Abstract
Diffuse large B-cell lymphoma (DLBCL) is a heterogeneous disease with variable outcomes. In this study, data of 84 DLBCL patients, who were tested EBV DNA in peripheral blood, were retrospectively analyzed. Patients were divided into three subgroups according to EBV copy number (EBV-CN) values: the negative (<500 copies/ml), low (500–104 copies/ml), and high EBV-CN group (≥104 copies/ml). The higher EBV-CN was associated with male and elderly patients. No significant difference was found between the three subgroups regarding immunophenotype, cytogenetic features, and molecular features. Patients of the high EBV-CN group had significantly worse progression-free and overall survival (OS) compared to other groups. After adjusting conventional risk factors, high EBV-CN was an independent prognostic factor for OS in multivariate analysis. Taken together, peripheral blood EBV-CN can predict outcomes of patients with DLBCL and 104 copies/ml is a more suitable boundary value than the traditional normal value in predicting prognosis.
Keywords:
Introduction
Epstein-Barr virus has been known as a human herpes virus since about five decades. It has been linked with lymphomas, initially with Burkitt’s lymphoma [Citation1], and then with other lymphoma types such as Hodgkin lymphoma (HL), natural killer/T-cell lymphoma, and diffuse large B-cell lymphoma (DLBCL) [Citation2–4]. DLBCL is the most common B cell lymphoma, accounting for 31% of adult NHLs [Citation5]. It is a heterogeneous disease and has been divided into several distinct subtypes. EBV-positive DLBCL was first defined as a provisional entity of a lymphoma occurring in older individuals (>50 years old) without any known immunodeficiency listed in 2008 WHO classification of tumors of hematopoietic and lymphoid tissues, and then reclassified as EBV-positive DLBCL, NOS (not otherwise specified), since this disease has been found in more and more younger immunocompetent patients [Citation6]. EBV-encoded small RNA (EBER) detection by in situ hybridization (EBER-ISH) is the standard method in diagnosing EBV positive DLBCL. Whole blood, plasma, or peripheral blood mononuclear cells (PBMCs) can be used for detection of EBV-DNA. Previous studies showed that EBV viral load of peripheral blood can be used as a biomarker in predicting prognosis in extranodal nasal-type NK/T-cell lymphoma (ENKTL), HL, and BL [Citation7–11]. Although several studies suggested that EBV DNA copy number may also be a potential prognostic biomarker of DLBCL [Citation12,Citation13], the correlation between EBV DNA copy number in PBMCs and DLBCL prognosis has not yet been clearly established and the stratification boundary value is controversial. Therefore, we performed this retrospective study to investigate the prognostic role of EBV DNA copy number in PBMCs in DLBCL patients.
Materials and methods
Patients and methods
In this study, we retrospectively collected clinical data of patients with DLBCL who had taken EBV copy number (EBV-CN) detection in PBMCs before treatment between March 2014 and January 2018. The eligibility criteria were: with newly diagnosed DLBCL, without human immunodeficiency virus infection, no previous history of any type of lymphoma, and not less than 18 years old. Eighty-four eligible patients were included. The DLBCL diagnosis was based on the 2016 World Health Organization (WHO) classification.
The follow-up information was available in 75 patients, among whom 68 were treated with anthracycline-based chemotherapy combined with rituximab as initial treatment, while two with the CHOP regimen (cyclophosphamide, doxorubicin, vincristine, and prednisolone) only and the other six with supportive care only. The study protocol was approved by the Institutional Review Board of Tongji Hospital.
Pathology
Pathologic evaluations were performed independently by two pathologists. Formalin-fixed paraffin-embedded tissue (FFPE) samples from lymph nodes were subjected to immunohistochemical staining using a panel of monoclonal antibodies against CD20 (L26, Dako, M0755, Nowy Sącz, Poland), CD79a (JCB117, Dako, M7050, Nowy Sącz, Poland), CD5 (4C7, Dako, M3641, Nowy Sącz, Poland), CD30 (Ber-H2, Dako, M0751, Nowy Sącz, Poland), CD10 (56C6, Dako, M7308, Nowy Sącz, Poland), BCL-2 (124, Dako, M0887, Nowy Sącz, Poland), BCL-6 (PG-B6p, Dako, M7211, Nowy Sącz, Poland), and Mum1 (MUM1p, Dako, M7259, Nowy Sącz, Poland). IHC analysis was performed using a Bond-III Autostainer (Leica Biosystems, Melbourne, Australia) under fully automated protocols as previous reported [Citation14]. The positivity cutoff value for these markers was as follows: 80% for CD20, 80% for CD79a, 10% for CD5, 10% for CD30, 10% for CD10, 30% for BCL2, 30% for BCL6, and 30% for MUM1.
According to the Hans algorithm, the patients were classified into GCB and non-GCB subtypes [Citation15]. EBER was identified by in situ hybridization (ISH) technique. Samples with EBER nuclear positivity in >30% of cells with malignant morphology were defined as EBER positive.
Fluorescence in situ hybridization (FISH)
FISH was performed on 4 μm FFPE tissue sections according to the manufacturer’s instructions, using the BCL2 break-apart probe (Vysis, 5NS1-20, Downers Grove, IL), C-MYC break-apart probe (Vysis, 1N63-20, Downers Grove, IL), BCL6 break-apart probe (Vysis, 1N23-20, Downers Grove, IL), and TP53/CEP17 probes (Vysis, 5N56-20, Downers Grove, IL). After treatment in 65 °C thermostat overnight, tissue sections were deparaffinized using xylene and then sequentially washed with 100%, 80%, and 70% ethanol. Slides were treated with configured protease K solution, followed by dehydrating in ethanol (70%, 80%, and 100%), after which the FISH probes were added before incubation for 16 h. Then slides were then washed for 1 min in 2× SSC/0.3% NP40 (SSC/Nonidet P40) at 70 °C and transferred to 2× SSC/0.1% NP40 at room temperature for 1 min, counterstained with DAPI, and observed under the microscope. Raw signals were collected and ratios calculated. The positive cutoff value used was 10% of suspected tumor cells for all FISH probes and was determined by studying a cohort of six normal control samples as previous reported [Citation16].
Quantification of EBV DNA load in PBMCs
Mononuclear cells were isolated from 2-ml pretreatment peripheral blood of each patient. PBMCs were isolated from K2EDTA-treated blood by Ficoll-Hypaque (Tianjin Hao Yang Biological Manufacture Co., Ltd., Tianjin, China) gradient centrifugation. Briefly, 2 ml of saline was added into a blood collection tube containing 2 ml of anticoagulated whole blood (1:1 dilution), and then the diluted blood was layered over 1 ml of Ficoll in a 15 ml conical tube. After centrifuging the tube at 1000×g for 30 min at 20 °C in a swinging-bucket rotor without brake, the fluffy white layer of PBMCs at the interphase was collected into a 1.5 ml centrifuge tube. After further centrifuging, the cell pellet was available for DNA extraction by using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany), according to manufacturer’s instructions. EBV-CN were quantitated by Real-Time PCR with a quantitative Epstein-Barr virus DNA detection kit (DAAN GENE, Guangzhou, China) with a detection sensitivity of 500 copies per ml. Peripheral blood EBV-CN less than 500 copies/ml was defined as negative.
Next-generation sequencing and mutation analysis
FFPE samples from lymph nodes from 41 DLBCL patients were subjected to targeted next-generation sequencing (NGS) using Ion AmpliSeq technology based on the Ion Torrent PGM platform, focusing on 20 candidate genes selected from previous studies [Citation17], including B2M, BCL2, BCL6, BRAF, CARD11, CD58, CD79B, CD83, CREBBP, EP300, EZH2, IRF4, KMT2D, MEF2B, MYC, MYD88, PIM1, PRDM1, TNFAIP3, and TP53. Ten nanograms of genomic DNA was used to prepare library by using Ion AmpliSeq Library kits 2.0 (Life Technologies, Carlsbad, CA). Then, the templates were prepared by transferring the libraries to the Ion OneTouch 2 system. Water-in-oil PCR was done on OneTouch 2 system. Sequencing was carried out on the Ion Personal Genome Machine™ System according to the protocol. Ion Reporter Server (5.0.2) and Annotate Variants plugin were used for data analysis. False positive mutations were eliminated by variant review with the Integrative Genomics Viewer, as previously described [Citation18]. Somatic cell variants corresponding to known variants present in the dbSNP or 1000 genomes program were discarded.
Statistical methods
Chi-square test or Fisher’s exact test was used to compare patient characteristics between groups. Overall survival (OS) was measured from the date of diagnosis until death or the last follow up. Progression-free survival (PFS) was calculated from the date of diagnosis until the first documented progress, relapse, death, or the last visit. Kaplan–Meier’s survival analysis was used to evaluate PFS and OS, and the log-rank test was used to assess differences. The independent risk factors were performed by using univariate analysis and multivariate analysis through the Cox proportional hazard model. Oncoplot of mutations was performed by R software v3.6.0 (‘maftools’ R package version 2.0.15). Statistical analyses were performed by using SPSS26.0 (SPSS Inc., Chicago, IL), two sides p values less than .05 were considered statistically significant.
Results
Patient characteristics
A total of 84 patients with DLBCL were included in this study, with a median age of 53 years (range, 46–62 years) and a male-to-female ratio was 0.87:1. The patients were divided into three subgroups according to EBV-CN, i.e. the negative (<500 copies/ml), low (500–104 copies/ml), and high EBV-CN group (≥104 copies/ml). Patients’ baseline characteristics are provided in . Among them, 46 patients were of negative EBV-CN, 27 patients of low EBV-CN, and 11 patients of high EBV-CN. Elderly and male patients had significantly higher EBV-CN compared to other patients (p= .049 and .003, respectively). Patients in the high EBV-CN group presented more extranodal involvement, higher ECOG score, and more non-GCB type, compared to the other two groups, though the difference was statistically non-significant.
Table 1. Baseline characteristics of patients according to EBV DNA copy number.
Immunophenotype
The pan B-cell antigens CD20 and CD79 were negative in only one patient. Compared to the EBV-CN negative and low groups, the EBV-CN high group had a higher positive rate of BCL2 (100%, 87%, and 83.8%, respectively), BCL6 (100%, 83.3%, and 85%, respectively), MUM1 (83.3%, 71.4%, and 68.6%, respectively), and EBER (25%, 11.1%, and 8.3%, respectively), and a lower positive rate of CD10 (0%, 26.1%, and 42.1, respectively); however, all these differences were statistically non-significant, except CD10 showing a marginal trend (p= .087; ).
Table 2. Pathologic characteristics of patients according to EBV DNA copy number.
Cytogenetic features
The prevalence of C-MYC rearrangement was higher in the EBV-CN high group than in the other two groups, but statistical significance was not reached (p= .083), both the C-MYC copy number variation and the bcl-2 copy number variation were not observed in the EBV-CN high group. In a word, no significant difference was found across the three EBV-CN groups for all the cytogenetic features investigated ().
Table 3. Tumor cytogenetic features of patients according to EBV DNA copy number.
Mutation profile of key genes
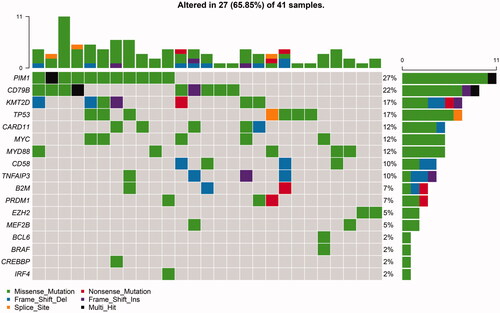
NGS was performed in 41 DLBCL patients (22 EBV-CN negative, 14 EBV-CN low, and five EBV-CN high). Among them, 27 patients (65.85%) presented at least one variant. Among 17 out of the 20 genes examined, we identified a total of 108 mutations. The genes mutated in at least 10% of the patients included PIMI (27%), CD79B (22%), KMT2D (17%), TP53 (17%), CARD11 (12%), MYC (12%), MYD88 (12%), CD58 (10%), and TNFAIP3 (10%) (). Gene mutation frequencies by subtype and in the overall cohort are shown in Figure S1. There was no significant difference between subgroups, though fewer gene mutations were observed in the EBV-CN high group.
Figure 1. Frequencies and distribution of gene mutations in patients with DLBCL. A total of 41 DLBCL cases were performed with next-generation sequencing. Twenty-seven patients (65.85%) presented at least one variant. The bar chart above presented mutation in each patient. The bar chart nearby presented mutation frequency of each gene.
Survival analysis
The median follow-up duration was 28.7 months (range, 9.9–50.8 months). By the latest follow-up visit, 19 patients were died, including seven patients in high (mortality rate: 63.6%), three patients in low (mortality rate: 12.5%), and nine patients in negative EBV-CN group (mortality rate: 22.5%).
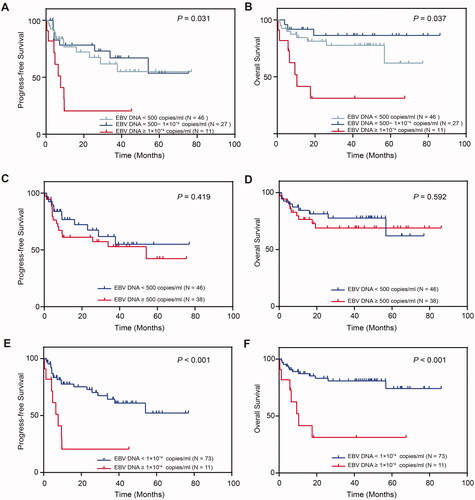
To further evaluate the prognostic significance of EBV-DNA copy number in PBMCs in DLBCL patients, we performed survival analysis for PFS and OS. As shown in , patients of the high EBV-CN group had significantly worse PFS and OS compared to the other two groups (p= .031 and .037, respectively), and hence we evaluated the prognosis value of 1 × 104 copies/ml and traditional positivity threshold (500 copies/ml), to confirm our hypothesis that threshold value of 1 × 104 copies/ml is more predictive. We found that there was no significant difference between EBV-CN negative and EBV-CN low-to-high groups (PFS, p= .419; OS, p= .592). In contrast, compared to the EBV-CN negative-to-low groups, the EBV-CN high group showed significantly worse prognosis (PFS, p< .001; OS, p< .001). It seems that cutting-off at 1 × 104 copies/ml can better predict the clinical prognosis of patients than the traditional positivity threshold (500 copies/ml).
Figure 2. Kaplan–Meier’s estimates of (A) progression-free survival and (B) overall survival for the negative (<500 copies/ml), low (500–104 copies/ml), and high EBV-CN group (≥104 copies/ml), (C) progression-free survival, and (D) overall survival for positive group (EBV-DNA ≥ 500 copies/ml) and negative group (EBV-DNA <500 copies/ml), (E) progression-free survival and (F) overall survival for high EBV-CN group (EBV-DNA ≥ 1 × 104 copies/ml) and not significantly increased EBV-CN group (EBV-DNA <1 × 104 copies/ml).
To adjust for the effects of potential confounding factors, we performed a Cox multivariate regression model analysis (). After adjusting for age, extranodal involvement, serum lactate dehydrogenase, ECOG score, Ann Arbor stage, chemotherapy, EBV-DNA copy number in PBMCs, high EBV-CN was an independent risk factor for poor prognosis of DLBCL (hazard ratio (HR)=5.841, 95% confidence interval (CI)= [1.660, 20.548], p=.006).
Table 4. Univariate and multivariate analyses of prognostic factors.
Discussion
Previous studies indicated the clinical significance of peripheral viral load in predicting the development and progression of EBV associated disease [Citation7,Citation8,Citation11,Citation19,Citation20]. In agreement with some previous studies, our data demonstrated that higher pretreatment EBV-DNA level in PBMCs correlates with male gender, older age, more extranodal involvement, higher ECOG score, and non-GCB type [Citation4,Citation19,Citation21]. In our study, we chose 500 copies/ml and 1 × 104 copies/ml as cutoff value, and we found that patients of the highest EBV copy number category (≥1 × 104 copies/ml) showed significantly poorer PFS and OS than all other patients (<1 × 104 copies/ml). In contrast, no difference in PFS or OS was observed between EBV-DNA positive and negative groups (dichotomized by 500 copies/ml). Furthermore, multivariate analysis has demonstrated high EBV-CN (≥1 × 104 copies/ml) as an independent risk factor of poor prognosis in DLBCL after adjusting the conventional risk factors.
EBER detection by in situ hybridization is the standard method in diagnosing EBV positive DLBCL, and its role on prognosis prediction in DLBCL has attracted considerable attention. In the era of chemoimmunotherapy (R-CHOP), the prognostic effect of EBV status on DLBCL patients is controversial. An Australian study found that EBV positivity was associated with worse survival although patients received R-CHOP [Citation22], while another study showed that there was no difference of the survival rates between the 28 EBV + DLBCL and the 695 EBV-negative patients when treated with R-CHOP [Citation23]. In our study, there was no difference in PFS and OS between five EBER + DLBCL and 41 EBER-negative DLBCL patients (p=.449 and .115, respectively, data not shown). It cannot be concluded that the ambiguous significance of EBER on DLBCL patients is due to the slightly higher cutoff value (30%). However, EBV copy number is accurate and easier to obtain, and we uncovered its predictive role in DLBCL patients seemingly better than EBER, which still needs to be confirmed by numerous prospective studies with larger sample size.
It is acknowledged that aging in humans has a close relation with impaired immune status, and evasion of immune system resulted from immunosenescence is thought to be an important mechanism in lymphomagenesis of EBV-positive DLBCL of the elderly [Citation24–27]. High EBV-DNA copy number in PBMCs is associated with poorer OS and older age as we have demonstrated. It is presumed that the pathogenesis of DLBCL with high EBV-DNA copy number in PBMCs is closely related with immunologic senescence caused by the aging process. Other evidence has shown that high load of EBV-DNA copy number in peripheral blood was more often detected in immunocompromised subjects [Citation28,Citation29]. That is probably due to host immune responses, including Natural killer cells and cytotoxic T cell, involving in viral clearance, which may play important roles in the control of tumor progression. However, the exact mechanisms of how peripheral EBV load affected the prognosis of DLBCL still needs more robust evidence in the future studies.
Our study evaluated the difference of immunophenotypic, cytogenetic, and molecular characteristics besides clinical characteristics and prognosis between subgroups. In our research, compared to negative-to-low EBV-CN groups, the high EBV-CN group presented increased BCL-2, BCL-6, and EBER expression, and more p53 deletion, c-myc rearrangement, and bcl-2 rearrangement. CD30 expression and polyploidy is more prevalent in low EBV-CN group than EBV-CN high and negative groups. The result is not entirely similar with previous studies about elderly EBV-positive DLBCL patients, which showed lower BCL-6, higher CD30 expression and less p53 deletion, c-myc rearrangement, and bcl-2 rearrangement [Citation26,Citation27,Citation30,Citation31]. The most frequently mutated genes were PIM1, CD79B, KMT2D, and TP53, which is similar with previous reports [Citation17]. A low number of genomic aberrations were observed in EBV-positive DLBCL of the elderly [Citation32], and in our study the high EBV-CN group had less mutations on the investigated genes. Besides, statistical significance was not observed regarding immunophenotype and cytogenetics between EBV-CN subgroups, suggesting the hypothesis that the oncogenic effects of EBV might associated with poorer prognosis.
Due to the less mutations in high EBV-CN group, molecular targeted therapy may not be a suitable treatment strategy for the patients with such poor prognosis. We presume that programmed cell death-1 (PD1) blockade may improve the prognosis of these patients by increasing the ability of immune surveillance. Grywalska et al. demonstrated that the expression of PD-1 and PD-L1 on host immune cells correlated positively with EBV DNA load in blood in patients diagnosed as chronic lymphocytic leukemia [Citation33]. One study illustrated that PD-1/CTLA-4 blockade could significantly increase EBV-specific T cell responses and inhibit Epstein-Barr virus-induced lymphoma growth in a cord blood humanized-mouse model [Citation34]. Quan et al. also demonstrated that PD-1 blockade could restore functions of T-cells in Epstein-Barr virus positive DLBCL [Citation35]. EBV associated DLBCL may be sensitive to PD-1 blockade therapy due to the increasing PD-1 and PD-L1 expression caused by EBV infection and the increasing T cell function.
What we need to know is that there are several limitations in the study. First, data on immunophenotype, cytogenetic features, and molecular features were not available for certain patients due to its retrospective nature. Second, 91% patients received anthracycline-based chemotherapy combined with rituximab as initial treatment. Third, the sample size was not large as it is a single center study. The results need further validation in prospective studies with a large number of patients. However, our study is the first study focusing on immunophenotypic, cytogenetic, and molecular characteristics of patients diagnosed as DLBCL with EBV DNA detection in peripheral blood.
In summary, based on our data, DLBCL patients with high pretreatment EBV-DNA copy number in PBMCs have poorer OS and PFS. These patients may need more effective treatment, such as possible addition of EBV-targeted therapy to conventional chemotherapy.
GLAL-2021-1047-File011.doc
Download MS Word (36.5 KB)GLAL-2021-1047-File010.doc
Download MS Word (37.5 KB)GLAL-2021-1047-File009.doc
Download MS Word (47.5 KB)GLAL-2021-1047-File008.jpg
Download JPEG Image (227.3 KB)Acknowledgements
We thank all the faculty members and staffs in the Clinical and Laboratory Unit of the Department of Hematology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology for their Clinical and technical support.
Disclosure statement
The authors have no conflict of interest.
Additional information
Funding
References
- Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet. 1964;283(7335):702–703.
- Siu LLP, Chan JKC, Kwong YL, et al. Natural killer cell malignancies: clinicopathologic and molecular features. Histol Histopathol. 2002;17(2):539–554.
- Pallesen G, Hamilton-Dutoit SJ, Rowe M, et al. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin's disease. Lancet. 1991;337(8737):320–322.
- Park S, Lee J, Ko YH, et al. The impact of Epstein-Barr virus status on clinical outcome in diffuse large B-cell lymphoma. Blood. 2007;110(3):972–978.
- Listed N. A clinical evaluation of the International Lymphoma Study Group Classification of non-Hodgkin's lymphoma. The Non-Hodgkin's Lymphoma Classification Project. Blood. 1997;89(11):3909–3918.
- Lynch RC, Gratzinger D, Advani RH. Clinical impact of the 2016 update to the WHO Lymphoma Classification. Curr Treat Options Oncol. 2017;18(10):60.
- Ito Y, Kimura H, Maeda Y, et al. Pretreatment EBV-DNA copy number is predictive of response and toxicities to SMILE chemotherapy for extranodal NK/T-cell lymphoma. Clin Cancer Res. 2012;18(15):4183–4190.
- Wang Z-Y, Liu Q-F, Wang H, et al. Clinical implications of plasma Epstein-Barr virus DNA in early-stage extranodal nasal-type NK/T-cell lymphoma patients receiving primary radiotherapy. Blood. 2012;120(10):2003–2010.
- Hohaus S, Santangelo R, Giachelia M, et al. The viral load of Epstein-Barr virus (EBV) DNA in peripheral blood predicts for biological and clinical characteristics in Hodgkin lymphoma. Clin Cancer Res. 2011;17(9):2885–2892.
- Gandhi MK, Lambley E, Burrows J, et al. Plasma Epstein-Barr virus (EBV) DNA is a biomarker for EBV-positive Hodgkin's lymphoma. Clin Cancer Res. 2006;12(2):460–464.
- Westmoreland KD, Montgomery ND, Stanley CC, et al. Plasma Epstein‐Barr virus DNA for pediatric Burkitt lymphoma diagnosis, prognosis and response assessment in Malawi. Int J Cancer. 2017;140(11):2509–2516.
- Okamoto A, Yanada M, Inaguma Y, et al. The prognostic significance of EBV DNA load and EBER status in diagnostic specimens from diffuse large B-cell lymphoma patients. Hematol Oncol. 2017;35(1):87–93.
- Tisi MC, Cupelli E, Santangelo R, et al. Whole blood EBV-DNA predicts outcome in diffuse large B-cell lymphoma. Leuk Lymphoma. 2016;57(3):628–634.
- Xu J, Li P, Chai J, et al. The clinicopathological and molecular features of sinusoidal large B-cell lymphoma. Mod Pathol. 2021;34(5):922–933.
- Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282.
- Yoon S, Jeon Y, Paik J, et al. MYC translocation and an increased copy number predict poor prognosis in adult diffuse large B-cell lymphoma (DLBCL), especially in germinal centre-like B cell (GCB) type. Histopathology. 2008;53(2):205–217.
- Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837.
- Robinson JT, Thorvaldsdóttir H, Wenger AM, et al. Variant review with the integrative genomics viewer. Cancer Res. 2017;77(21):e31–e34.
- Chen Y, Zheng X, Chen B, et al. The clinical significance of Epstein-Barr virus DNA in peripheral blood mononuclear cells in patients with non-Hodgkin lymphoma. Leuk Lymphoma. 2017;58(10):2349–2355.
- Gallagher A, Armstrong AA, MacKenzie J, et al. Detection of Epstein‐Barr virus (EBV) genomes in the serum of patients with EBV‐associated Hodgkin's disease. Int J Cancer. 1999;84(4):442–448.
- Morales D, Beltran B, De Mendoza FH, et al. Epstein-Barr virus as a prognostic factor in de novo nodal diffuse large B-cell lymphoma. Leuk Lymphoma. 2010;51(1):66–72.
- Keane C, Tobin J, Gunawardana J, et al. The tumour microenvironment is immuno‐tolerogenic and a principal determinant of patient outcome in EBV‐positive diffuse large B‐cell lymphoma. Eur J Haematol. 2019;103(3):200–207.
- Chi YO, Li L, Xu-Monette ZY, et al. Prevalence and clinical implications of Epstein-Barr virus infection in de novo diffuse large B-Cell lymphoma in Western countries. Clin Cancer Res. 2014;20(9):2338–2349.
- Nikolich-Žugich J. Ageing and life-long maintenance of T-cell subsets in the face of latent persistent infections. Nat Rev Immunol. 2008;8(7):512–522.
- Berglund M, Thunberg U, Amini R-M, et al. Evaluation of immunophenotype in diffuse large B-cell lymphoma and its impact on prognosis. Mod Pathol. 2005;18(8):1113–1120.
- Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951.
- Ziarkiewicz M, Wołosz D, Dzieciątkowski T, et al. Epstein-Barr virus-positive diffuse large B cell lymphoma in the experience of a tertiary medical center in Poland. Arch Immunol Ther Exp. 2016;64(2):159–169.
- Sangam K, Kumar Y, Minz RW, et al. Patients with plasma cell disorders have high EBV DNA in peripheral blood than the general population. Pathol Oncol Res. 2020;26(4):2789–2794.
- Kanakry JA, Hegde AM, Durand CM, et al. The clinical significance of EBV DNA in the plasma and peripheral blood mononuclear cells of patients with or without EBV diseases. Blood. 2016;127(16):2007–2017.
- Ok CY, Ye Q, Li L, et al. Age cutoff for Epstein-Barr virus-positive diffuse large B-cell lymphoma-is it necessary? Oncotarget. 2015;6(16):13933–13945.
- Montes-Moreno S, Odqvist L, Diaz-Perez JA, et al. EBV-positive diffuse large B-cell lymphoma of the elderly is an aggressive post-germinal center B-cell neoplasm characterized by prominent nuclear factor-kB activation. Mod Pathol. 2012;25(7):968–982.
- Ok CY, Papathomas TG, Medeiros LJ, et al. EBV-positive diffuse large B-cell lymphoma of the elderly. Blood. 2013;122(3):328–340.
- Grywalska E, Pasiarski M, Sosnowska-Pasiarska B, et al. Programmed cell death 1 expression and Epstein-Barr virus infection in chronic lymphocytic leukaemia: a prospective cohort study. Cancer Manag Res. 2019;11:7605–7618.
- Ma S-D, Xu X, Jones R, et al. PD-1/CTLA-4 blockade inhibits Epstein-Barr virus-induced lymphoma growth in a cord blood humanized-mouse model. PLoS Pathog. 2016;12(5):e1005642.
- Quan L, Chen X, Liu A, et al. PD-1 blockade can restore functions of T-cells in Epstein-Barr virus-positive diffuse large B-cell lymphoma in vitro. PLoS One. 2015;10(9):e0136476.
