Abstract
Molecular medicine has entered a high-tech age that provides curative treatments of complex genetic diseases through genetically engineered cellular medicinal products. Their clinical implementation requires the ability to stably integrate genetic information through gene transfer vectors in a safe, effective and economically viable manner. The latest generation of Sleeping Beauty (SB) transposon vectors fulfills these requirements, and may overcome limitations associated with viral gene transfer vectors and transient non-viral gene delivery approaches that are prevalent in ongoing pre-clinical and translational research. The SB system enables high-level stable gene transfer and sustained transgene expression in multiple primary human somatic cell types, thereby representing a highly attractive gene transfer strategy for clinical use. Here we review several recent refinements of the system, including the development of optimized transposons and hyperactive SB variants, the vectorization of transposase and transposon as mRNA and DNA minicircles (MCs) to enhance performance and facilitate vector production, as well as a detailed understanding of SB’s genomic integration and biosafety features. This review also provides a perspective on the regulatory framework for clinical trials of gene delivery with SB, and illustrates the path to successful clinical implementation by using, as examples, gene therapy for age-related macular degeneration (AMD) and the engineering of chimeric antigen receptor (CAR)-modified T cells in cancer immunotherapy.
Introduction: vectors in gene therapy and genetic engineering
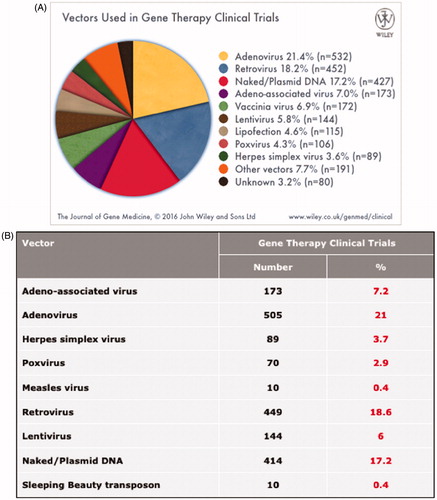
The ability to deliver natural or synthetic genes into human somatic cell types provides the technological basis for gene therapy to treat inherited and acquired genetic diseases, and enables genetic engineering of cellular medicinal products that are endowed with novel properties and functions for use as diagnostic and therapeutic tools in medicine. The vast majority (∼70%) of gene delivery systems used in ongoing clinical trials of gene therapy are based on viral vectors, and only a small proportion of trials utilizes non-viral delivery systems, mainly non-integrating naked plasmid DNA, that only provide transient gene expression (Gene Therapy Clinical Trials Worldwide, http://www.abedia.com/wiley/vectors.php ().
Figure 1. Viral and non-viral vectors in currently running gene therapy clinical trials. A) Percent contribution of different vector systems of currently running clinical trials. B) List of some of the most popular vector systems employed in clinical trials. The list also contains the currently running trials with the Sleeping Beauty transposon (see colour version of this figure at www.tandfonline.com/ibmg).
This article will focus on non-viral stable gene transfer through the Sleeping Beauty (SB) transposon as a viable and ready-to-use alternative to viral gene transfer vectors. We will review basic principles, and feature the key scientific and technologic advances that have been made to position SB transposition as a preferred gene delivery system in translational and clinical medicine. The conceptual appeal of SB transposition for gene delivery is illustrated by examples of clinical applications in gene therapy and genetic engineering, and complemented by a concise review of regulatory aspects for clinical implementation.
Viral gene transfer vectors
Viral vector systems are attractive for gene delivery, because viruses have evolved the ability to cross through cellular membranes by infection, thereby delivering nucleic acids to target cells. However, some viral vectors, including those derived from adenoviruses or adeno-associated viruses (AAVs), are not equipped for chromosomal integration, and thus remain largely episomal. Especially in cycling cells, episomal DNA is gradually lost requiring re-administration in vivo in order to sustain a desired level of transgene expression over time. However, repeated delivery can provoke immune responses against vector-encoded proteins [reviewed in (Hartman et al., Citation2008)], which can manifest as loss of the desired therapeutic effect and potentially therapy-related adverse events. In contrast, retroviral vectors integrate their therapeutic cargo into the genome, and thus have the potential to confer long-term transgene expression. Indeed, hematopoietic stem cell (HSC)-based gene therapy with integrating viral vectors has clearly provided therapeutic benefit in primary immunodeficiencies (including SCID-X1, ADA-SCID), thalassemia and leukodystrophies (Aiuti et al., Citation2013a; Biffi et al., Citation2013; Cartier et al., Citation2009; Cavazzana-Calvo et al., Citation2010). A concern with retroviral vectors is that chromosomal integration may be associated with genotoxicity and mutagenic effects elicited by insertion of the vector into or near proto-oncogenes (Baum et al., Citation2004; Deichmann et al., Citation2007; Hacein-Bey-Abina et al., Citation2003; Hacein-Bey-Abina et al., Citation2008; Kustikova et al., Citation2005). Such risk is especially pronounced with gammaretroviral vectors based on the murine leukemia virus (MLV) that preferentially integrate into transcriptional regulatory elements of active genes (Cattoglio et al., Citation2010; Cavazza et al., Citation2013; De Ravin et al., Citation2014); in fact, severe adverse events associated with vector integration have been observed in clinical trials for SCID-X1 (Hacein-Bey-Abina et al., Citation2003; Hacein-Bey-Abina et al., Citation2008; Howe et al., Citation2008; Thrasher et al., Citation2006), X-CGD (Ott et al., Citation2006) and WAS (Braun et al., Citation2014).
Furthermore, HIV-derived lentiviral vectors are potential mutagens due to their biased insertion into transcription units (Cavazza et al., Citation2013). Although lentiviral vectors appear to be safer than gammaretroviral vectors in gene therapy, recent studies indicate that some HIV integrations into genes associated with cancer or cell cycle regulation may confer a survival advantage of HIV-infected cells, and thus a clonal imbalance of HIV integrations in AIDS patients (Maldarelli et al., Citation2014; Wagner et al., Citation2014). A potential technical hurdle that can set limitations to vector design for clinical use of retroviral vectors is that large transgenes may inhibit viral reverse transcription and packaging. Finally, the high costs associated with manufacture of clinical-grade retroviral vector batches seem incompatible with their use as gene delivery systems in large patient cohorts in routine medical practice.
As a result, significant efforts have been made to craft novel gene transfer vectors that exceed the qualities of currently available viral vectors in: i) efficacy: i.e. achieve high-level stable gene transfer at low toxicity to the host cell; ii) safety: i.e. induce low levels of genotoxicity and possess a “safe” integration profile, with a high proportion of integrations into genomic safe harbors (GSHs); iii) economic viability: i.e. are associated with acceptable cost per treatment, and scalable/exportable vector production to serve large numbers of patients.
Non-viral gene transfer vectors
Non-viral vector systems, including nucleic acid vectors such as plasmid DNA, generally suffer from inefficient cellular delivery, pronounced cellular toxicity and limited duration of transgene expression due to the lack of genomic insertion and resulting degradation and/or dilution of the vector in transfected cell populations. Recent developments of non-viral delivery techniques, including liposomal formulations, nanoparticles, advanced electroporation methods such as nucleofection and cell-penetrating peptides can significantly enhance transfer of nucleic acids into therapeutically relevant cell types. However, just like non-integrating viral systems, non-viral vectors do not provide long-term nuclear maintenance and transgene expression in dividing cell types such as stem cells. One class of non-viral vector system that unites favorable characteristics of integrating viral vectors (i.e. stable chromosomal integration and long-lasting transgene expression) with those of non-viral delivery systems (i.e. lower immunogenicity, enhanced safety profile and reduced costs of GMP manufacture) is transposon-based gene delivery systems.
Non-viral gene transfer using the Sleeping Beauty transposon
Jumping genes: transposon basics
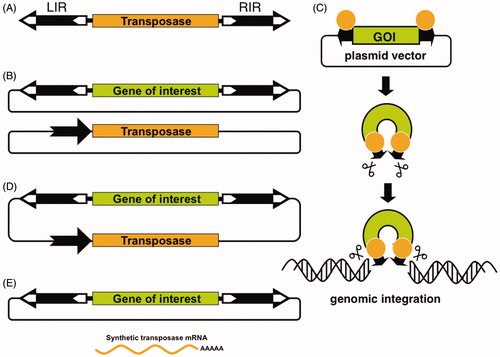
DNA transposons are genetic elements with the ability to change their positions within the genome. The prevalent mode of transposition in the genome is via a cut-and-paste mechanism. In nature, these elements exist as mobile (“jumping”) units of DNA containing a transposase gene flanked by terminal inverted repeats (TIRs) that carry transposase binding sites (). Importantly, it is possible to separate the two functional components of the transposon (the TIRs and the transposase) in the form of bi-component vector systems. Transposon-based vectors enable incorporation of virtually any DNA sequence of interest between the transposon TIRs and mobilization by trans-supplementing the transposase in the form of an expression plasmid (), very much like non-autonomous viral vectors are produced in packaging cell lines. In the transposition process, the transposase enzyme mediates the excision of the element from its donor plasmid, followed by reintegration of the transposon into a chromosomal locus (). This feature makes transposons natural and easily controllable DNA delivery vehicles that can be used as tools for versatile applications in genetic engineering, including gene therapy.
Figure 2. The Sleeping Beauty transposon system. A) Autonomous transposable elements consist of terminal inverted repeats (LIR = left inverted repeat; RIR = right inverted repeat, black arrows) that flank the transposase gene (yellow). B) Bi-component, trans-arrangement transposon vector system for delivering transgenes that are maintained in plasmids. One component contains a gene of interest (green) between the transposon TIRs carried by a plasmid vector, whereas the other component is a transposase expression plasmid, in which the black arrow represents the promoter driving expression of the transposase. C) The transposon carrying a gene of interest (GOI) is excised from the donor plasmid and is integrated at a chromosomal site by the transposase. D) One-vector or cis-arrangement, in which the transposase expression cassette and the GOI are located on the same plasmid. E) Plasmid-based transposon cassettes can be mobilized by transposase supplied as mRNA (see colour version of this figure at www.tandfonline.com/ibmg).
Discovery of the Sleeping Beauty transposon
Based on ancient, inactive transposon sequences isolated from fish genomes, an active transposon was reconstructed, and named SB after the Grimm brothers’ famous fairy tale, because it was literally awakened after a long evolutionary “sleep” (Ivics et al., Citation1997). SB was the first transposon ever shown capable of efficient transposition in vertebrate cells, thereby enabling new avenues for genetic engineering, including gene therapy [reviewed in (Boehme et al., Citation2015; Hackett et al., Citation2010; Citation2011; Ivics et al., Citation2009; Izsvak et al., Citation2010; Ivics & Izsvak, Citation2010, Citation2011; Narayanavari et al., Citation2016; Vandendriessche et al., Citation2009)].
The advantage of SB transposon-based gene delivery is that it combines the favorable features of viral vectors with those of naked DNA molecules. Namely, owing to permanent genomic insertion of transgene constructs (), transposition-mediated gene delivery can lead to sustained and efficient transgene expression in preclinical animal models (Hackett et al., Citation2010). However, in contrast to viral vectors, transposon vectors can be maintained and propagated as plasmid DNA (), which makes them simple and inexpensive to manufacture, an important consideration for implementation and scale-up in clinical practice. Further advantages of SB as gene-transfer system compared to viral vectors include its lower immunogenicity in vivo (Yant et al., Citation2000), the greater capacity for genetic cargo (Zayed et al., Citation2004) and its superior biosafety profile (Ivics et al., Citation2007; Moldt et al., Citation2007; Vandendriessche et al., Citation2009; Walisko et al., Citation2008). Because transposition proceeds through a cut-and-paste mechanism that only involves DNA, transposon vectors are not prone to incorporating mutations by reverse transcription [that are generated in retroviral stocks at reasonable frequencies (Menendez-Arias, Citation2009)], and can tolerate larger and more complex transgenes.
Performance facts of Sleeping Beauty transposition
Cargo capacity
There is an inverse correlation between the size of the SB transposon unit and the efficiency at which transposition takes place (Fischer et al., Citation1999; Izsvák et al., Citation2000; Karsi et al., Citation2001; Lampe et al., Citation1998), and such effect was also observed with the insect piggyBac (PB) transposon (Wang et al., Citation2014) and with the bacterial transposons IS1 (Chandler et al., Citation1982) and Tn10 (Morisato et al., Citation1983). There are at least two molecular mechanisms that can explain this phenomenon. First, in order to form a catalytically primed transposon/transposase complex (also called transpososome), the two ends of the transposon must be held together in close physical proximity by transposase molecules in a complex often called a synaptic complex or a paired-end complex (PEC) (Ivics & Izsvak, Citation2015). Formation of the PEC is a strict requirement for transposon excision (Wang et al., Citation2016b). It is conceivable that there might be a limitation in PEC formation in case the transposon ends are far apart, which can manifest in less efficient transposition of larger transposons. A strategy to partially rescue transposition of large transgene constructs is to shorten the length of DNA that connects the transposon ends outside the transposon (Izsvák et al., Citation2000). In case of a plasmid vector carrying the transposon unit, this means shortening the plasmid backbone. By placing the transposon TIRs next to one another, bacterial artificial chromosomes (BACs) over 100 kb in length have been demonstrated to transpose at reasonable efficiencies in human ES cells (Rostovskaya et al., Citation2012; Rostovskaya et al., Citation2013). The second mechanism limiting transposition of large constructs is a suicidal transpositional mechanism called autointegration (Wang et al., Citation2014). Autointegration means that following excision the transposon ends attack a TA dinucleotide inside the transposon, as opposed to a TA site in the target genome. This will abort the reaction and effectively kill the transposing element. Since larger transposons contain more potential target sites, they could be particularly attractive targets for autointegration. Indeed, increasing size was found to sensitize SB transposition for autointegration (Wang et al., Citation2014).
Although larger transposons are expected to transpose less efficiently, there does not seem to exist an absolute upper size limit for transposition, or at least this limit has not been experimentally reached. This is in sharp contrast to popular viral gene delivery systems, where packaging into infectious viral particles sets a strict limitation to the size of cargo to be incorporated. Indeed, recombinant vectors based on AAV are not competent to package vector genomes >5 kb (Hirsch et al., Citation2016; Salganik et al., Citation2015), and both retroviral and lentiviral vectors undergo a severe loss of titer beyond a vector size of ∼8–10 kb (Matrai et al., Citation2010; Sinn et al., Citation2005).
Transgene expression level and stability
Any transgene vector system should ideally provide long-term expression of transgenes. By using classical, plasmid-based, non-viral delivery approaches, expression from the extrachromosomal plasmid rapidly declines following delivery. Transgenes delivered by non-viral approaches often form long, repeated arrays (concatemers) that are targets for transcriptional silencing by heterochromatin formation. In addition, long-term expression of transgenes delivered by retroviruses has been shown to be compromised by transcriptional silencing (Jahner et al., Citation1982; Pannell & Ellis, Citation2001). It was shown that the zinc finger protein ZFP809 bridges the integrated proviral MLV genome and the tripartite motif-containing 28 transcriptional co-repressor in embryonic stem cells (Wolf & Goff, Citation2009). Thus, sequence elements in the vector itself can predispose the cargo for silencing. Cut-and-paste DNA transposition results in a single copy of the transgene per insertion locus; thus, concatemer-induced gene silencing is by definition a non-issue. Indeed, Grabundzija et al. found that transposon insertions delivered by the SB system only rarely (<2% of all insertions) undergo silencing in HeLa cells (Grabundzija et al., Citation2010), indicating that it is unlikely that certain sequence motifs in the transposon vector are recognized by mediators of silencing in the cell.
An additional factor that may provoke transgene silencing is the cargo DNA, particularly the type of promoter used to drive expression of the gene of interest. Indeed, it was previously shown that transgene constructs delivered into mouse cells using SB transposition can be subject to epigenetic regulation by CpG methylation. A determinant of epigenetic modifications of the integrating transposon vector was found to be the cargo transgene construct itself, with viral promoter elements playing a major role (Garrison et al., Citation2007). However, with careful promoter choice, several studies have established that SB-mediated transposition provides long-term expression in vivo. Notably, stable transgene expression from SB vectors was seen in mice after gene delivery in the liver (Aronovich et al., Citation2009; Kren et al., Citation2009; Ohlfest et al., Citation2005b; Yant et al., Citation2000), lung (Belur et al., Citation2003; Liu et al., Citation2004), brain (Ohlfest et al., Citation2005a) and blood after hematopoietic reconstitution in vivo (Mátés et al., Citation2009; Xue et al., Citation2009). Thus, it appears that SB transposon vectors have the capacity to provide long-term expression of transgenes both in vitro and in vivo.
Sleeping Beauty transposition in pre-clinical models
In ex vivo gene delivery, the therapeutic gene vector is introduced into a selected cell population that had been isolated from a donor, and the genetically engineered cells are transplanted into a patient. Depending on whether the donor is the patient itself or another person, we differentiate between autologous or allogeneic cell products, respectively. Unlike viruses, transposons are not equipped to cross cell membranes through infection. Thus, it is necessary to combine the transposon vectors composed of naked nucleic acids (DNA and mRNA) with technologies capable of efficient delivery of these non-viral vectors into cells. Since the efficiency of transposition is dependent on the efficiency of uptake of the introduced nucleic acids into the cell nuclei, delivery is a rate-limiting factor in transposition, and is thus of paramount importance. In principle, any technology developed for transferring nucleic acids into cells can be combined with transposon vectors. Unfortunately, there is no generally applicable method, and procedures must be established for each cell type. In hard-to-transfect cells, including primary human cell types, delivery of transposon-based vectors can be significantly facilitated by nucleofection, a procedure based on electroporation that transfers nucleic acids directly into the nucleus. Indeed, nucleofection facilitated transposition in CD34+ HSCs (Hollis et al., Citation2006; Izsvak et al., Citation2009; Mátés et al., Citation2009; Sumiyoshi et al., Citation2009; Xue et al., Citation2009), primary T cells (Gogol-Doring et al., Citation2016; Huang et al., Citation2008, Citation2010; Singh et al., Citation2008) and human embryonic stem cells (Orban et al., Citation2009; Wilber et al., Citation2007). Importantly, in the context of the hematopoietic system, this ex vivo gene delivery procedure apparently did not compromise the potential of transposon-marked CD34+ cells to differentiate normally into the erythroid, megakaryocytic, granulocyte/monocyte/macrophage (Mátés et al., Citation2009) as well as into the CD4+CD8+ T, CD19+ B, CD56+CD3− NK, and CD33+ myeloid lineages (Xue et al., Citation2009). The robustness and feasibility of this non-viral, transposon-based procedure significantly facilitates clinical realization of ex vivo stem cell therapy for the treatment of hematopoietic disorders and cancer, and is already successfully applied in humans (Kebriaei et al., Citation2016).
Refinement of Sleeping Beauty for clinical applications
There were three major areas of development and refinement of SB technology bearing paramount importance for clinical translation: i) increasing gene transfer rate and lowering toxicity; ii) defining suitable vectors to encode transposase and transposon; iii) establishing a biosafety profile that satisfied requirements for clinical use.
Enhancing Sleeping Beauty’s performance index
Hyperactive SB transposases
In evolutionary terms, the SB transposon was a successful element with the ability of colonizing several fish genomes millions of years ago (Ivics et al., Citation1996). However, even successful transposons have not been selected for the highest possible activity. On the contrary, there is likely a strong selective pressure to avoid insertional mutagenesis of essential genes of their host. In an attempt to derive hyperactive transposase variants for advanced genetic engineering, amino acid substitutions spanning almost the entire SB transposase polypeptide have been screened for eliciting a change in catalytic activity. These amino acid replacements were conducted either by systematic alanine-scanning (Wang et al., Citation2016b; Yant et al., Citation2004), by “transplanting” single amino acids or small (2–7 aa) blocks of amino acids from related transposases (Baus et al., Citation2005; Geurts et al., Citation2003; Zayed et al., Citation2004), and by replacement of selected amino acid residues based on charge (Zayed et al., Citation2004). These approaches generated transposase variants with i) no change in activity; ii) reduced activity or iii) a relatively modest increase of transposition activity. A second-generation SB transposase called SB11 contains five amino acid replacements (selected based on a phylogenetic comparison to active Tc1/mariner transposases) over the first-generation transposase (Geurts et al., Citation2003). SB11 is about 3-fold more active than the first-generation SB transposase, and has been primarily employed in currently running clinical trials based on CAR-engineered T cells (Kebriaei et al., Citation2016) ().
Surprisingly, some combinations of hyperactive mutations were found to result in a significant reduction of activity. Nevertheless, screening of a library of possible combinations yielded SB transposase variants with significantly enhanced activities (Mátés et al., Citation2009). The most hyperactive SB transposase version currently available, SB100X, displays a ∼100-fold hyperactivity when compared to the originally resurrected transposase (Mátés et al., Citation2009). The hyperactivity of SB100X cannot be explained by altered transposase stability, nor by increased binding to the transposon TIRs; instead, the particular combination of mutations in SB100X appears to affect the folding properties of the transposase (Mátés et al., Citation2009). The SB100X transposase enables highly efficient germline transgenesis in relevant mammalian models, including mice, rats, rabbits and pigs (Ivics et al., Citation2014a, Citation2014b, Citation2014c). Moreover, the use of the SB100X system yielded robust gene transfer efficiencies into human HSCs (Mátés et al., Citation2009; Xue et al., Citation2009), mesenchymal stem cells, muscle stem/progenitor cells (myoblasts), iPSCs (Belay et al., Citation2010) and T cells (Jin et al., Citation2011). These cells are relevant targets for stem cell biology and for regenerative medicine and gene- and cell-based therapies of complex genetic diseases. Thus, the SB100X hyperactive transposase holds great promise for of ex vivo and in vivo gene therapies.
Optimized transposons and transposon donor vectors
The first-generation SB transposon vector (called pT) was based on a naturally occurring sequence originally isolated from the Tanichthys albonubes genome (Ivics et al., Citation1997). As with the transposase, although to a lesser extent, mutagenesis of the SB TIR sequences has been undertaken with the aim to increase the efficiency of transposition. For example, flanking transgenes by two left TIRs of SB (instead of the canonical arrangement of one left TIR (LIR) and one right TIR (RIR) ()) was shown to enhance transposition by ∼3-fold, likely due to the presence of a transpositional enhancer sequence located in LIR (Izsvák et al., Citation2002). Replacement of four base pairs in RIR and flanking both TIRs by a doublet of TAs (i.e. by TATA sequences as opposed to the canonical TA sites) resulted in ∼3-fold increase in transposition over pT (Cui et al., Citation2002). Incorporation of a multi-cloning-site consisting of several unique restriction enzymes sites available for cloning genes of interests into the optimized transposon sequences led to the pT2-generation SB transposon vector, which has become the most popular SB transposon vector in the scientific community. A combination of the 2-left-TIR and 2-TA arrangements led to a vector called pT3 (Yant et al., Citation2004). Finally, optimization of SB transposase binding to its internal binding sites within the transposon TIRs by cyclic amplification and selection of targets (CASTing) recently yielded pT4 with modest hyperactivity (Wang et al., Citation2016b). In addition to enhancing transposition per se, SB vectors of enhanced utility have been devised, for example by including cassettes for constitutive or inducible expression of any gene of interest in combination with different antibiotic selection markers and fluorescent reporters (Kowarz et al., Citation2015).
Vectorization of Sleeping Beauty components
Plasmid DNA to encode SB transposase and transposon
The typical setup for delivery of the SB transposon system into cells is supplying the two components of the vector system (i.e. the engineered transposon carrying a gene of interest and a transposase source) as conventional plasmids. Although both components can be placed on a single plasmid (Mikkelsen et al., Citation2003) (), this arrangement has not become popular, mainly because it does not allow careful titration of transposase expression required for optimal transposition at a certain transposon dose. Because optimal transposon-to-transposase ratio can substantially vary dependent on cell type, most investigators opt for the use of the 2-plasmid vector system (). Two recent developments that address both the efficiency and safety of SB gene delivery are the use of mRNA-encoded SB100X and minicircle (MC) vectors to encode transposase/transposon. Both are key milestones toward the clinical use of SB transposition.
mRNA as a transient source of SB transposase
The transposase expression plasmid that is typically used to provide a transposase source in cultured cell lines can be replaced by mRNA synthesized in in vitro transcription reactions (). Co-delivery of SB transposase-encoding mRNA with an SB transposon plasmid to somatic cells was originally tested in a mammalian cell line in vitro and in the mouse liver in vivo, using an early version of the SB transposase, SB11 (described above) (Wilber et al., Citation2006, Citation2007). By applying mRNA for intracellular delivery in therapeutically relevant cells ex vivo, some hurdles of gene transfer typical for DNA-based vectors can be avoided. For example, nucleofection of primary human cells, including HSCs and T cells, with mRNA was shown to cause significantly reduced cellular toxicity as compared to nucleofection with plasmid DNA (Monjezi et al., Citation2016; Wiehe et al., Citation2007). Second, upon transfection, mRNA translocates into the cytoplasm where it is readily available for the host translational machinery and protein production. Finally, the implementation of an mRNA source for transient delivery of the SB transposase increases the biosafety of this approach, as mRNA does not bear the risk of chromosomal integration. In contrast, it is known that transfection of plasmid DNA is associated with a small, but non-negligible chance of spontaneous, illegitimate vector integration into the host genome (Wang et al., Citation2004). Genomic integration of the SB transposase coding sequence into the genome represents a finite risk in a gene therapy application, because such event could lead to genomic instability due to prolonged and uncontrollable transposase expression resulting in continuous remobilization of the already integrated SB transposon.
Minicircle DNA – minimalistic transposase and transposon expression cassettes
A recent addition to the development of SB vectors with enhanced utility for clinical applications is the use of MC vectors as carriers of the SB transposon components. The MC technology allows for a significant reduction of SB vector size by removing most of the backbone sequences from parental plasmids (Sharma et al., Citation2013). The presence of bacterial backbone elements is typically required for vector propagation in Escherichia coli, but completely redundant or even undesired for clinical applications. The first evident advantage of using MC vectors over plasmids is related to increased cell survival rates following nucleofection of human T cells (Monjezi et al., Citation2016) (described in detail below in “Cancer immunotherapy with tumor-reactive CAR T-cells”). Unmethylated CpG motifs, which are highly enriched in the bacterial backbone of exogenously delivered plasmids, were shown to trigger strong inflammatory responses through toll-like receptor-9 (Chuang et al., Citation2002; Hemmi et al., Citation2000) and/or interferon induction (Huerfano et al., Citation2013). Activation of these cellular sensors might be a conceivable explanation for the observed cytotoxicity and thus for the loss of a large fraction of cells transfected with plasmid DNA from the rest of the proliferating cell culture. Indeed, removal of CpG motifs from plasmid DNA vectors was demonstrated to reduce inflammatory responses upon pulmonary gene delivery (Hyde et al., Citation2008), and vector CpG methylation was able to lower immune responsiveness towards “non-self” DNA and led to delayed clearance of transfected cells in vivo (Reyes-Sandoval & Ertl, Citation2004). Along with decreased levels of cytotoxicity, nucleofection of T cells with SB transposon components supplied as MCs resulted in enhanced transient gene delivery and more efficient stable genome modification as compared to conventional plasmid vectors (Monjezi et al., Citation2016) (described in detail below in “Cancer immunotherapy with tumor-reactive CAR T-cells”).
The MC components are likely more efficient in transfection than plasmids because, due to their smaller size, they cross cellular membranes more efficiently than plasmids (Chabot et al., Citation2013; Darquet et al., Citation1997). Second, MC vectors may support enhanced transcription of transgene cassettes. Indeed, enhanced and sustained transgene expression has been seen in episomal gene therapy applications with MC vectors with concomitant gene silencing commencing rapidly after hydrodynamic injection of parental plasmid vectors in vivo (Chen et al., Citation2003; Osborn et al., Citation2011). Covalent linkage of bacterial DNA sequences to a eukaryotic expression cassette has been suggested to facilitate the spreading of repressive chromatin formed primarily on the bacterial backbone, leading in turn to rapid loss of transgene expression from plasmid DNA vectors (Chen et al., Citation2004, Citation2008). Finally, the elevated levels of transposition observed with MC vectors are likely supported, at least in part, by the relatively short, ∼200 bp distance between the SB transposon ends in the MC-based transposon vector, owing to the depletion of the bacterial plasmid backbone. Indeed, SB transposition was shown to be far more efficient when the length of DNA sequence outside the transposon unit was shortened, likely by aiding transposon/transposase complex formation, as discussed above (Izsvák et al., Citation2000). In addition to efficacy, the MC technology also offers biosafety advantages over conventional plasmid DNA vectors. Namely, the absence of bacterial plasmid backbone elements in therapeutic vectors is highly relevant in clinical applications, because antibiotic resistance genes included in a therapeutic cell product may raise safety concerns.
Establishing a biosafety profile
Genome-wide distribution of Sleeping Beauty insertions
With any vector that integrates into chromosomes in a semi-random manner comes the potential risk of insertional mutagenesis leading to transcriptional activation or inactivation of cellular genes (Baum et al., Citation2004). Such genotoxic effects can have devastating consequences for the cell and the whole organism, including the development of cancer, as discussed above in “Viral gene transfer vectors”.
SB displays considerable specificity in target site selection at the primary DNA sequence level in that TA dinucleotides are near-obligate target sites (Vigdal et al., Citation2002). A palindromic AT-repeat consensus sequence with bendability and hydrogen bonding potential was found to constitute preferred target sites (Vigdal et al., Citation2002). It was later shown that a characteristic deformation of the DNA sequence may be a recognition signal for target selection (Liu et al., Citation2005). Although characterization of the target site selection properties of different vector systems still falls short of predicting the actual risk of insertional oncogenesis in a clinical trial, it is highly useful for ranking the different vector types and designs according to their genotoxic potential (Naldini, Citation2011). We have previously undertaken a comparative study addressing target site selection properties of the SB and PB transposons as well as MLV-derived gammaretroviral and HIV-derived lentiviral systems in primary human CD4+ T cells. Our bioinformatic analyzes included mapping against the T cell genome with respect to proximity to genes, transcriptional start sites (TSSs), CpG islands, DNaseI hypersensitive sites, chromatin marks and transcriptional status of genes. The SB transposon displayed the least deviation from random with respect to genome-wide distribution: no apparent bias was seen for either heterochromatin marks or euchromatin marks and only a weak correlation with transcriptional status of targeted genes was detected (Gogol-Doring et al., Citation2016). Our compiled datasets also allowed us to rank these vector systems with respect to their projected relative “safety” based on the frequencies of integration into GSHs that were previously proposed to satisfy five criteria: (i) distance of at least 50 kb from the 5’-end of any gene, (ii) distance of at least 300 kb from any cancer-related gene, (iii) distance of at least 300 kb from any microRNA (miRNA), (iv) location outside a transcription unit and (v) location outside ultraconserved regions of the human genome (Papapetrou et al., Citation2011; Sadelain et al., Citation2012). Our analyzes collectively established a favorable integration profile of the SB transposon, suggesting that SB might be safer for therapeutic gene delivery than the integrating viral vectors that are currently used in clinical trials. Importantly, no SB-associated adverse effects have been observed in preclinical animal studies (Fernando & Fletcher, Citation2006; Hackett et al., Citation2010; Ivics & Izsvak, Citation2006; Izsvak et al., Citation2010).
Vector-associated transcriptional activity
Integration of a vector into a gene or its regulatory elements can knock out the gene, overexpress the gene or alter its spatio/temporal expression pattern. The major underlying mechanism, by which insertional mutagenesis manifests in gene therapy appears to be transcriptional trans-activation of oncogenes (Baum et al., Citation2004; Deichmann et al., Citation2007) by the retroviral long terminal repeats (LTRs) that carry strong enhancer/promoter elements. Due to their intrinsic preference for integrating into TSSs (see above), MLV-derived gammaretroviral vectors are especially prone to transcriptionally upregulate endogenous genes. Indeed, it has been demonstrated that MLV-based gammaretroviral insertions, although they target GSHs >2-fold more frequently than HIV-based lentiviral vectors (Gogol-Doring et al., Citation2016), were approximately 3-fold more likely to trigger transformation of primary HSCs in a cell-based immortalization assay (Modlich et al., Citation2009). This suggests that an MLV insertion next to a TSSs tends to be more genotoxic than an HIV insertion in a gene body, in line with the expectation that gain-of-function due to oncogene upregulation has a more profound effect on cellular homeostasis than loss-of-function due to knockout mutations (unless the affected gene is haplo-insufficient). To ameliorate vector-induced insertional oncogenesis, second-generation, self-inactivating (SIN) vectors have been developed for both gammaretroviral and lentiviral vectors. These vectors lack transcriptional enhancer/promopter elements in their LTRs (Schambach et al., Citation2006, Citation2007), and therefore they are expected to have an enhanced safety profile in clinical applications. Indeed, SIN vectors result in reduced clonal immortalization in cell-based genotoxicity assays (Modlich et al., Citation2009).
Unlike the LTRs of retroviruses, the TIRs of SB vectors have negligible enhancer/promoter activity (Moldt et al., Citation2007; Walisko et al., Citation2008). The left TIR of SB is separated from the transposase coding sequence by a 5’-UTR of 160 bp stretch of DNA with no apparent function in the transposition reaction (Zayed et al., Citation2004). As measured by transient reporter assays, transcription driven by the 5’-UTR of SB is ∼18-fold higher than transcription of a promoter-less sequence, and ∼5-fold higher than transcription driven by a TATA-box minimal promoter (Walisko et al., Citation2008). Importantly, the 5’-UTR is not present in SB transposon vectors (because it is dispensable for transposition). Thus, contemporary SB vectors are transcriptionally neutral, and therefore it is expected that their mutagenic potential will largely depend on their cargo (including the transcriptional regulatory elements that drive transgene expression).
Beyond the genomic integration pattern and the transcriptional activities associated with a given vector, a third determinant of risk stemming from vector integration is the type of cell, in which therapeutic gene transfer is executed. In contrast to HSC-based gene therapy, leukemia was never observed in preclinical animal models or clinical trials involving gene transfer into peripheral blood-derived T lymphocytes (Newrzela et al., Citation2008; Recchia et al., Citation2006). Thus, mature T cells seem to be less susceptible to transformation by genotoxic events than are HSCs. It is likely that other types of terminally differentiated cells, including retinal pigment epithelium (RPE) cells isolated from the eye are also more refractory to oncogenic transformation than stem cells (Balaggan et al., Citation2012), thereby presenting attractive targets for Phase I clinical trials.
Clinical application of Sleeping Beauty gene delivery
Overview
The past few years have seen a steadily growing interest in applying SB transposition in gene therapy to provide innovative and potentially curative treatments for genetic disorders [reviewed in (Boehme et al., Citation2015; Essner et al., Citation2005; Hackett et al., Citation2005, Citation2010, Citation2011; Ivics & Izsvak, Citation2006, Citation2011; Izsvak & Ivics, Citation2004; Izsvak et al., Citation2010)]. Prime examples for the use of SB in gene therapy include the treatment of hematologic disorders, lysosomal storage diseases, pulmonary disorders, dermatologic diseases, a variety of metabolic disorders, neurologic disorders, muscle disorders and cancer ().
Table 1. Preclinical studies with Sleeping Beauty gene transfer in disease models.
In addition, important steps have been made toward SB-mediated gene transfer in the eye to treat neovascular age-related macular degeneration (AMD), which will be discussed in detail in “Sleeping Beauty non-viral gene delivery for gene therapy of neovascular age-related macular degeneration”, as an example for successful clinical translation. Further, a medical revolution is currently taking place in the field of hematology and oncology with the successful clinical use of genetically modified tumor-reactive T cells that are equipped with synthetic chimeric antigen receptors (CARs) or transgenic T-cell receptors (TCRs). Clinical-proof-of-concept has been obtained with both CAR-modified T cells in advanced hematologic malignancies and TCR-transgenic T cells in hematologic and solid tumors (Davila et al., Citation2014; Kochenderfer et al., Citation2015; Maude et al., Citation2014; Morgan et al., Citation2006; Rapoport et al., Citation2015, Turtle et al., Citation2016a, Citation2016b; Turtle & Maloney, Citation2016). Intriguingly, the concept of endowing T cells with a novel specificity through CAR- and TCR-gene transfer is already being adopted to also treat infections and autoimmune diseases and will establish novel treatment paradigms in medicine. As a prime example, the use of SB-mediated gene transfer to engineer CAR-modified T cells for anti-cancer therapy will be discussed in detail in “Cancer immunotherapy with tumor-reactive CAR T-cells”.
Sleeping Beauty non-viral gene delivery for gene therapy of neovascular age-related macular degeneration
TargetAMD is an international consortium of universities, research institutes and commercial organizations funded by the European Commission to execute a Phase Ia/IIb clinical trial for the treatment of neovascular AMD by transplantation of genetically modified, autologous retinal pigment epithelial (RPE) or iris pigment epithelial (IPE) cells that overexpress pigment epithelial-derived factor (PEDF) to the subretinal space of the eye. Specifically, RPE or IPE cells isolated from the peripheral retina or obtained from an iris biopsy of a patient will be transfected with an SB transposon vector carrying a PEDF expression cassette and transplanted back into the same patient during one surgical session lasting about 60 min.
AMD is a major cause of acquired irreversibleblindness in adults
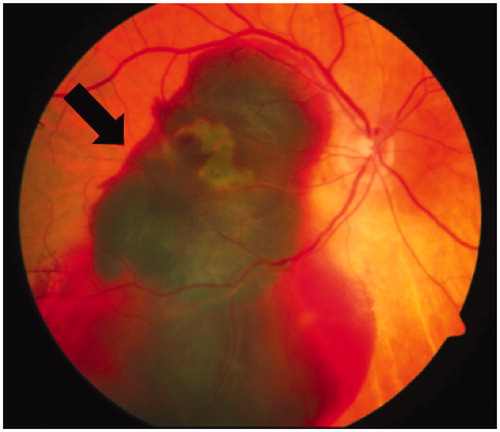
AMD refers to the degeneration of RPE cells in the macula area (), which is essential for central vision. With over 13 million people affected, AMD is the fourth most common cause of blindness after cataract, retinopathy of prematurity, and glaucoma in the world and the leading cause of irreversible blindness in people aged >50 years in developed countries (Velez-Montoya et al., Citation2014). There are two forms of AMD: avascular AMD (avAMD) or geographic atrophy (GA) and neovascular AMD (nvAMD), which is characterized by the subretinal invasion of choroidal vessels. Whereas avAMD is a slow progressing disorder, in which photoreceptor degeneration follows RPE cell degeneration, nvAMD progresses rapidly to blindness following RPE cell degeneration. No treatments are available for avAMD.
Figure 3. Neovascular age-related macular degeneration (nvAMD). In nvAMD choroidal neovascularization (CNV) due to an imbalance of pro- and anti-angiogenic factors like VEGF and PEDF leads to severe hemorrhages damaging the retinal pigment epithelium (RPE) and the neural retina. The funduscopic image illustrates such a bleeding (arrow) in advanced nvAMD (see colour version of this figure at www.tandfonline.com/ibmg).
Limited clinical benefit of AMD fromantibody-mediated VEGF blockade
Since overexpression of vascular endothelial growth factor (VEGF) has been shown to be responsible for the development of subretinal neovascularization underlying the development of nvAMD (Ohno-Matsui et al., Citation2001; Velez-Montoya et al., Citation2014), its current treatment, which is effective in 30–40% of patients, is based on monthly, life-long, intravitreal injections of inhibitors of VEGF. The three most commonly used anti-VEGFs are bevacizumab (Avastin®, Genentech, San Francisco, CA), its Fab fragment ranibizumab (Lucentis®, Genentech) and aflibercept (Eylea®, Regeneron, Tarrytown, NY), which is a recombinant fusion protein consisting of the extracellular binding domains of human VEGF receptors 1 and 2 fused to the Fc portion of the human IgG1 immunoglobulin.
The two major limitations of anti-VEGFs are the lack of improvement of vision in 60–70% of patients and adverse effects (Nesmith et al., Citation2014) including more rapid progression of GA (Enslow et al., Citation2016), increased intraocular pressure, retinal detachment, endophthalmitis, photoreceptor cell death and thinning of the inner neuronal layer of the retina (Saint-Geniez et al., Citation2008; Scott & Bressler, Citation2013). In addition to the adverse effects, the issues associated with logistics of blind or low-vision patients traveling to a clinic on a monthly basis result in a significant number of patients discontinuing treatment, which has been reported to be from 57% over five years (Boulanger-Scemama et al., Citation2015) to as high as 71% within 24 months (Lad et al., Citation2014). Therefore, it is evident that new approaches are required for the efficient treatment of nvAMD.
One-shot AMD treatment: Sleeping Beauty-based PEDF gene therapy
The evidence that nvAMD is caused by RPE cell degeneration has engendered the hypothesis that the condition could be treated by replacing the degenerated RPE cells with healthy RPE cells, which synthesize and secrete the anti-angiogenic and neuroprotective factor pigment epithelium-derived factor (PEDF) in vivo. Administration of recombinant PEDF is not feasible because of its short half-life, and transplantation of RPE or IPE cells, as a substitute for degenerated RPE cells, has not resulted in significant improvement in visual acuity in nvAMD patients (Aisenbrey et al., Citation2006; Binder et al., Citation2004; Falkner-Radler et al., Citation2011; Lappas et al., Citation2004), indicating that the replacement cells did not express anti-angiogenic factors at sufficient levels to overcome the pathologic overexpression of VEGF.
Therefore, a desirable alternative to overcome the difficulties associated with frequent, life-long intravitreal injections would be a treatment modality that introduces an inhibitor of neovascularization to the retina that would last for the life of the patient. To meet such treatment modality, Campochiaro and colleagues (Campochiaro et al., Citation2006) delivered a PEDF transgene cassette to the retina of nvAMD patients using an adenoviral vector, and reported significant improvement in 25% of patients after 12 weeks and no harmful side effects. However, no follow-up to the trial has been reported. Currently, four clinical trials are ongoing (www.clinicaltrials.gov): i) trial NCT01494805 (Avalanche Biotechnologies, Menlo Park, CA), phase I/II, that employs a recombinant adeno-associated viral (rAAV) vector encoding sFlt-1, a splice variant of VEGF receptor 1, by subretinal delivery to nvAMD patients; ii) trial NCT01024998 (Genzyme, Cambridge, MA), phase I, that delivers an rAAV vector encoding sFLT01, a fusion protein that consists of the VEGF binding domain of sFlt-1 fused to the Fc portion of human IgG1, by a single intravitreal injection in nvAMD patients; iii) trial NCT01301443 (Oxford Biomedica, Oxford, UK), phase I, that delivers a lentiviral vector encoding endostatin and angiostatin, both inhibitors of VEGF (RetinoStat®), subretinally to nvAMD patients; and iv) trial NCT01678872 (Oxford Biomedica), phase I, that is a 15-year trial to determine safety and efficacy of subretinal RetinoStat® encoded in a lentiviral vector in nvAMD patients enrolled in the NCT01301443 trial.
As discussed earlier, viral vectors, although efficient at gene delivery, have limitations in human use; since adenoviral and rAAV vectors remain episomal and readministration may elicit immune responses (Basner-Tschakarjan & Mingozzi, Citation2014; Dijkhuizen et al., Citation1998; Hermens & Verhaagen, Citation1997; Tse et al., Citation2015), while lentiviral and gammaretroviral vectors have a preference for transgene integration into transcriptionally active genomic regions that could result in insertional mutagenesis and aberrant expression of endogenous genes (Cesana et al., Citation2012; Hacein-Bey-Abina et al., Citation2008; Hargrove et al., Citation2008) with potentially detrimental consequences for the patient.
In nvAMD, choroidal blood vessel growth into the subretinal space not only disorganizes the normal architecture of the retina, but leads to RPE cell degeneration and loss of vision. The use of vectors to deliver inhibitors of neovascularization to the subretinal space of nvAMD patients will benefit only those patients that have normally functioning RPE cells. However, due to advanced RPE cell degeneration, 60–70% of nvAMD patients do not benefit from anti-VEGF treatment, and thus replacement of the degenerated RPE cells appears essential to regain vision. Since transplantation of RPE or IPE cells does not have a significant effect on choroidal neovascularization (CNV), the hallmark of nvAMD, and does not improve vision in nvAMD patients, we have hypothesized that transplantation of genetically modified RPE or IPE expressing PEDF at supra-physiological levels would not only replace the degenerated cells, but at the same time could inhibit CNV, prevent the continued degeneration of RPE cells, and restore vision in nvAMD patients (.
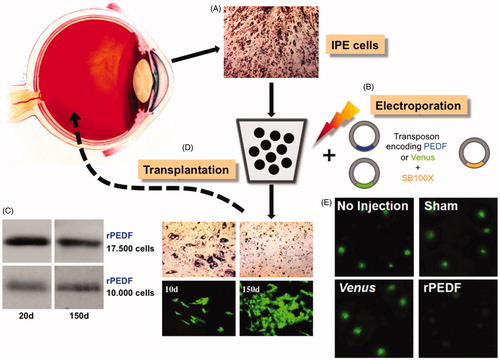
Figure 4. Gene therapy to treat neuroretinal degeneration. A) The anatomy of the human eye is shown. Image source: National Eye Institute, NIH. A small biopsy is taken from the patient’s iris for the isolation of ∼10.000 IPE cells that are taken in culture in an appropriate buffer. B) The IPE cells are electroporated ex vivo with components of the SB transposon system: a transposon carrying the therapeutic PEDF transgene or a Venus reporter and the SB100X transposase. C) Stable transfection and gene expression in primary human IPE cells. For the analysis of Venus reporter gene expression, human IPE cells were isolated post mortem, and taken in culture for four weeks before transfection with the transposon system components. For the analysis of PEDF expression, human IPE cells were isolated from donor tissue, and taken in culture for 8–12 weeks before transfection with the transposon system components. Expression of Venus (micrographs) and secretion of human PEDF (Western blot) at early (10 and 20 days) and late (150 days) indicate stable gene transfer and transgene expression. D) Genetically engineered IPE cells expressing and secreting human PEDF are grafted onto the patient’s retina. E) PEDF-transfected cells significantly reduced CNV in rats in vivo and corneal neovascularization in rabbits (Johnen et al., Citation2015; Kuerten et al., Citation2015). Shown here are retinal flat mounts of rats treated with laser to provoke CNV. Lesions are stained with FITC-conjugated isolectin B4 at day 14 post-transplantation. In non-injected (No Injection), sham-treated (Sham) animals and after transplantation of Venus-transfected cells (Venus) lesions are unaltered, whereas after transplantation with PEDF-transfected cells (rPEDF) lesion size was significantly reduced (see colour version of this figure at www.tandfonline.com/ibmg).
Local delivery of PEDF through SB-engineered autologous RPE/IPE cells
Because the inhibition of VEGF must be constant and the proper balance between angiogenic and anti-angiogenic activities must be maintained for the life of the patients for an effective gene therapy of nvAMD, stable transgene integration and continuous PEDF expression are critical for re-acquiring vision. To avoid the risks accompanied by gene delivery mediated by viral vectors, the TargetAMD consortium has been pursuing the use of the hyperactive SB100X system for efficient delivery of a human PEDF transgene cassette to cultured and freshly isolated RPE and IPE cells (). Importantly, cells transfected with a PEDF-transposon and the SB100X transposase have been found to express recombinant PEDF long-term for the duration of 16 months that the cells have been in culture () (Johnen et al., Citation2012). Genome-wide insertion site analysis in human RPE cells established a close-to-random insertion profile of SB and a frequency of inserting into GSHs similar to that found in human T cells (Gogol-Doring et al., Citation2016; Monjezi et al., Citation2016; Thumann et al., Citation2017). Importantly, no insertions were observed in genes critical to RPE cell function, including the angiogenic factor VEGF-A, the visual cycle protein CRALBP, the lysosomal enzyme cathepsin D, the tight-junction protein ZO-1 and the cytokeratin KRT8. SB100X-mediated stable gene transfer of PEDF in the human RPE cell line ARPE-19 cells, followed by subconjunctival transplantation in a rabbit model significantly suppressed and prevented neovascularization (Kuerten et al., Citation2015). To establish whether RPE cells expressing elevated levels of PEDF inhibit CNV, 10.000 rat RPE cells transfected with the PEDF gene using SB100X and secreting approximately 2 ng PEDF/day were transplanted to the subretinal space of rats in which CNV was induced by laser rupture of Bruch’s membrane (Grossniklaus, et al., Citation2010). A marked reduction of neovascularization was observed at 7 and 14 d post-transplantation, with the area of neovascularization reduced by 50% () (Johnen et al., Citation2015).
It has been demonstrated that, by using SB transposon plasmids in conjunction with the SB100X transposase, the PEDF gene can be efficiently delivered to IPE cells; however, although safer than viral vectors, plasmids carry antibiotic resistance genes, e.g. the pT2 SB transposon vector (Cui et al., Citation2002) carries an ampicillin-resistance gene, which is necessary for plasmid maintenance in E. coli, and thus for efficient manufacturing of the plasmid vector. The presence of antibiotic resistance markers in gene therapy vectors is a matter of concern, since there is a risk that the antibiotic resistance genes could elicit resistance to pathogenic bacteria by horizontal gene transfer and residues of antibiotics could contaminate the final product, placing patients with severe hypersensitivity to antibiotics at risk (Solensky, Citation2003). In fact, regulatory agencies recommend that antibiotic resistance genes be eliminated from gene therapy treatments whenever possible. Thus, the TargetAMD consortium resorted to apply a variant of MC technology in free of antibiotic markers (pFAR) miniplasmids (Marie et al., Citation2010). Like MCs, pFAR-based vactors lack antibiotic-resistance genes, thereby significantly enhancing the safety profile of nonviral gene delivery in clinical settings.
TargetAMD: Phase I clinical trial of SB-based PEDF gene therapy launched in 2017
The ultimate goal of TargetAMD is to perform a Phase Ia/IIb clinical trial in which IPE cells will be isolated from an iris biopsy of a nvAMD patient, transfected with the PEDF gene and transplanted to the subretinal space of the same patient during a single surgical session lasting approximately one hour (. Since the isolation of IPE or RPE cells from a patient’s biopsy yields a limited number of cells, and since safety is a major concern of any therapy, and of gene therapy in particular, we have developed a protocol for the efficient delivery of the PEDF gene in as few as 5.000–10.000 freshly isolated IPE and RPE cells from human donor eyes by vectorizing the PEDF-encoding SB transposon and the SB100X transposase in pFAR miniplasmids (Marie et al., Citation2010; Thumann et al., Citation2017). Using this protocol, a standard operating procedure has been established that i) consistently shows highly efficient transfer of the PEDF gene in RPE and IPE cells obtained from donor eyes, ii) enables expression of recombinant PEDF at high levels of recombinant protein in cultured PEDF-transfected cells, and iii) allows for sustained transgene expression (for over one year that the cells have been in culture) in genetically engineered cells. The robustness of this procedure is coupled with salient safety features including a close-to-random transgene integration profile of the SB transposon in human IPE and RPE cells and the lack of antibiotic resistance genes in vector components. Based on the results described here and approval by the Swissmedic regulatory agency, TargetAMD will begin patient recruitment for the phase Ia/IIb clinical trial in the spring of 2017 and expects to complete the first European in-man clinical trial using SB transposon and pFAR technologies.
Cancer immunotherapy with tumor-reactive CAR T-cells
Adoptive immunotherapy with tumor-reactive T cells that are engineered by gene transfer to express a synthetic CAR is emerging as an effective and potentially curative treatment for advanced malignancies. CARs are designer molecules comprised of an extracellular antigen binding domain, most commonly the variable light and heavy chains of a monoclonal antibody, a spacer and transmembrane region that anchors the receptor on the T-cell surface and provides reach and flexibility for binding the target epitope, and an intracellular signaling module, most commonly CD3 zeta and one or more costimulatory domains, that mediate T-cell activation after antigen binding (Sadelain et al., Citation2013; Srivastava & Riddell, Citation2015) (. The genetic information required for encoding a CAR is ∼1–2 kb depending on binding domain configuration, spacer length and the number of costimulatory moieties.
Figure 5. Chimeric antigen receptors (CARs) are synthetic designer molecules. A) The CAR is a synthetic molecule that can be designed using specialized software, a gene encoding the desired aminoacid sequence synthesized and introduced into cells by an electroporation unit. B) CARs consist of an extracellular antigen binding domain (e.g. the variable light and heavy chains of a monoclonal antibody shown in yellow), a spacer and transmembrane region and an intracellular signaling domain (e.g. CD3 zeta and one or more costimulatory domains shown in blue). C) Expression of the CAR enables T cells (yellow) to recognize surface molecules on tumor cells (grey) and exert their anti-tumor effector function such as the release of cytolytic granules (triangles) and cytokines (circles). D) To manufacture a CAR T-cell product, white blood cells are obtained from the patient or a matched donor, CD8+ killer and CD4+ helper T cells isolated, the genetic information for the CAR introduced, CAR-modified T cells expanded and administered to the patient (see colour version of this figure at www.tandfonline.com/ibmg).
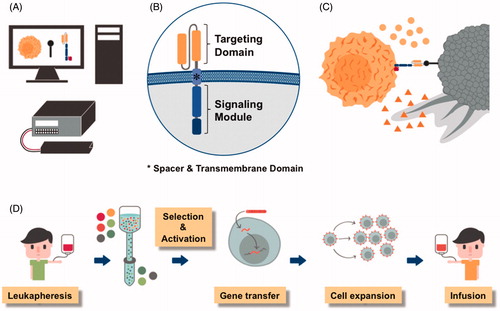
A medical revolution: CAR T-cell immunotherapy for B-cell malignancies
The most advanced clinical development is the use of CARs specific for the B-lineage marker CD19 that is expressed on B-cell acute and chronic lymphocytic leukemia and B-cell lymphoma. Several groups have reported up to 90% complete remissions in patients with chemotherapy- and radiotherapy-refractory B-cell acute lymphoblastic leukemia (ALL) and >60% complete remissions in patients with non-Hodgkin lymphoma (NHL) after administration of autologous (i.e. patient-derived) CD19-CAR T cells (Davila et al., Citation2014; Kochenderfer et al., Citation2015; Lee et al., Citation2015; Maude et al., Citation2014; Turtle et al., Citation2016a, Citation2016b; Turtle & Maloney, Citation2016) (). These results are considered by many to be a medical breakthrough, given the advanced disease stage and failure of conventional treatments in many patients that were included in these clinical trials. Side effects of CD19-CAR T-cell therapy are a consequence of the strong anti-tumor immune response and include tumor lysis syndrome due to the rapid destruction of a large number of tumor cells, cytokine release syndrome due to the rapid release of cytokines by CAR T cells and other immune cells, and the depletion of normal B-cells due to their physiologic expression of CD19.
Table 2. Clinical trials with CD19-CAR T cells.
Viral gene transfer vectors dominate pre-clinical and clinical studies with CAR T cells
The overwhelming majority of pre-clinical work and clinical trials that reported efficacy of CD19-CAR T-cell therapy has employed gammaretroviral and lentiviral vectors to stably integrate the genetic information of the CD19-CAR in patient’s T cells (). As a consequence, the CAR is not only expressed in T cells that are modified and infused to the patient, but also in subsequent generations of daughter T cells after cell division and propagation. The use of stable gene transfer systems is consistent with the concept and conducive to the ambition of CAR T-cell immunotherapy that administration of a small number of patient-derived CAR T cells is sufficient to achieve engraftment and proliferation in vivo until all tumor cells have been cleared and persistence as memory CAR T cells that protect the patient from tumor relapse long-term.
The potential to achieve transient CAR expression in T cells through transfection of mRNA has been demonstrated and results in CAR surface expression for up to several days which can be sufficient to induce an anti-tumor effect, especially when high doses of CAR T cells are administered (Beatty & Moon, Citation2014; Caruso et al., Citation2016). However, because of the rapid decline in CAR expression, multiple subsequent administrations are required to sustain the therapeutic effect. A problem with administering sequential doses of CAR T cells is the induction of immune responses to immunogenic epitopes that may be harbored in the targeting domain (that are often of murine or other “foreign” origin), and fusion sites in the synthetic CAR molecule. Such immune responses may lead to rapid and sometimes fulminant CAR T-cell rejection with loss of the therapeutic effect (Maus et al., Citation2013).
Sleeping Beauty and CAR T cells: clinicalproof-of-concept obtained
The potential to use SB transposition to integrate the genetic information of the CAR into T cells has first been explored by the group of Cooper et al. (Singh et al., Citation2008). It was demonstrated that SB transposase can be provided either as plasmid DNA or mRNA in combination with a plasmid-encoded CAR transposon and introduced into T cells by electroporation to yield functional CD19-CAR T cells. Consistent with observations in other mammalian cell types, the use of SB11 and hyperactive SB100X accomplished higher rates of gene transfer than first-generation SB transposase (Jin et al., Citation2011; Singh et al., Citation2013). The same group has also provided the successful clinical debut of SB-engineered CD19-CAR T cells, and recently reported results of two pilot clinical trials in 26 patients with ALL and NHL who had undergone autologous (n = 7, ClinicalTrials.gov Identifier 00968760) or allogeneic (n = 19, ClinicalTrials.gov Identifier NCT01497184) HSC transplantation (HSCT) prior to CAR T-cell therapy (Kebriaei et al., Citation2016). From these clinical studies, it was concluded that the administration of SB-engineered CD19-CAR T cells is safe, and may provide additional tumor control in patients after HSCT. The SB transposition strategy pursued in this trial was relatively “basic”, and comprised the nucleofection of plasmid-encoded SB11 transposase and a plasmid-encoded, pT-based CAR transposon, followed by ex vivo propagation of CAR-modified T cells for ∼28 days (four stimulation cycles with artificial antigen presenting cells). These studies are the first CAR T cell clinical trials that rely on non-viral SB-based gene transfer, and provide proof-of-concept for the utility of SB transposition in CAR T-cell engineering. However, because CD19-CAR T cells were administered as an adjuvant therapy after HSCT, the presented data are somewhat less spectacular than the dramatic and durable anti-tumor responses with sometimes year-long persistence of CD19-CAR T cells that have been reported in other clinical trials that used viral gene transfer vectors and administered CD19-CAR T cells outside the HSCT setting (Maude et al., Citation2014; Turtle et al., Citation2016a, Citation2016b). We would like to point out, however, that comparisons between clinical trials of CAR therapy conducted by different groups are difficult due to multiple variations in study design and clinical parameters of their patient population. Further, there are significant differences between trials in the specific design of CD19-CAR constructs, the CAR T-cell manufacturing process, CAR T-cell dose and subset composition, all of which are factors that we and others have shown significantly affect clinical safety and efficacy (Hudecek et al., Citation2015; Sommermeyer et al., Citation2015).
In our view, the use of SB transposition is a preferable gene transfer strategy for CAR T-cell engineering. The clinical trials reported by Kebriaei et al. (Citation2016) provide a first benchmark; however, manufacturing and clinical performance of SB-engineered CD19-CAR T-cell products may be enhanced by the use of: i) an SB transposase (SB100X) and transposon (pT2) with enhanced transposition rate (see above in “Enhancing Sleeping Beauty?s performance index”); ii) improved vectorization and transfection techniques that achieve higher gene transfer rates at lower toxicity, and enable iii) a short ex vivo manufacturing process with optimal fitness of CAR T cells at the time of infusion.
Enhanced CAR T-cell engineering with minicircle-encoded SB100X and CAR transposon
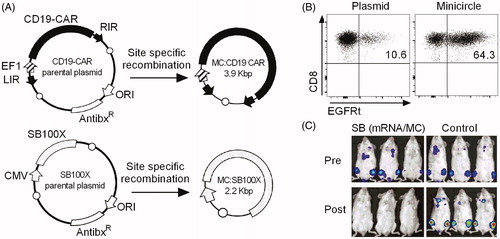
We have recently shown that vectorization of SB100X and a pT2-based CAR transposon as MC DNA () dramatically improves transposition efficacy (Monjezi et al., Citation2016). Our experience with CD19-CAR T cells is that the gene transfer rate is ∼5-fold higher with the use of MCs compared to conventional plasmids (). Importantly, despite higher transposition rates, the toxicity of transfecting MCs into T cells is lower compared to standard DNA plasmids (Monjezi et al., Citation2016). As a consequence, therapeutic doses of CD19-CAR T cells from 1 × 10e6 input T cells can be obtained within as short as 14 days with our MC-based SB approach, and the manufacturing process can be further trimmed to only a few days if higher input T-cell numbers are utilized. Similarly effective, and preferable from a regulatory point of view, is the use of mRNA-encoded SB100X in combination with a MC-encoded CAR transposon. CD19-CAR T cells that we engineered with the mRNA-MC (and the MC-MC) combination confer potent anti-tumor responses in vitro and in vivo (), and are equally functional as CD19-CAR T cells prepared by lentiviral transduction (Monjezi et al., Citation2016).
Figure 6. Anti-tumor function of CD19-CAR T cells engineered by Sleeping Beauty transposition. A) Schematic of minicircle (MC) DNA vectors encoding SB100X transposase and CD19-CAR transposon. MCs are prepared from parental conventional plasmids through site-specific intramolecular recombination. MCs contain exclusively the transgene and its promoter, but no bacterial origin of replication and antibiotic resistance genes. EF1, elongation factor-1 alpha promoter; CMV, cytomegalovirus promoter; ORI, bacterial origin of replication; AntibxR, antibiotic resistance gene; LIR, left inverted repeat; RIR, right inverted repeat; open circle = recombination site. B) CAR expression in CD8+ killer T cells after transfection of MC- vs. plasmid-encoded SB100X transposase and CD19-CAR transposon. Flow cytometric analysis was performed on day 14 and shows a significantly higher percentage of CAR-modified T cells after transfection of MCs compared to plasmids. C) Anti-tumor efficacy of CD19-CAR T cells prepared by SB-transposition in a murine xenograft model of systemic B-cell lymphoma. Immunodeficient NSG mice were inoculated with firefly-luciferase expressing Raji lymphoma cells and treated with 5 × 10e6 CD19-CAR T cells (1:1 ratio of CD8+ and CD4+ T cells, 2.5 × 10e6 each) that had been generated by transfection with SB100X mRNA and CD19-CAR MC or control unmodified T cells. Bioluminescence images were obtained before T-cell infusion (pre, upper row) and 7 days after T-cell infusion (post, lower row) and show complete lymphoma eradication in the SB CD19-CAR T-cell group, whereas control mice present with progressive disease. A–C) modified from Monjezi et al. (Citation2016) (see colour version of this figure at www.tandfonline.com/ibmg).
We have undertaken a careful analysis of genomic insertion sites and found a close-to-random insertion profile of CAR transposons after mobilization from MCs, consistent with previous studies where SB transposition had occurred from conventional pT2 transposon donor plasmids (Gogol-Doring et al., Citation2016). A substantial fraction and indeed higher number of CAR transposon insertions compared to lentiviral integrants had occurred in GSHs of the T-cell genome that are neither expected to cause genotoxicity, nor to induce malignant transformation of their host cells as has been described for gammaretroviral vectors after gene therapy in HSCs (Hacein-Bey-Abina et al., Citation2003; Monjezi et al., Citation2016; Sadelain et al., Citation2012). Importantly, no case of malignant transformation has thus far been reported in clinical trials of CD19-CAR T-cell therapy that employed viral (and non-viral) gene transfer vectors. However, in some patients that achieved remission of their disease, the therapeutic effect was apparently mediated by single CD19-CAR T-cell clones that expanded in vivo to comprise a significant proportion of the patient’s peripheral T-cell repertoire. Based on the available data, the potential for malignant transformation after CAR gene transfer in human T cells is anticipated to be very low. However, especially once larger cohorts of patients are treated, non-viral SB transposition may be preferable to gammaretroviral and lentiviral gene transfer vectors due to its ‘safer’ genomic integration profile.
Clinical trial with CAR T cells engineered by Sleeping Beauty transposition from minicircle vectors
A clinical trial with CD19-CAR T cells that are engineered by SB transposition from MC vectors is in preparation at the Universitätsklinikum Würzburg, Germany, to obtain clinical proof-of-concept for this novel approach. In this trial, a CD19-CAR construct will be employed that has been selected from pre-clinical analyzes due to optimal anti-tumor function, and has been validated in clinical trials of CD19-CAR therapy that relied on lentiviral gene transfer (Hudecek et al., Citation2015; Turtle et al., Citation2016a). Further, cell products that contain equal proportions of CAR-modified CD8+ killer and CD4+ helper T cells will be formulated, based on our previous work which showed this is advantageous to the use of cell products with random subset composition (Sommermeyer et al., Citation2015). The key advantages of CAR T-cell products with defined subset composition are that i) lower total numbers of CAR T cells are sufficient for clinical efficacy which reduces the risk for side effects and shortens the manufacturing process; and ii) because of less product-to-product variability between patients, the time to and level of CAR T-cell engraftment and proliferation is more consistent and predictable, and allows establishing dose-response relationships as well as immune pharmacokinetic and pharmacodynamic parameters for CAR T cells as medicinal products. We have determined the copy number of the CD19-CAR transposon in both CD8+ and CD4+ T cells, and found it to be in an acceptable range (n = 6 and n = 5, respectively), similar to that obtained with gammaretroviral and lentiviral vectors (Monjezi et al., Citation2016).
For future clinical trials, there is a pre-clinical development pipeline with CARs specific for target antigens in other hematologic and solid tumors, including a SLAMF7-specific CAR in multiple myeloma, and a ROR1-specific CAR in lymphoma, breast and lung cancer (Berger et al., Citation2015; Hudecek et al., Citation2013). Importantly, the MC-encoded SB transposon can accommodate additional genetic cargo without compromising transposition efficacy. Our CAR transposon includes an EGFRt marker that was developed in Mike Jensen’s laboratory and serves multiple functions, e.g. to select CAR-modified T cells to further refine the infusion product, and to detect CAR T cells in blood and tissue samples (Wang et al., Citation2011). Further, CD19-CAR_EGFRt T cells can be depleted through administration of an anti-EGFR antibody to mitigate toxicity or terminate the therapeutic effect after tumor clearance. We have recently shown in a pre-clinical model that depletion of CD19-CAR_EGFRt T cells after lymphoma eradication reverses B-cell aplasia as the major long-term side effect and adds another layer of safety to CD19-CAR T-cell therapy (Paszkiewicz et al., Citation2016).
Regulatory considerations for clinical trial approval
Currently, the main focus of clinical development of the SB transposon/transposase system is on ex vivo gene delivery into primary cells. Thus, from a regulatory point of view, the actual investigational medicinal product (IMP) administered to humans are the genetically modified cells, which in the European Union (EU) are classified as gene therapy medicinal product (GTMP) in most cases. In the EU, GTMPs belong to the group of advanced therapy medicinal products (ATMPs). Basic guidance on the regulatory requirements for this type of products regarding quality and non-clinical aspects, as well as on clinical development is given in the “Guideline on quality, non-clinical and clinical aspects of medicinal products containing genetically modified cells” (EMA/CAT/GTWP/671639/2008), which was released in 2012. However, in this guideline only limited information is provided on the new technologies for genetic modification of cells, such as the transposon/transposase system.
For genetically modified cells, both the cells to be modified as well as the vector encoding the transposon and the therapeutic gene are considered to be starting materials, and need to be manufactured according to the principles of GMP. However, in case of primary cell donation, procurement and testing of the cells can be performed outside of a GMP environment. The principles of testing quality parameters during manufacturing and for release of the final product are, for the cellular component, similar to most cell therapies, irrespective whether the cells are genetically modified or not, but may depend on the cell type employed.
One important aspect that is specific for transposon-based genetic modification of cells is to monitor the integration profile of the transposon and the number of vector copies integrated into the host genome. As discussed above, genomic integration may have functional consequences, and such impact on the genetic stability of the cells and their proliferation and differentiation capacity needs to be analyzed and documented. In addition, potential toxic effects by the transposase need to be taken into account. In many cases the potency of such products is directly linked to the number or percentage of gene-modified cells as well as to the levels of expression of the therapeutic sequence. It is important to determine these parameters already in early clinical phases as release criteria for each batch. Increasing clinical experience would then allow the development of biological assays indicative for the functional activity of the therapy and related to the clinical efficacy.
In contrast to classical integrating vectors such as retro- or lentiviral vectors, which deliver and insert the therapeutic sequence into the host genome in a “one-shot” process, the transposon in theory is able to be repeatedly excised and integrated at different loci in case of persistent transposase activity. Therefore, a critical issue is the presence of the transposase at the time of administration of the IMP. Delivery of the transposase as protein might thus be preferable to delivery via transposase-encoding mRNA or DNA; however, this will most probably be at the expense of efficacy of the genetic modification. Currently, transcriptionally regulated transposase expression systems are under evaluation, to allow for highly efficient delivery of the transposase gene and concurrent switch-off of gene expression prior to administration of the cell to the patient (Cocchiarella et al., Citation2016). In such a case, full function of the system including control of leakage needs to be demonstrated before clinical application.
As for all other gene modifying systems, the risk of insertional oncogenesis due to semi-random genomic integration is an important regulatory concern prior to human use. Transposons integrate their payload, similar to retro- and lentiviruses, in a more or less random fashion bearing the risk of activating a proto-oncogene or inactivating a tumor suppressor gene, as discussed above. However, additional factors may contribute to the risk of oncogenesis, such as the presence and strength of transcriptional regulatory elements, the therapeutic gene, the vector dose, the target cell population, and the indication to be treated (Aiuti et al., Citation2013b). Thus, the need and, if so, the extent of integration studies, genotoxicity and tumorigenicity studies strongly depends on the above parameters employed in context of the transposon/transposase delivery system. The current clinical database available for retro- or lentiviral vector-transduced T cells used in cancer immunotherapy, for example, justifies that integration studies for this type of therapy are omitted, taking into account also the life-threatening nature of the disease with poor treatment alternatives. For the transposon/transposase system, clinical experience is more limited, and thus a decision on the need for integration and/or tumorigenicity studies is on a case-by-case basis, and should follow a risk-based approach. In any case, use of transient delivery of the transposase in the form of mRNA or recombinant protein is preferred over transposase coding sequences in the form of DNA to limit transposase activity to reduce the risk of insertional mutagenesis.
Perspective: Sleeping Beauty – a possible new gold standard for gene transfer!
The first clinical application of the SB system is currently ongoing using autologous T cells gene-modified with SB vectors carrying a CAR to render the T cells cytotoxic specifically toward CD19-positive lymphoid tumors (Huang et al., Citation2008; Kebriaei et al., Citation2016; Singh et al., Citation2008). Lymphocytes represent a suitable initial platform for testing new gene transfer systems, as T cells can be genetically modified using viral and non-viral approaches without apparent resulting genotoxicity. The advantage of using the SB system for genetic modification includes i) the ease and reduced cost associated with manufacturing of clinical-grade, plasmid-based vectors compared with recombinant viral vectors, ii) scalability: SB vectors can be manufactured in any quantity, iii) easy quality control for clinical use, and iii) indefinite storage with absolute fidelity.
A major hurdle in ex vivo delivery of the transposon components into relevant primary cell types is the toxicity of contemporary transfection/electroporation protocols. In situations, where target cells are scarce and/or culturing and expansion of the transfected cells is impossible or cannot be solved without compromising cell identity and grafting potential, cytotoxicity of the transfection procedures is a serious issue that may undermine clinical applications. However, recent experimental data indicate that by employing transposon cassettes vectorized as MCs and by providing the transposase in the form of mRNA cellular toxicity can be reduced (Monjezi et al., Citation2016), thereby positioning clinical applications with SB well within reach, at least in the area of T cell engineering.
Alternatively, the development of hybrid vector systems combining the natural ability of viruses to traverse cell mebranes with efficient genomic insertion mediated by the SB system can be a promising strategy. Indeed, components of the SB transposon have been incorporated into integrase-defective lentiviral particles that showed efficient gene transfer in a range of human cell types and an insertion profile favorable to conventional lentiviral vectors (Moldt et al., Citation2011; Staunstrup et al., Citation2009; Vink et al., Citation2009). Hybrid adenovirus/SB vectors (Boehme et al., Citation2016; Yant et al., Citation2002) have been used to efficiently deliver SB transposon vectors expressing FIX into the liver in a hemophilic dog model (Hausl et al., Citation2010). Recently, a new approach was established based on a hybrid adenovirus/SB vector targeted to CD46, a receptor expressed on HSCs. The system has been shown to yield up to 10% stable gene transfer in HSCs following in vivo transduction, thereby alleviating the need for myeloablation and transplantation (Richter et al., Citation2016). Retroviral vectors disabled in generating a cDNA copy of the retroviral vector have been shown to deliver the SB transposase mRNA into target cells with an impressive efficiency (Galla et al., Citation2011). Herpes simplex virus vectors with a tropism to infect neural progenitor cells have been used to target SB insertions in the central nervous system in an in utero gene delivery system in the mouse (Bowers et al., Citation2006; de Silva et al., Citation2010). Finally, similar to adenovirus, baculoviral vectors can efficiently transduce many cell types, but can only provide transient expression in the absence of stable genomic integration. Hybrid baculovirus/SB vectors have been shown to confer prolonged transgene expression in human cells both in vitro and in vivo in intratumoral injections in xenograft mice (Luo et al., Citation2012), as well as in vivo in the mouse eye (Turunen et al., Citation2014). Note that, although these hybrid virus/SB transposon vectors may provide advantage from a technical point of view (non-toxic delivery of the SB vector components into target cells), their application would lose all the advantages of a fully non-viral system that we outlined above.
Ex vivo modification of T cells is currently limited to a small number of centers with the required infrastructure and expertise in the logistics of cell isolation, activation, gene transfer, expansion and cryopreservation steps. To simplify procedures and widen applicability for clinical therapies, automation of these procedures is being developed. The CliniMACS Prodigy® (Miltenyi Biotec, Bergisch Gladbach, Germany) device delivers automated cell processing in a closed GMP-compliant system, and has recently been adapted for lentiviral transduction of T cells (Mock et al., Citation2016). The advantage of the system is that it can standardize the complete manufacturing process of cellular products. Future generations of the system adapted to non-viral vector delivery may advance manufacturing of SB-engineered cellular products.
Alternative technologies for genetic engineering in clinically relevant cell types are rapidly advancing. Designer nucleases, including zinc finger nucleases (ZFNs), transcription activator like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeat (CRISPR)/Cas nucleases are excellent tools for genome engineering and gene editing (Doudna & Charpentier, Citation2014). The common advantage of these technologies is that they enable site-specific engineering of the genome, thereby allowing introduction of facile and highly specific changes in the genetic code. These technologies are making their way toward therapeutic applications (Yu et al., Citation2016); for example, site-specific gene addition at reasonable efficiencies has recently been demonstrated by mRNA encoding ZFN and a homology template delivered as AAV in both HSCs (Wang et al., Citation2015) and T cells (Wang et al., Citation2016a). One key difference that sets designer nucleases apart from a transposon is that they are specialized and highly active at cutting the genome at a specific sequence, but transgene addition at the cut site is dependent on double strand break repair mechanisms of the cell. In contrast, transposons are gene integration “machines” that evolved to insert themselves into the genome; an attribute that forms the basis of applying them for robust gene transfer.
Finally, the sequence “space” of the SB transposase remains largely unexplored for hyperactive phenotypes. Importantly, the crystal structure of SB100X’s catalytic domain has recently been solved (Voigt et al., Citation2016). The structure was highly informative with respect to rationalizing the biochemical basis of hyperactivity of already existing SB mutants, and can likely be used in the future for structure-based design of next generation SB transposases for therapeutic gene delivery.
Acknowledgments
M.H. is a member of the “Young Scholar Program” (Junges Kolleg) of the Bavarian Academy of Sciences (Bayerische Akademie der Wissenschaften).
Disclosure statement
M.H., Z.Iz. and Z.Iv. are inventors on patents related to Sleeping Beauty gene transfer technology.
Additional information
Funding
References
- Aisenbrey S, Lafaut BA, Szurman P, et al. (2006). Iris pigment epithelial translocation in the treatment of exudative macular degeneration: a 3-year follow-up. Arch Ophthalmol 124:183–8.
- Aiuti A, Biasco L, Scaramuzza S, et al. (2013a). Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 341:1233151.
- Aiuti A, Cossu G, De Felipe P, et al. (2013b). The committee for advanced therapies’ of the European medicines agency reflection paper on management of clinical risks deriving from insertional mutagenesis. Hum Gene Ther Clin Dev 24:47–54.
- Aronovich EL, Bell JB, Belur LR, et al. (2007). Prolonged expression of a lysosomal enzyme in mouse liver after sleeping beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J Gene Med 9:403–15.
- Aronovich EL, Bell JB, Khan SA, et al. (2009). Systemic correction of storage disease in MPS I NOD/SCID mice using the sleeping beauty transposon system. Mol Ther 17:1136–44.
- Balaggan KS, Duran Y, Georgiadis A, et al. (2012). Absence of ocular malignant transformation after sub-retinal delivery of rAAV2/2 or integrating lentiviral vectors in p53-deficient mice. Gene Ther 19:182–8.
- Basner-Tschakarjan E, Mingozzi F. (2014). Cell-mediated immunity to AAV vectors, evolving concepts and potential solutions. Front Immunol 5:350.
- Baum C, Kalle CV, Staal FJT, et al. (2004). Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol Ther 9:5–13.
- Baus J, Liu L, Heggestad AD, et al. (2005). Hyperactive transposase mutants of the sleeping beauty transposon. Mol Ther 12:1148–56.
- Beatty GL, Moon EK. (2014). Chimeric antigen receptor T cells are vulnerable to immunosuppressive mechanisms present within the tumor microenvironment. Oncoimmunology 3:e970027.
- Belay E, Matrai J, Acosta-Sanchez A, et al. (2010). Novel hyperactive transposons for genetic modification of induced pluripotent and adult stem cells: a nonviral paradigm for coaxed differentiation. Stem Cells (Dayton, Ohio) 28:1760–71.
- Belcher JD, Vineyard JV, Bruzzone CM, et al. (2010). Heme oxygenase-1 gene delivery by Sleeping Beauty inhibits vascular stasis in a murine model of sickle cell disease. J Mol Med 88:665–75.
- Belur LR, Frandsen JL, Dupuy AJ, et al. (2003). Gene insertion and long-term expression in lung mediated by the sleeping beauty transposon system. Mol Ther 8:501–7.
- Belur LR, Podetz-Pedersen KM, Sorenson BS, et al. (2011). Inhibition of angiogenesis and suppression of colorectal cancer metastatic to the liver using the sleeping beauty transposon system. Mol Cancer 10:14.
- Berger C, Sommermeyer D, Hudecek M, et al. (2015). Safety of targeting ROR1 in primates with chimeric antigen receptor-modified T cells. Cancer Immunol Res 3:206–16.
- Biffi A, Montini E, Lorioli L, et al. (2013). Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341:1233158.
- Binder S, Krebs I, Hilgers RD, et al. (2004). Outcome of transplantation of autologous retinal pigment epithelium in age-related macular degeneration: a prospective trial. Invest Ophthalmol Vis Sci 45:4151–60.
- Boehme P, Doerner J, Solanki M, et al. (2015). The sleeping beauty transposon vector system for treatment of rare genetic diseases: an unrealized hope? Curr Gene Ther 15:255–65.
- Boehme P, Zhang W, Solanki M, et al. (2016). A high-capacity adenoviral hybrid vector system utilizing the hyperactive sleeping beauty transposase SB100X for enhanced integration. Mol Ther Nucleic Acids 5:e337.
- Boulanger-Scemama E, Querques G, About F, et al. (2015). Ranibizumab for exudative age-related macular degeneration: a five year study of adherence to follow-up in a real-life setting. J Fr Ophtalmol 38:620–7.
- Bowers WJ, Mastrangelo MA, Howard DF, et al. (2006). Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol Ther 13:580–8.
- Braun CJ, Boztug K, Paruzynski A, et al. (2014). Gene therapy for Wiskott-Aldrich syndrome–long-term efficacy and genotoxicity. Sci Transl Med 6:227ra33.
- Campochiaro PA, Nguyen QD, Shah SM, et al. (2006). Adenoviral vector-delivered pigment epithelium-derived factor for neovascular age-related macular degeneration: results of a phase I clinical trial. Hum Gene Ther 17:167–76.
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. (2009). Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326:818–23.
- Caruso HG, Torikai H, Zhang L, et al. (2016). Redirecting T-cell specificity to EGFR using mRNA to self-limit expression of chimeric antigen receptor. J Immunother 39:205–17.
- Cattoglio C, Pellin D, Rizzi E, et al. (2010). High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116:5507–17.
- Cavazza A, Moiani A, Mavilio F. (2013). Mechanisms of retroviral integration and mutagenesis. Hum Gene Ther 24:119–31.
- Cavazzana-Calvo M, Payen E, Negre O, et al. (2010). Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 467:318–22.
- Cesana D, Sgualdino J, Rudilosso L, et al. (2012). Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations. J Clin Invest 122:1667–76.
- Chabot S, Orio J, Schmeer M, et al. (2013). Minicircle DNA electrotransfer for efficient tissue-targeted gene delivery. Gene Ther 20:62–8.
- Chandler M, Clerget M, Galas DJ. (1982). The transposition frequency of IS1-flanked transposons is a function of their size. J Mol Biol 154:229–43.
- Chen Z-Y, He C-Y, Ehrhardt A, Kay MA. (2003). Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther 8:495–500.
- Chen Z-Y, Riu E, He C-Y, et al. (2008). Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther 16:548–56.
- Chen ZJ, Kren BT, Wong PY, et al. (2005). Sleeping Beauty-mediated down-regulation of huntingtin expression by RNA interference. Biochem Biophys Res Commun 329:646–52.
- Chen ZY, He CY, Meuse L, Kay MA. (2004). Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther 11:856–64.
- Chuang T-H, Lee J, Kline L, et al. (2002). Toll-like receptor 9 mediates CpG-DNA signaling. J Leukoc Biol 71:538–44.
- Cocchiarella F, Latella MC, Basile V, et al. (2016). Transcriptionally regulated and nontoxic delivery of the hyperactive sleeping beauty transposase. Mol Ther Methods Clin Dev 3:16038.
- Cui Z, Geurts AM, Liu G, et al. (2002). Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol 318:1221–35.
- Darquet AM, Cameron B, Wils P, et al. (1997). A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther 4:1341–9.
- Davila ML, Riviere I, Wang X, et al. (2014). Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med 6:224–5.
- De Ravin SS, Su L, Theobald N, et al. (2014). Enhancers are major targets for murine leukemia virus vector integration. J Virol 88:4504–13.
- De Silva S, Mastrangelo MA, Lotta LT Jr, et al. (2010). Extending the transposable payload limit of sleeping beauty (SB) using the herpes simplex virus (HSV)/SB amplicon-vector platform. Gene Ther 17:424–31.
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, et al. (2007). Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest 117:2225–32.
- Deniger DC, Pasetto A, Tran E, et al. (2016). Stable, nonviral expression of mutated tumor neoantigen-specific T-cell receptors using the sleeping beauty transposon/transposase system. Mol Ther 24:1078–89.
- Dijkhuizen PA, Pasterkamp RJ, Hermens WT, et al. (1998). Adenoviral vector-mediated gene delivery to injured rat peripheral nerve. J Neurotrauma 15:387–97.
- Doudna JA, Charpentier E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096.
- Enslow R, Bhuvanagiri S, Vegunta S, et al. (2016). Association of anti-VEGF injections with progression of geographic atrophy. Ophthalmol Eye Dis 8:31–2.
- Escobar H, Schowel V, Spuler S, et al. (2016). Full-length dysferlin transfer by the hyperactive sleeping beauty transposase restores dysferlin-deficient muscle. Mol Ther Nucleic Acids 5:e277.
- Essner JJ, Mcivor RS, Hackett PB. (2005). Awakening gene therapy with sleeping beauty transposons. Curr Opin Pharmacol 5:513–9.
- Eyjolfsdottir H, Eriksdotter M, Linderoth B, et al. (2016). Targeted delivery of nerve growth factor to the cholinergic basal forebrain of Alzheime’s disease patients: application of a second-generation encapsulated cell biodelivery device. Alzheimers Res Ther 8:30.
- Falkner-Radler CI, Krebs I, Glittenberg C, et al. (2011). Human retinal pigment epithelium (RPE) transplantation: outcome after autologous RPE-choroid sheet and RPE cell-suspension in a randomised clinical study. Br J Ophthalmol 95:370–5.
- Fernando S, Fletcher BS. (2006). Sleeping beauty transposon-mediated nonviral gene therapy. BioDrugs 20:219–29.
- Fischer SE, Van Luenen HG, Plasterk RH. (1999). Cis requirements for transposition of Tc1-like transposons in C. elegans. Mol Gen Genet 262:268–74.
- Galla M, Schambach A, Falk CS, et al. (2011). Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Res 39:7147–60.
- Garrison BS, Yant SR, Mikkelsen JG, Kay MA. (2007). Postintegrative gene silencing within the sleeping beauty transposition system. Mol Cell Biol 27:8824–33.
- Geurts AM, Yang Y, Clark KJ, et al. (2003). Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther 8:108–17.
- Gogol-Doring A, Ammar I, Gupta S. (2016). Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the piggyBac transposon in primary human CD4(+) T cells. Mol Ther 24:592–606.
- Grabundzija I, Irgang M, Mates L, et al. (2010). Comparative analysis of transposable element vector systems in human cells. Mol Ther 18:1200–9.
- Grossniklaus HE, Kang SJ, Berglin L. (2010). Animal models of choroidal and retinal neovascularization. Prog Retin Eye Res 29:500–19.
- Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. (2008). Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest 118:3132–42.
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. (2003). LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415–9.
- Hackett PB, Ekker SC, Largaespada DA, Mcivor RS. (2005). Sleeping beauty transposon-mediated gene therapy for prolonged expression. Adv Genet 54:189–232.
- Hackett PB Jr., Aronovich EL, Hunter D, et al. (2011). Efficacy and safety of Sleeping Beauty transposon-mediated gene transfer in preclinical animal studies. Curr Gene Ther 11:341–9.
- Hackett PB, Largaespada DA, Cooper LJN. (2010). A transposon and transposase system for human application. Mol Ther 18:674–83.
- Hargrove PW, Kepes S, Hanawa H, et al. (2008). Globin lentiviral vector insertions can perturb the expression of endogenous genes in beta-thalassemic hematopoietic cells. Mol Ther 16:525–33.
- Hartman ZC, Appledorn DM, Amalfitano A. (2008). Adenovirus vector induced innate immune responses: impact upon efficacy and toxicity in gene therapy and vaccine applications. Virus Res 132:1–14.
- Hausl MA, Zhang W, Muther N, et al. (2010). Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol Ther 18:1896–906.
- He C-X, Shi D, Wu W-J, et al. (2004). Insulin expression in livers of diabetic mice mediated by hydrodynamics-based administration. World J Gastroenterol 10:567–72.
- Hemmi H, Takeuchi O, Kawai T, et al. (2000). A toll-like receptor recognizes bacterial DNA. Nature 408:740–5.
- Hermens WT, Verhaagen J. (1997). Adenoviral vector-mediated gene expression in the nervous system of immunocompetent Wistar and T cell-deficient nude rats: preferential survival of transduced astroglial cells in nude rats. Hum Gene Ther 8:1049–63.
- Hirsch ML, Wolf SJ, Samulski RJ. (2016). Delivering transgenic DNA eExceeding the carrying capacity of AAV vectors. Methods Mol Biol 1382:21–39.
- Hollis RP, Nightingale SJ, Wang X, et al. (2006). Stable gene transfer to human CD34(+) hematopoietic cells using the sleeping beauty transposon. Exp Hematol 34:1333–43.
- Howe SJ, Mansour MR, Schwarzwaelder K, et al. (2008). Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest 118:3143–50.
- Huang X, Guo H, Kang J, et al. (2008). Sleeping beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther 16:580–9.
- Huang X, Guo H, Tammana S, et al. (2010). Gene transfer efficiency and genome-wide integration profiling of sleeping beauty, Tol2, and piggyBac transposons in human primary T cells. Mol Ther 18:1803–13.
- Hudecek M, Lupo-Stanghellini MT, Kosasih PL, et al. (2013). Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin Cancer Res 19:3153–64.
- Hudecek M, Sommermeyer D, Kosasih PL, et al. (2015). The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol Res 3:125–35.
- Huerfano S, Ryabchenko B, Forstová J. (2013). Nucleofection of expression vectors induces a robust interferon response and inhibition of cell proliferation. DNA Cell Biol 32:467–79.
- Hyde SC, Pringle IA, Abdullah S, et al. (2008). CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat Biotechnol 26:549–51.
- Ivics Z, Garrels W, Mátés L, et al. (2014a). Germline transgenesis in pigs by cytoplasmic microinjection of Sleeping Beauty transposons. Nat Protoco 9:810–27.
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. (1997). Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91:501–10.
- Ivics Z, Hiripi L, Hoffmann OI, et al. (2014b). Germline transgenesis in rabbits by pronuclear microinjection of Sleeping Beauty transposons. Nat Protoc 9:794–809.
- Ivics Z, Izsvak Z. (2006). Transposons for gene therapy! Curr Gene Ther 6:593–607.
- Ivics Z, Izsvak Z. (2010). The expanding universe of transposon technologies for gene and cell engineering. Mob DNA 1:25.
- Ivics Z, Izsvak Z. (2011). Nonviral gene delivery with the sleeping beauty transposon system. Hum Gene Ther 22:1043–51.
- Ivics Z, Izsvak Z. (2015). Sleeping beauty transposition. Microbiol Spectr 3:MDNA3.
- Ivics Z, Izsvák Z, Minter A, Hackett PB. (1996). Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci. USA 93:5008–13.
- Ivics Z, Katzer A, Stuwe EE, et al. (2007). Targeted sleeping beauty transposition in human cells. Mol Ther 15:1137–44.
- Ivics Z, Li MA, Mates L, et al. (2009). Transposon-mediated genome manipulation in vertebrates. Nat Methods 6:415–22.
- Ivics Z, Mates L, Yau TY, et al. (2014c). Germline transgenesis in rodents by pronuclear microinjection of sleeping beauty transposons. Nat Protoc 9:773–93.
- Izsvak Z, Chuah MK, Vandendriessche T, Ivics Z. (2009). Efficient stable gene transfer into human cells by the sleeping beauty transposon vectors. Methods 49:287–97.
- Izsvak Z, Hackett PB, Cooper LJN, Ivics Z. (2010). Translating sleeping beauty transposition into cellular therapies: victories and challenges. Bioessays 32:756–67.
- Izsvak Z, Ivics Z. (2004). Sleeping beauty transposition: biology and applications for molecular therapy. Mol Ther 9:147–56.
- Izsvák Z, Ivics Z, Plasterk RH. (2000). Sleeping beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol 302:93–102.
- Izsvák Z, Khare D, Behlke J, et al. (2002). Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in sleeping beauty transposition. J Biol Chem 277:34581–8.
- Jahner D, Stuhlmann H, Stewart CL, et al. (1982). De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298:623–8.
- Jin Z, Maiti S, Huls H, et al. (2011). The hyperactive sleeping beauty transposase SB100X improves the genetic modification of T cells to express a chimeric antigen receptor. Gene Ther 18:849–56.
- Johnen S, Djalali-Talab Y, Kazanskaya O, et al. (2015). Antiangiogenic and neurogenic activities of sleeping beauty-mediated PEDF-transfected RPE cells in vitro and in vivo. Biomed Res Int 2015:863845.
- Johnen S, Izsvak Z, Stocker M, et al. (2012). Sleeping beauty transposon-mediated transfection of retinal and iris pigment epithelial cells. Invest Ophthalmol Vis Sci 53:4787–96.
- Karsi A, Moav B, Hackett P, Liu Z. (2001). Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Mar Biotechnol 3:241–5.
- Kebriaei P, Singh H, Huls MH, et al. (2016). Phase I trials using sleeping beauty to generate CD19-specific CAR T cells. J Clin Invest 126:3363–76.
- Kochenderfer JN, Dudley ME, Kassim SH, et al. (2015). Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J Clin Oncol 33:540–9.
- Kowarz E, Loscher D, Marschalek R. (2015). Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J 10:647–53.
- Kren BT, Unger GM, Sjeklocha L, et al. (2009). Nanocapsule-delivered sleeping beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J Clin Invest 119:2086–99.
- Kuerten D, Johnen S, Harmening N, et al. (2015). Transplantation of PEDF-transfected pigment epithelial cells inhibits corneal neovascularization in a rabbit model. Graefes Arch Clin Exp Ophthalmol 253:1061–9.
- Kustikova O, Fehse B, Modlich U, et al. (2005). Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science (New York, N.Y.) 308:1171–4.
- Lad EM, Hammill BG, Qualls LG, et al. (2014). Anti-VEGF treatment patterns for neovascular age-related macular degeneration among medicare beneficiaries. Am J Ophthalmol 158:537–43 e2.
- Lampe DJ, Grant TE, Robertson HM. (1998). Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149:179–87.
- Lappas A, Foerster AM, Weinberger AW, et al. (2004). Translocation of iris pigment epithelium in patients with exudative age-related macular degeneration: long-term results. Graefes Arch Clin Exp Ophthalmol 242:638–47.
- Latella MC, Cocchiarella F, De Rosa L, et al. (2016). Correction of recessive dystrophic epidermolysis bullosa by transposon-mediated integration of COL7A1 in transplantable patient-derived primary keratinocytes. J Invest Dermatol, in press.
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, et al. (2015). T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385:517–28.
- Liu G, Geurts AM, Yae K, et al. (2005). Target-site preferences of sleeping beauty transposons. J Mol Biol 346:161–73.
- Liu L, Liu H, Visner G, Fletcher BS. (2006a). Sleeping beauty-mediated eNOS gene therapy attenuates monocrotaline-induced pulmonary hypertension in rats. FASEB J 20:2594–6.
- Liu L, Mah C, Fletcher BS. (2006b). Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice using an endothelial-targeted Sleeping Beauty transposon. Mol Ther 13:1006–15.
- Liu L, Sanz S, Heggestad AD, et al. (2004). Endothelial targeting of the sleeping beauty transposon within lung. Mol Ther 10:97–105.
- Luo WY, Shih YS, Hung CL, et al. (2012). Development of the hybrid sleeping beauty: baculovirus vector for sustained gene expression and cancer therapy. Gene Ther 19:844–51.
- Magnani CF, Turazzi N, Benedicenti F, et al. (2016). Immunotherapy of acute leukemia by chimeric antigen receptor-modified lymphocytes using an improved sleeping beauty transposon platform. Oncotarget 7:51581–97.
- Maldarelli F, Wu X, Su L, et al. (2014). HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–83.
- Marie C, Vandermeulen G, Quiviger M, et al. (2010). pFARs, plasmids free of antibiotic resistance markers, display high-level transgene expression in muscle, skin and tumour cells. J Gene Med 12:323–32.
- Mátés L, Chuah MK, Belay E, et al. (2009). Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet, 41:753–61.
- Matrai J, Chuah MK, Vandendriessche T. (2010). Recent advances in lentiviral vector development and applications. Mol Ther 18:477–90.
- Maude SL, Frey N, Shaw PA, et al. (2014). Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 371:1507–17.
- Maus MV, Haas AR, Beatty GL, et al. (2013). T cells expressing chimeric antigen receptors can cause anaphylaxis in humans. Cancer Immunol Res 1:26–31.
- Menendez-Arias L. (2009). Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses 1:1137–65.
- Mikkelsen JG, Yant SR, Meuse L, et al. (2003). Helper-Independent sleeping beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol Ther 8:654–65.
- Mock U, Nickolay L, Philip B, et al. (2016). Automated manufacturing of chimeric antigen receptor T cells for adoptive immunotherapy using CliniMACS prodigy. Cytotherapy 18:1002–11.
- Modlich U, Navarro S, Zychlinski D, et al. (2009). Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther 17:1919–28.
- Moldt B, Miskey C, Staunstrup NH, et al. (2011). Comparative genomic integration profiling of sleeping beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells. Mol Ther 19:1499–510.
- Moldt B, Yant SR, Andersen PR, et al. (2007). Cis-acting gene regulatory activities in the terminal regions of sleeping beauty DNA transposon-based vectors. Hum Gene Ther 18:1193–204.
- Monjezi R, Miskey C, Gogishvili T, et al. (2016). Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia 31:186–94.
- Montini E, Held PK, Noll M, et al. (2002). In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol Ther 6:69.
- Morgan RA, Dudley ME, Wunderlich JR, et al. (2006). Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 314:126–9.
- Morisato D, Way JC, Kim HJ, Kleckner N. (1983). Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell 32:799–807.
- Muses S, Morgan JE, Wells DJ. (2011). Restoration of dystrophin expression using the sleeping beauty transposon. PLoS Curr 3:RRN1296.
- Naldini L. (2011). Ex vivo gene transfer and correction for cell-based therapies. Nat Rev Genet 12:301–15.
- Narayanavari SA, Chilkunda SS, Ivics Z, Izsvak Z. (2016). Sleeping beauty transposition: from biology to applications. Crit Rev Biochem Mol Biol 52:18–44.
- Nesmith BL, Ihnen M, Schaal S. (2014). Poor responders to bevacizumab pharmacotherapy in age-related macular degeneration and in diabetic macular edema demonstrate increased risk for obstructive sleep apnea. Retina 34:2423–30.
- Newrzela S, Cornils K, Li Z, et al. (2008). Resistance of mature T cells to oncogene transformation. Blood 112:2278–86.
- Ohlfest JR, Demorest ZL, Motooka Y, et al. (2005a). Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the sleeping beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol Ther 12:778–88.
- Ohlfest JR, Frandsen JL, Fritz S, et al. (2005b). Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood 105:2691–8.
- Ohno-Matsui K, Morita I, Tombran-Tink J, et al. (2001). Novel mechanism for age-related macular degeneration: an equilibrium shift between the angiogenesis factors VEGF and PEDF. J Cell Physiol 189:323–33.
- Orban TI, Apati A, Nemeth A, et al. (2009). Applying a “double-feature” promoter to identify cardiomyocytes differentiated from human embryonic stem cells following transposon-based gene delivery. Stem Cells 27:1077–87.
- Ortiz-Urda S, Lin Q, Yant SR, et al. (2003). Sustainable correction of junctional epidermolysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther 10:1099–104.
- Osborn MJ, Mcelmurry RT, Lees CJ, et al. (2011). Minicircle DNA-based gene therapy coupled with immune modulation permits long-term expression of α-L-Iduronidase in mice with mucopolysaccharidosis type I. Mol Ther 19:450–60.
- Ott MG, Schmidt M, Schwarzwaelder K, et al. (2006). Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 12:401–9.
- Pan XJ, Ma ZZ, Zhang QJ, et al. (2012). Sleeping beauty transposon system is a reliable gene delivery tool for hereditary tyrosinaemia type 1 disease gene therapy: size of the foreign gene decides the timing of stable integration into the host chromosomes. J Int Med Res 40:1850–9.
- Pannell D, Ellis J. (2001). Silencing of gene expression: implications for design of retrovirus vectors. Rev Med Virol 11:205–17.
- Papapetrou EP, Lee G, Malani N, et al. (2011). Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol 29:73–8.
- Paszkiewicz PJ, Frassle SP, Srivastava S, et al. (2016). Targeted antibody-mediated depletion of murine CD19 CAR T cells permanently reverses B cell aplasia. J Clin Invest 126:4262–72.
- Peng PD, Cohen CJ, Yang S, et al. (2009). Efficient nonviral sleeping beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther 16:1042–9.
- Rapoport AP, Stadtmauer EA, Binder-Scholl GK, et al. (2015). NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med 21:914–21.
- Recchia A, Bonini C, Magnani Z, et al. (2006). Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci USA 103:1457–62.
- Reyes-Sandoval A, Ertl HC. (2004). CpG methylation of a plasmid vector results in extended transgene product expression by circumventing induction of immune responses. Mol Ther 9:249–61.
- Richter M, Saydaminova K, Yumul R, et al. (2016). In vivo transduction of primitive mobilized hematopoietic stem cells after intravenous injection of integrating adenovirus vectors. Blood 128:2206–17.
- Rostovskaya M, Fu J, Obst M, et al. (2012). Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res 40:e150.
- Rostovskaya M, Naumann R, Fu J, et al. (2013). Transposon mediated BAC transgenesis via pronuclear injection of mouse zygotes. Genesis 51:135–41.
- Sadelain M, Brentjens R, Riviere I. (2013). The basic principles of chimeric antigen receptor design. Cancer Discov 3:388–98.
- Sadelain M, Papapetrou EP, Bushman FD. (2012). Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer 12:51–8.
- Saint-Geniez M, Maharaj AS, Walshe TE, et al. (2008). Endogenous VEGF is required for visual function: evidence for a survival role on muller cells and photoreceptors. PLoS One 3:e3554.
- Salganik M, Hirsch ML, Samulski RJ. (2015). Adeno-associated virus as a mammalian DNA vector. Microbiol Spectr 3:MDNA3-0052-2014.
- Schambach A, Bohne J, Chandra S, et al. (2006). Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol Ther 13:391–400.
- Schambach A, Galla M, Maetzig T, et al. (2007). Improving transcriptional termination of self-inactivating gamma-retroviral and lentiviral vectors. Mol Ther 15:1167–73.
- Scott AW, Bressler SB. (2013). Long-term follow-up of vascular endothelial growth factor inhibitor therapy for neovascular age-related macular degeneration. Curr Opin Ophthalmol 24:190–6.
- Sharma N, Cai Y, Bak RO, et al. (2013). Efficient sleeping beauty DNA transposition from DNA minicircles. Mol Ther Nucleic Acids 2:e74.
- Singh H, Figliola MJ, Dawson MJ, et al. (2013). Manufacture of clinical-grade CD19-specific T cells stably expressing chimeric antigen receptor using sleeping beauty system and artificial antigen presenting cells. PLoS One 8:e64138.
- Singh H, Manuri PR, Olivares S, et al. (2008). Redirecting specificity of T-cell populations for CD19 using the sleeping beauty system. Cancer Res 68:2961–71.
- Sinn PL, Sauter SL, Mccray PB Jr. (2005). Gene therapy progress and prospects: development of improved lentiviral and retroviral vectors–design, biosafety, and production. Gene Ther 12:1089–98.
- Solensky R. (2003). Hypersensitivity reactions to beta-lactam antibiotics. Clin Rev Allergy Immunol 24:201–20.
- Sommermeyer D, Hudecek M, Kosasih PL, et al. (2015). Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia 30:492–500.
- Song J, Kim C, Ochoa ER. (2009). Sleeping beauty-mediated suicide gene therapy of hepatocellular carcinoma. Biosci Biotechnol Biochem 73:165–8.
- Srivastava S, Riddell SR. (2015). Engineering CAR-T cells: design concepts. Trends Immunol 36:494–502.
- Staunstrup NH, Moldt B, Mates L, et al. (2009). Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol Ther 17:1205–14.
- Sumiyoshi T, Holt NG, Hollis RP, et al. (2009). Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells, using the sleeping beauty transposon system. Hum Gene Ther 20:1607–26.
- Thrasher AJ, Gaspar HB, Baum C, et al. (2006). Gene therapy: X-SCID transgene leukaemogenicity. Nature 443:E5–6. discussion E6–7.
- Thumann G, Harmening N, Prat-Souteyrand C, et al. (2017). Engineering of PEDF-expressing primary pigment epithelial cells by the sleeping beauty transposon system delivered by pFAR4 miniplasmids. Mol Ther Nucleic Acids 6:302–14.
- Tse LV, Moller-Tank S, Asokan A. (2015). Strategies to circumvent humoral immunity to adeno-associated viral vectors. Expert Opin Biol Ther 15:845–55.
- Turtle CJ, Hanafi LA, Berger C, et al. (2016a). CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 126:2123–38.
- Turtle CJ, Hanafi LA, Berger C, et al. (2016b). Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci Transl Med 8:355ra116.
- Turtle CJ, Maloney DG. (2016). Clinical trials of CD19-targeted CAR-modified T cell therapy; a complex and varied landscape. Expert Rev Hematol 9:719–21.
- Turunen TA, Kurkipuro J, Heikura T, et al. (2016). Sleeping beauty transposon vectors in liver-directed gene delivery of LDLR and VLDLR for gene therapy of familial hypercholesterolemia. Mol Ther 24:620–35.
- Turunen TA, Laakkonen JP, Alasaarela L, et al. (2014). Sleeping beauty-baculovirus hybrid vectors for long-term gene expression in the eye. J Gene Med 16:40–53.
- Vandendriessche T, Ivics Z, Izsvak Z, Chuah MKL. (2009). Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood 114:1461–8.
- Velez-Montoya R, Oliver SC, Olson JL, et al. (2014). Current knowledge and trends in age-related macular degeneration: genetics, epidemiology, and prevention. Retina 34:423–41.
- Vigdal TJ, Kaufman CD, Izsvák Z, et al. (2002). Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol 323:441–52.
- Vink CA, Gaspar HB, Gabriel R, et al. (2009). Sleeping beauty transposition from nonintegrating lentivirus. Mol Ther 17:1197–204.
- Voigt F, Wiedemann L, Zuliani C, et al. (2016). Sleeping beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat Commun 7:11126.
- Wagner TA, Mclaughlin S, Garg K, et al. (2014). HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345:570–3.
- Walisko O, Schorn A, Rolfs F, et al. (2008). Transcriptional activities of the sleeping beauty transposon and shielding its genetic cargo with insulators. Mol Ther 16:359–69.
- Wang J, Declercq JJ, Hayward SB, et al. (2016a). Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res 44:e30.
- Wang J, Exline CM, Declercq JJ, et al. (2015). Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol 33:1256–63.
- Wang X, Chang WC, Wong CW, et al. (2011). A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118:1255–63.
- Wang X, Sarkar DP, Mani P, et al. (2009). Long-term reduction of jaundice in Gunn rats by nonviral liver-targeted delivery of sleeping beauty transposon. Hepatology 50:815–24.
- Wang Y, Pryputniewicz-Dobrinska D, Nagy EE, et al. (2016b). Regulated complex assembly safeguards the fidelity of sleeping beauty transposition. Nucleic Acids Res 45:311–26.
- Wang Y, Wang J, Devaraj A, et al. (2014). Suicidal autointegration of sleeping beauty and piggyBac transposons in eukaryotic cells. PLoS Genet 10:e1004103.
- Wang Z, Troilo PJ, Wang X, et al. (2004). Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther 11:711–21.
- Wiehe JM, Ponsaerts P, Rojewski MT, et al. (2007). mRNA-mediated gene delivery into human progenitor cells promotes highly efficient protein expression. J Cell Mol Med 11:521–30.
- Wilber A, Frandsen JL, Geurts JL, et al. (2006). RNA as a source of transposase for sleeping beauty-mediated gene insertion and expression in somatic cells and tissues. Mol Ther 13:625–30.
- Wilber A, Wangensteen KJ, Chen Y, et al. (2007). Messenger RNA as a source of transposase for sleeping beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol Ther 15:1280–7.
- Wolf D, Goff SP. (2009). Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–4.
- Xiao J, Meng XM, Huang XR, et al. (2012). miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther 20:1251–60.
- Xue X, Huang X, Nodland SE, et al. (2009). Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive sleeping beauty transposon system. Blood 114:1319–30.
- Yant SR, Ehrhardt A, Mikkelsen JG, et al. (2002). Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol 20:999–1005.
- Yant SR, Meuse L, Chiu W, et al. (2000). Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet 25:35–41.
- Yant SR, Park J, Huang Y, et al. (2004). Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol 24:9239–47.
- Yu KR, Natanson H, Dunbar CE. (2016). Gene editing of human hematopoietic stem and progenitor cells: promise and potential hurdles. Hum Gene Ther, in press.
- Zayed H, Izsvak Z, Walisko O, Ivics Z. (2004). Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther 9:292–304.
- Zhu J, Kren BT, Park CW, et al. (2007). Erythroid-specific expression of beta-globin by the sleeping beauty transposon for Sickle cell disease. Biochemistry 46:6844–58.
