Monitoring of minimal residual disease (MRD) has become routine clinical practice in frontline treatment of virtually all childhood acute lymphoblastic leukemia (ALL) and many adult ALL patients [Citation1]. MRD diagnostics has proven to be the strongest prognostic factor, allowing for risk group assignment into different treatment arms, resulting in significant treatment reduction or mild or strong intensification. Within the Dutch Childhood Oncology Group (DCOG) ALL10 protocol, MRD-based low-risk patients received significant treatment reduction, resulting in an excellent outcome with very few side effects [Citation2].
Given the significant treatment reduction of MRD-negative patients within the DCOG-ALL10 protocol, the strict criteria of the MRD-based low-risk group of the original I-BFM-SG study [Citation3] have been retained to define MRD negativity, using at least two different types of sensitive immunoglobulin (IG)-T-cell receptor (TR) polymerase chain reaction (PCR) targets, thereby avoiding or reducing oligoclonality problems and related false negativity [Citation3–5]. However, false negativity may also be caused by mixing up patient samples [Citation6,Citation7], resulting in under-treatment of the corresponding patient and increased relapse risk. We used short tandem repeat (STR) analysis to evaluate whether sample mix-up occurred in low-risk (LR) ALL patients treated within the DCOG-ALL10 protocol.
The DNA samples from 30 LR patients used for MRD diagnostics at diagnosis, day 33 and day 78 were evaluated by STR analysis using the PowerPlex® 16 system (Promega, Leiden, the Netherlands). This kit evaluates 15 tetranucleotide repeat loci on different chromosomal positions. In addition, diagnostic and day 33 samples from 20 intermediate risk (IR; MRD positive at day 33, therefore confirmed not to be mixed up) were used for comparison.
In five out of 30 LR patients (16.7%), differences were observed between the diagnostic sample and the follow-up sample(s) in one or more evaluated STR ( and ). Of note, for the LR patients, the STR patterns in the two follow-up samples were always identical, suggesting that sample mix up was unlikely. This was supported by the finding that also in 5 out of 20 (25%) IR patients differences in one or more STRs were observed (). Therefore, instead of a results of sample mix up, the differences in STR pattern more likely are related to differences between leukemic cells (predominant in the diagnostic sample) and normal cells (predominant in the follow-up samples). We therefore evaluated these 10 cases with STR differences with respect to their cytogenetic findings. Indeed, in seven out of 10 cases, the difference in STR pattern could be explained by the chromosomal abnormalities in the leukemic cells (). Three cases (LR-02; LR-30 and IR-16), however, could not be explained by the cytogenetic data. For these cases, both the leukemic cells and normal cells from the (viably frozen) diagnostic sample were separated by FACS-sorting and STR was performed on the sorted cell populations. In all three cases, the STR pattern in the sorted normal cells from diagnosis was similar to the STR pattern in the follow-up samples, whereas the STR pattern of the sorted leukemic cells was different (and identical to the STR pattern of the nonsorted diagnostic sample) (). These data therefore further confirm that the different STR pattern between diagnosis and follow-up is not related to sample mix up but indeed reflects the genetic abnormalities present in the leukemic cells. MLPA analysis (SALSA MLPA P335 ALL-IKZF1 probe mix; MRC-Holland, Amsterdam, the Netherlands) was performed in these three patients. In patient LR-30, a deletion of the ETV6 gene on 12p13 was observed by FISH, whereas by STR analysis one allele for VWF on 12p12 was lost, suggesting a deletion both comprising 12p12 and 12p13. In the two other patients, MLPA analysis did not explain the STR data. It may be expected that also in patient IR-16 the missing THO STR is due to a small chromosomal deletion not detected by routine cytogenetics. We have no explanation for the shift in the VWF STR in patient LR-02, but genetic changes in the leukemic cells apparently occurred in this locus.
Figure 1. STR results of patient IR-16. (A) Diagnostic sample (top row) versus follow-up (bottom row). In the diagnostic sample, an STR for THO (11p) seems missing as compared to the follow-up sample. (B) Nonsorted cells (top row) versus sorted ALL cells (middle row) and sorted normal cells (bottom row) of the same patient clearly show specific loss of a STR in the leukemic cells.
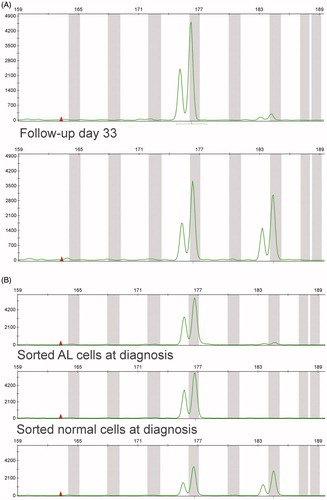
Table 1. Results of STR analysis and possible explanation by cytogenetic findings.
Our data show that sample mix up had not occurred in the LR patients evaluated in this study. In addition, our data show that the interpretation of STR data from samples obtained from leukemic patients, comparably to patients with other malignancies [Citation8–10], is not always straight-forward but can be hampered by changes in the STR pattern caused by genetic changes in the malignant cells. Results of subsequent STR analyses should therefore be interpreted with care.
Potential conflict of interest
Disclosure forms provided by the authors are available with the full text of this article online at https://doi.org/10.1080/10428194.2017.1382699.
ICMJE Forms for Disclosure of Potential Conflicts of Interest
Download Zip (4 MB)References
- van Dongen JJ, van der Velden VH, Bruggemann M, et al. Minimal residual disease diagnostics in acute lymphoblastic leukemia: need for sensitive, fast, and standardized technologies. Blood. 2015;125:3996–4009.
- Pieters R, de Groot-Kruseman H, Van der Velden V, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch childhood oncology group. JCO. 2016;34:2591–2601.
- van Dongen JJM, Seriu T, Panzer-Grumayer ER, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet. 1998;352:1731–1738.
- van der Velden VH, Cazzaniga G, Schrauder A, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21:604–611.
- van der Velden VH, van Dongen JJ. MRD detection in acute lymphoblastic leukemia patients using Ig/TCR gene rearrangements as targets for real-time quantitative PCR. Methods Mol Biol. 2009;538:115–150.
- de Leeuw N, Hehir-Kwa JY, Simons A, et al. SNP array analysis in constitutional and cancer genome diagnostics–copy number variants, genotyping and quality control. Cytogenet Genome Res. 2011;135:212–221.
- Hunt JL. Identifying cross contaminants and specimen mix-ups in surgical pathology. Adv Anat Pathol. 2008;15:211–217.
- Clark JR, Scott SD, Jack AL, et al. Monitoring of chimerism following allogeneic haematopoietic stem cell transplantation (HSCT): technical recommendations for the use of short tandem repeat (STR) based techniques, on behalf of the United Kingdom National External Quality Assessment Service for Leucocyte Immunophenotyping Chimerism Working Group. Br J Haematol. 2015;168:26–37.
- Much M, Buza N, Hui P. Tissue identity testing of cancer by short tandem repeat polymorphism: pitfalls of interpretation in the presence of microsatellite instability. Hum Pathol. 2014;45:549–555.
- Poetsch M, Petersmann A, Woenckhaus C, et al. Evaluation of allelic alterations in short tandem repeats in different kinds of solid tumors-possible pitfalls in forensic casework. Forensic Sci Int. 2004;145:1–6.
