maternal pre-pregnancy BMI and gestational weight gain with the infant gut microbiome? Int J Obes. 44(1):23–32. doi:10.1038/s41366-018-0273-0.
- Skrypnik D, Bogdanski P, Zawiejska A, Wender-Ozegowska E. 2019. Role of gestational weight gain, gestational diabetes, breastfeeding, and hypertension in mother-to-child obesity transmission. Pol Arch Intern Med. 129(4):267–275.
- Soderborg TK, Borengasser SJ, Barbour LA, Friedman JE. 2016. Microbial transmission from mothers with obesity or diabetes to infants: an innovative opportunity to interrupt a vicious cycle. Diabetologia. 59(5):895–906. doi:10.1007/s00125-016-3880-0.
- Soderborg TK, Clark SE, Mulligan CE, Janssen RC, Babcock L, Ir D, Young B, Krebs N, Lemas DJ, Johnson LK, et al. 2018. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun. 9(1):4462. doi:10.1038/s41467-018-06929-0.
- Sommer F, Bäckhed F. 2013. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 11(4):227–238. doi:10.1038/nrmicro2974.
- Stagg AJ. 2018. Intestinal dendritic cells in health and gut inflammation. Front Immunol. 9:2883. doi:10.3389/fimmu.2018.02883.
- Stanislawski MA, Dabelea D, Wagner BD, Iszatt N, Dahl C, Sontag MK, Knight R, Lozupone CA, Eggesbø M. 2018. Gut microbiota in the first 2 years of life and the association with body mass index at age 12 in a norwegian birth cohort. mBio. 9(5):e01751–18. doi:10.1128/mBio.01751-18.
- Sugino KY, Hernandez TL, Barbour LA, Kofonow JM, Frank DN, Friedman JE. 2022. A maternal higher-complex carbohydrate diet increases bifidobacteria and alters early life acquisition of the infant microbiome in women with gestational diabetes mellitus. Front Endocrinol. 13:921464. doi:10.3389/fendo.2022.921464.
- Sun T, Nguyen A, Gommerman JL. 2020. Dendritic cell subsets in intestinal immunity and inflammation. J Immunol. 204(5):1075–1083. doi:10.4049/jimmunol.1900710.
- Sureshchandra S, Doratt BM, Mendza N, Varlamov O, Rincon M, Marshall NE, Messaoudi I. 2023. Maternal obesity blunts antimicrobial responses in fetal monocytes. Elife. 12:e81320. doi:10.7554/eLife.81320.
- Swiatczak B, Rescigno M. 2012. How the interplay between antigen presenting cells and microbiota tunes host immune responses in the gut. Semin Immunol. 24(1):43–49. doi:10.1016/j.smim.2011.11.004.
- Tamburini S, Shen N, Wu HC, Clemente JC. 2016. The microbiome in early life: implications for health outcomes. Nat Med. 22(7):713–722. doi:10.1038/nm.4142.
- Tanaka M, Nakayama J. 2017. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Inter Offic J Japan Soc Allergol. 66(4):515–522. doi:10.1016/j.alit.2017.07.010.
- Tezuka H, Ohteki T. 2019. Regulation of IgA production by intestinal dendritic cells and related cells. Front Immunol. 10:1891. doi:10.3389/fimmu.2019.01891.
- Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature. 535(7610):65–74. doi:10.1038/nature18847.
- Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, Roberts LK, Wong CHY, Shim R, Robert R, et al. 2015. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 6(1):7320. doi:10.1038/ncomms8320.
- Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, Becker AB, Mandhane PJ, Turvey SE, Subbarao P, Sears MR, et al. 2018. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. JAMA Pediatr. 172(4):368–377. doi:10.1001/jamapediatrics.2017.5535.
- Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, Guttman DS, Becker AB, Mandhane PJ, Turvey SE, et al. 2017. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome. 5(1):40. doi:10.1186/s40168-017-0254-x.
- Turta O, Selma-Royo M, Kumar H, Collado MC, Isolauri E, Salminen S, Rautava S. 2022. Maternal intrapartum antibiotic treatment and gut microbiota development in healthy term infants. Neonatology. 119(1):93–102. doi:10.1159/000519574.
- Valiathan R, Ashman M, Asthana D. 2016. Effects of ageing on the immune system: infants to elderly. Scand J Immunol. 83(4):255–266. doi:10.1111/sji.12413.
- van den Berg JP, Westerbeek EA, van der Klis FR, Berbers GA, van Elburg RM. 2011. Transplacental transport of IgG antibodies to preterm infants: a review of the literature. Early Hum Dev. 87(2):67–72. doi:10.1016/j.earlhumdev.2010.11.003.
- van den Elsen LWJ, Garssen J, Burcelin R, Verhasselt V. 2019. Shaping the gut microbiota by breastfeeding: the gateway to allergy prevention? Front Pediatr. 7:47. doi:10.3389/fped.2019.00047.
- Ventura M, Milani C, Lugli GA, van Sinderen D. 2019. Health benefits conferred by the human gut microbiota during infancy. Microb Biotechnol. 12(2):243–248. doi:10.1111/1751-7915.13334.
- Waldrop SW, Niemiec S, Wood C, Gyllenhammer LE, Jansson T, Friedman JE, Tryggestad JB, Borengasser SJ, Davidson EJ, Yang IV, et al. 2024. Cord blood DNA methylation of immune and lipid metabolism genes is associated with maternal triglycerides and child adiposity. Obesity (Silver Spring. 32(1):187–199. doi:10.1002/oby.23915.
- Walters KE, Martiny JBH. 2020. Alpha-, beta-, and gamma-diversity of bacteria varies across habitats. PLoS ONE. 15(9):e0233872. doi:10.1371/journal.pone.0233872.
- Wampach L, Heintz-Buschart A, Fritz JV, Ramiro-Garcia J, Habier J, Herold M, Narayanasamy S, Kaysen A, Hogan AH, Bindl L, et al. 2018. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat Commun. 9(1):5091. doi:10.1038/s41467-018-07631-x.
- Wang K, Dong H, Qi Y, Pei Z, Yi S, Yang X, et al. 2017. Lactobacillus casei regulates differentiation of Th17/Treg cells to reduce intestinal inflammation in mice. Can J Vet Res. 81(2):122–128.
- Wang L, Zhu L, Qin S. 2019. Gut microbiota modulation on intestinal mucosal adaptive immunity. Journal of Immunology Research. 2019:1–10. doi:10.1155/2019/4735040.
- Wang Q, McLoughlin RM, Cobb BA, Charrel-Dennis M, Zaleski KJ, Golenbock D, Tzianabos AO, Kasper DL. 2006. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J Exp Med. 203(13):2853–2863. doi:10.1084/jem.20062008.
- Wang W, Yan Y, Yu F, Zhang W, Su S. 2023. Role of oral and gut microbiota in childhood obesity. Folia Microbiol. 68(2):197–206. doi:10.1007/s12223-023-01033-3.
- Wang Y, Xu H, Zheng X, Wei H, Sun R, Tian Z. 2007. High expression of NKG2A/CD94 and low expression of granzyme B are associated with reduced cord blood NK cell activity. Cell Mol Immunol. 4(5):377–382.
- Weaver CT, Elson CO, Fouser LA, Kolls JK. 2013. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol Mech Dis. 8(1):477–512. doi:10.1146/annurev-pathol-011110-130318.
- Wiertsema SP, van Bergenhenegouwen J, Garssen J, Knippels LMJ. 2021. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients. 13(3):886. doi:10.3390/nu13030886.
- Wilson RM, Marshall NE, Jeske DR, Purnell JQ, Thornburg K, Messaoudi I. 2015. Maternal obesity alters immune cell frequencies and responses in umbilical cord blood samples. Pediatric Allergy Immunology. 26(4):344–351. doi:10.1111/pai.12387.
- Wilson RM, Messaoudi I. 2015. The impact of maternal obesity during pregnancy on offspring immunity. Mol Cell Endocrinol. 418(0 2):134–142. doi:10.1016/j.mce.2015.07.028.
- Xu J, Lawley B, Wong G, Otal A, Chen L, Ying TJ, Lin X, Pang WW, Yap F, Chong Y-S, et al. 2020. Ethnic diversity in infant gut microbiota is apparent before the introduction of complementary diets. Gut Microbes. 11(5):1362–1373. doi:10.1080/19490976.2020.1756150.
- Yamazaki K, Kamada N. 2024. Exploring the oral-gut linkage: interrelationship between oral and systemic diseases. Mucosal Immunol. 17(1):147–153. doi:10.1016/j.mucimm.2023.11.006.
- Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, Sun J, Pan F, Zhou J, Zhang W, et al. 2020. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 11(1):4457. doi:10.1038/s41467-020-18262-6.
- Yao Y, Cai X, Ye Y, Wang F, Chen F, Zheng C. 2021. The role of microbiota in infant health: from early life to adulthood. Front Immunol. 12:708472. doi:10.3389/fimmu.2021.708472.
- Yap PSX, Chong CW, Ahmad Kamar A, Yap IKS, Choo YM, Lai NM, Teh CSJ. 2021. Neonatal intensive care unit (NICU) exposures exert a sustained influence on the progression of gut microbiota and metabolome in the first year of life. Sci Rep. 11(1):1353. doi:10.1038/s41598-020-80278-1.
- Yoo JY, Groer M, Dutra SVO, Sarkar A, McSkimming DI. 2020. Gut microbiota and immune system interactions. Microorganisms. 8(12):1587. doi:10.3390/microorganisms8101587.
- Yu JC, Khodadadi H, Malik A, Davidson B, Salles ÉdSL, Bhatia J, Hale VL, Baban B. 2018. Innate immunity of neonates and infants. Front Immunol. 9:1759. doi:10.3389/fimmu.2018.01759.
- Zheng M, Zhu J. 2022. Innate Lymphoid Cells and Intestinal Inflammatory Disorders. IJMS. 23(3):1856. doi:10.3390/ijms23031856.
- Zhuang L, Chen H, Zhang S, Zhuang J, Li Q, Feng Z. 2019. Intestinal microbiota in early life and its implications on childhood health. Genomics Proteomics Bioinformatics. 17(1):13–25. doi:10.1016/j.gpb.2018.10.002.
- Zoratti E, Panzer A, Sitarik A, Jones K, Wegienka G, Havstad S, Lukacs N, Boushey H, Johnson CC, Ownby D, et al. 2020. Prenatal indoor dog exposure and early life gut microbiota in the microbes, asthma, allergy and pets birth cohort. Journal of Allergy and Clinical Immunology. 145(2):AB185. doi:10.1016/j.jaci.2019.12.325.