Abstract
Common kernel density estimators (KDE) are generalised, which involve that assumptions on the kernel of the distribution can be given. Instead of using metrics as input to the kernels, the new estimators use parameterisable pseudometrics. In general, the volumes of the balls in pseudometric spaces are dependent on both the radius and the location of the centre. To enable constant smoothing, the volumes of the balls need to be calculated and analytical expressions are preferred for computational reasons. Two suitable parametric families of pseudometrics are identified. One of them has common KDE as special cases. In a few experiments, the proposed estimators show increased statistical power when proper assumptions are made. As a consequence, this paper describes an approach, where partial knowledge about the distribution can be used effectively. Furthermore, it is suggested that the new estimators are adequate for statistical learning algorithms such as regression and classification.
Introduction
The multivariate kernel density estimator (KDE) with a fixed bandwidth is given by
Several authors have suggested a variable bandwidth version of Equation (2), where the two main approaches are the balloon estimator and the sample smoothing estimator. A comparison between KDE with fixed and variable estimators is given by CitationTerrel and Scott (1992).
The balloon estimator lets to be equal to the distance to the kth nearest neighbour of
. The estimator is given by
The sample smoothing estimator was suggested by CitationBreiman, Meisel, and Purcell (1977). Here, the bandwidth parameter is independent of the query point, but dependent on the sample points. The estimator is
where
is the distance to the k-nearest neighbour of
.
To improve readability, we recollect the following definitions.
Definition 1.1
The kernel of a function is an equivalence relation =f given by
where
and
are elements in X.
Definition 1.2
A pseudometric on a set X is a function (distance function) which for all
in X satisfies the following conditions:
1.
(non-negativity ).
]
(symmetry ).
]
(triangle inequality).
If we add the requirement of identity, which is if and only if
then d is a metric as well.
Definition 1.3
A pseudometric space M is an ordered pair (X, d). A closed ε- ball on M that is centred at is defined as
which is equivalent to the definition of a closed ε-ball in a metric space. Note that ε is called the radius of the ball.
This definition of pseudometric is exactly the same as the definition of a semimetric in the field of nonparametric functional data analysis (CitationFerraty and Vieu 2006). To avoid confusion in this paper, we will keep referring to pseudometrics, even though most of the literature in nonparametric functional data analysis uses the term semimetric.
In nonparametric functional data analysis, the explanatory variables are functional and most of the work has been done on regression and classification. An overview of the field is given in the book by CitationFerraty and Vieu (2006). In the case of density estimation, CitationDelaigle and Hall (2010) proved that a probability density function generally does not exist for functional data, but developed notions of density and mode, when functional data are considered in the space determined by the eigenfunctions of the principal components. In the paper by CitationFerraty, Kudraszow, and Vieu (2012), an infinite dimensional analogue of a density estimator is discussed, when the small ball probability associated with the functional data can be approximated as a product of two independent functions; one depending on the centre of the ball and one depending on the radius.
In this paper, we consider only finite dimensional pseudometric spaces, where balls with finite radius have finite volumes. Such spaces exist as a specification of the theory of nonparametric functional data analysis (CitationFerraty et al. 2012). The theory in this paper is distinguished from that of nonparametric functional data analysis, because we consider pseudometric spaces, where volumes of balls with fixed radius are allowed to be dependent on locations of centres. In all previous work on using pseudometrics for functional data, the volumes of the balls are always considered as independent on the locations of the centres. This generalisation opens up for applications that are less linked to functional data and more related to restricted nonparametric and semiparametric estimators on real (non-functional) data.
To be more accurate, the parameters of the pseudometric precisely describe the kernel, or more roughly the level sets of the distribution. This is important for visualisation and interpretation of the estimated distribution. For instance, the complete multi-dimensional density that is estimated with an estimator using a pseudometric of family one (described later in this paper) can be described by a two-dimensional plot only.
The choice of the family of pseudometrics describes the constraints on the level sets of the distributions that can be modelled. In case of fitting parameters, it is therefore important that the level sets of the actual distribution are not in violation with these constraints. This is somewhat analogous to parametric models, where serious misbehaviour can be observed, if the distribution has a different shape than any of the members in the family of models.
An example of a similar approach can be found in a paper by CitationLiescher (2005), where the level sets of the distributions were constrained to be of elliptic shape. The elliptic shape of the level sets was constrained by a transformation that was performed prior to applying the nonparametric density estimator. It was shown that the convergence rates were independent of the number of dimensions, except in the neighbourhood of the mean, and thereby overcoming the so-called curse of dimensionality. The estimators in this paper can be seen as a generalisation of the estimators of CitationLiescher (2005), because a wider family of associated distributions is allowed. Convergence rates are not explicitly given in this paper, but simulations show that the performance close to the mode coincides with the common KDE and improves quickly away from the mode, which is similar to the results obtained by CitationLiescher (2005).
In the context of reducing the curse of dimensionality of nonparametric density estimators, it is also worth mentioning the projection pursuit density estimator. This method was first introduced in CitationFriedman, Stuezle, and Schroeder (1984), and a parametric extension was given by CitationWelling, Zemel, and Hinton (2003). In projection pursuit one projects the explanatory variables into principal directions and fits one-dimensional smooth density functions to these projections. The resulting density is the product of these densities. A special case is the flexible Bayesian estimator, where the projection matrix is identity. In essence, the flexible Bayesian method is assuming independence between the explanatory variables. The estimator will be used later in this paper and is plug-in
where K is a univariate kernel, the bandwidth is h, xj is the jth coordinate of
and xij is the jth coordinate in sample i. Consistency is proved under the assumption of independence (CitationJohn and Langley 1995). This is a nonparametric variation of the old and well-known naive-Bayesian estimator, where the individual distributions are assumed Gaussian. A similarity between the estimators in this paper and the projection pursuit density estimators is that the resulting density can be visualised in lower dimensional spaces. In projection pursuit, it is enough to show the one-dimensional (ridge) functions along with the principal directions to understand the full distribution.
The proposed estimators have less in common with semiparametric models, where part of the model is fully parametric. For instance models which are a product of a parametric and a nonparametric component, such as in CitationNaito (2004) and references therein. Another example is when some of the explanatory variables are modelled nonparametrically, while the rest are modelled parametrically, such as in CitationHoti and Holmström (2003).
In the next section, the underlying assumptions of the KDE with fixed and variable bandwidths are discussed. This is followed by formal definitions of the new estimators. A framework for finding suitable parametric families of pseudometrics is outlined in Section 3 and two families are identified. Some experiments are given in Section 4, where the new KDE are compared with common KDE on samples drawn from common distributions. In Section 5, open questions and ideas of how to use the new estimators on various problems are elaborated on, before the work is concluded.
2. Assumptions for kernel density estimation and definition of new estimators
It is convenient to start our discussion by investigating the k-nearest neighbour method, because it is conceptually easier than the other KDE. If we generalise Equation (4) for using any metric d, then
For the sake of discussion, a random variable Y is defined as a k-nearest neighbour estimate at a particular . Each k-nearest neighbour estimate requires drawing a new training sample and this is why Y is a random variable. The variance of Y is a consequence of which sample is chosen (because this determines the location of the kth-nearest neighbour), while a great portion of the bias can be explained by the shape of the ball.
This is potentially more clear if we realise that Equation (5) can be derived from two assumptions that were not stated in CitationLoftsgaarden and Quesenberry (1965). The first assumption is that
This argumentation can be transferred to all KDE with both fixed and variable bandwidths, because these methods are essentially weighting the contribution of each sample as a function of
. For the KDE with fixed bandwidth, the balloon estimator and the sample smoothing estimator, respectively, we could write
where
,
and
are nonnegative, strictly decreasing, one-sided univariate kernels that integrate to one, when integrated over
. If we let the random variable Y be defined as any of the kernel density estimates at a particular
, then the bias of Y is highly related to the shape of the balls in combination with the shape of the f near
.
2.1. Definition of the k-nearest pseudo-neighbour estimator
A generalisation of the k-nearest neighbour estimator is proposed as
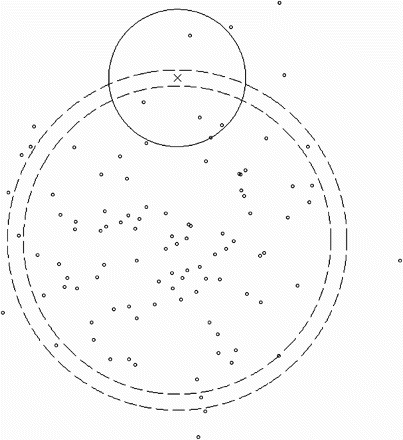
where dp is a pseudometric. illustrates the principles of k-nearest neighbour estimator as of CitationLoftsgaarden and Quesenberry (1965) together with the k-nearest pseudo-neighbour estimator. This particular pseudometric is a special case of the parametric families of pseudometrics that are outlined later in this paper.
Figure 1. Description of the k-nearest pseudo-neighbour estimator. The scatter plot shows a training population with 100 points that are i.i.d and drawn from a two-dimensional normal distribution. A cross is marked at a query point and in k-nearest neighbour estimation, the kth-nearest neighbour is found by comparing the Euclidean distance to all other points. The smallest Euclidean ball that encapsulates the 5th-nearest neighbours is shown as a solid circle. Clearly, f is not very constant inside this ball. In k-nearest pseudo-neighbour estimation, the kth-nearest neighbour is found by comparing the distance in the pseudometric space from the query point to all other points. The interior between the two dashed lines represents a ball in this pseudometric space, where the query point is in the centre. The radius of the ball (not of the dashed circles) is the distance to the 5th-pseudo-nearest neighbour. Obviously, f is far more constant inside this ball. Consequently, improved statistical power is achieved when the equivalence classes with respect to the kernel of the distribution are assumed to be circles around the mean.
It is postulated that a proper choice of a pseudometric can reduce Equation (7) far more than a metric, because balls in pseudometric spaces span a richer variety of shapes. The key idea is to find a parametric family of pseudometrics and select parameters by either Bayesian inference or learning.
It is a crucial point that the parameters of a pseudometric family are typically associated with various sample statistics and associations between the explanatory variables. In the example shown in , the centre of the dashed circles is a parameter of the pseudometric. This parameter can be estimated by the sample mean.
2.2. Definition of pseudometric KDE
A challenge with using pseudometrics is that the volumes of the balls with constant radius are generally dependent on the locations of the centres. This effect is neutralised by defining the equivalent Euclidean distance, which is the function
The idea is to weight the contributions of the ’s according to the equivalent Euclidean distances from the query point. This ensures that the level of smoothing is constant with respect to volume, which is the case for KDE with fixed bandwidths.
The pseudometric kernel density estimator (PKDE) is suggested as
If we choose where
and a bandwidth h, then Equation (1) becomes a special case of Equation (9), when
and normal kernels are used. The only difference is that the kernel in Equation (9) takes a scalar argument, while the kernel in Equation (1) takes a vector. A similar argumentation can be made for any kernel and this is why the PKDE is a generalisation of the standard KDE.
In the case of variable bandwidths, the pseudometric balloon estimator is suggested as
where
and
is the k-nearest pseudo-neighbour of
(nearest in the sense dp). Notice that if we choose dp to be Euclidean and a kernel that has uniform density on the Euclidean ball, the k-nearest neighbour method falls out as a special case.
Moreover, the pseudometric sample smoothing estimator is suggested as
where
and
is the k-nearest pseudo-neighbour of
.
3. Choosing appropriate parametric families of pseudometrics
A clear challenge with using the new estimators is proposing suitable families of pseudometrics. It is therefore instructive to define the equivalence relation , where
if and only if
. While discussing the usefulness of a family of pseudometrics, it makes sense to ask which density functions are invariant to
, and the next two definitions are motivated by this.
Definition 3.1
A probability density function is said to be an associated distribution of a pseudometric (or metric) dp, denoted as when
is invariant to
. Invariance to
means that
implies that
for all
in X.
Definition 3.2
An associated distribution satisfies the strong condition if
is equal to
where
is the kernel of
. This is the case when
for all
in X.
3.1. Volumes of generalised balls
Two new families of pseudometrics are proposed, including the associated distributions and the derivations of the analytical expressions for the volumes of their balls. Before this, a definition of the closed linearly transformed generalised balls and a derivation of an analytical expression for the volumes are needed. contains a list of symbols that are consistently used in rest of this paper.
Table 1. List of important symbols.
Definition 3.3
The closed linearly transformed generalised ε-ball that is centred in is denoted
where
Here,
is a vector of the positive pis, while p is the harmonic average of the pi′s (i.e.
). Moreover,
and
where A is a square transformation matrix with nonzero determinant.
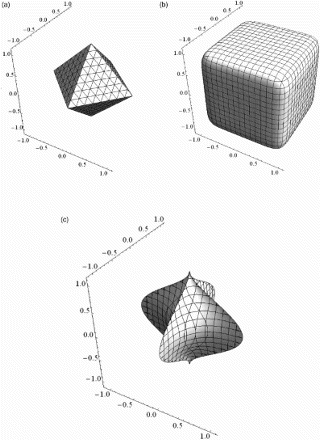
In general is a semimetric, because the triangle inequality does not hold when at least one of the pis are less than 1. Depending on the pis, the generalised balls describe a wide range of multi-dimensional geometrical objects, where spheres, cubes, cylinders and stars are special cases. A few three-dimensional objects are shown in .
In articles by CitationWang (2005) and CitationGao (2013), the volume of the generalised unit ball was derived.
Theorem 3.4
The volume of the closed generalised unit ball is
The proof by Wang used induction and was motivated by an idea from CitationFolland (2001), while the proof by Gao used properties of the exponential distribution. An alternative proof of Equation (10) is included in the appendix.
Theorem 3.5
The volume of the closed linearly transformed generalised ε-ball that is centred in is
This means that and the absolute value of the determinant of
determine the overall scaling of the volume. This is a consequence of choosing the exponent in
to be 1/p.
3.2. Pseudometric family of type one
Definition 3.6
The pseudometric family of type one is given by
where
is a chosen point (for instance a mean or a median).
The parameters that can be trained for this pseudometric are ,
and
. In the family of pseudometric spaces (
), the balls can be viewed as generalisations of the spherical shell and two examples are shown in .
Figure 3. Balls in the pseudometric spaces (ℝ3, dp1, I, 0, p). One octant is removed in both figures, so it is possible to see more of the boundaries of the balls. (a) and (b)
.

3.2.1. Associated distributions
Theorem 3.7
The family of associated distributions is equal to a function g(u), which satisfies
If we choose g(u) to be an exponential function a exp(−u), then the coefficient a can be found from Equation (11). When inserting for g(u), the integral part of Equation (11) becomes , so that
The parametric family of associated distributions is therefore a linear transformation of the m-dimensional generalised normal distribution (also known as the exponential power distribution)
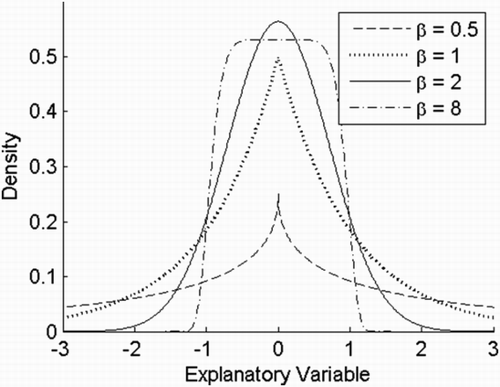
To interpret this result, a description of the univariate case is given by
where more details are given in . Also, if we choose
to be diagonal with 1/αis on the diagonal then
In this case all dimensions are independent and the αis are the scale factors and pis are the shape factors.
Figure 4. The univariate generalised normal distribution is a parametric family of symmetric unimodal distributions that includes all normal and Laplace distributions, and as limiting cases it includes all continuous uniform distributions on bounded intervals of the real line. It includes a shape factor β and a scale factor α. Various choices of shape factors are shown in this figure, where the scale factor is 1 and the mean is 0.
3.2.2. Volumes of the balls
Theorem 3.8
The volumes of the balls in the family of pseudometric spaces are given by
where O is the set of positive odd numbers that are smaller than or equal to m.
The continuity of the ball at is seen, when noting the fact that
and
. In the case when
, the volume is a polynomial with respect to either ε or
. The same is true when
, but the polynomial is different. This demonstrates how the volumes of the balls are dependent on the locations of the centres, and this is why it was necessary to define the equivalent Euclidean distance in Equation (8).
3.3. Pseudometric family of type two
Definition 3.9
The pseudometric family of type two is proposed as
where q is an integer larger than or equal to one, I is identity,
and
. The notations
means
and
is the upper left sub-matrix of A with size
. Also we have m1+m2=m.
The parameters that can be trained here is q, ,
and
, but also which xis are put into
(it does not have to be the m1 first). In the limiting case when
The ball is as usual defined by
If m1 is less or equal to 1,
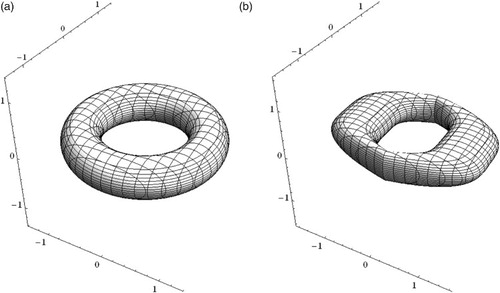
is the familiar m-dimensional ball. The familiar torus and a deformation of a torus are other special cases that are shown in .
3.3.1. Associated distributions
In terms of discussing the associated distributions for , it is an important observation that the equivalence classes of
are all topological manifolds of dimension m1−1, while the equivalence classes of
are (m−1)-manifolds. This means that
can never be equal to
, unless m1 is equal to m.
This dimension reduction may be desirable with regards to improving robustness, because the family of associated distributions spans wider. On the other side, it may reduce statistical power because AV defined in Equation (7) may not be minimised properly.
Theorem 3.10
The parametric family of associated distributions is equal to a function
which satisfies
where
.
If we also assume that the are all independent of
(and vice versa), then
. Hence, g1(u) is subject to the constraint
while
is a probability density function that integrates to 1, when integrated over
. Clearly, the family of associated distributions
has the family of associated distributions
and the family of all distributions as special cases.
3.3.2. Volumes of the balls
Theorem 3.11
The volumes of the balls in the family of pseudometric spaces are given by
where
Here, Bt(a, b) is the incomplete beta function,
and O is the set of positive odd numbers that are smaller than or equal to m1.
In the case when , the volume is a polynomial with respect to either ε or
. The same is true when
, but the polynomial is different.
Theorem 3.12
The volumes of the balls in the family of pseudometric spaces in the limiting case when
are given by
where O is the set of positive odd numbers that are smaller than or equal to m1.
The continuity of the ball at is seen, when noting the fact that
and
. This formula is simpler than the general formula given in Theorem 3.11 and it may be desirable in the context of reducing computational time.
3.4. Bandwidth selection
In CitationRosenblatt (1956), approximations for the bias, the variance, the mean squared error and the mean integrated squared error were derived in the univariate case. It was shown that by minimising the asymptotic mean integrated squared error (AMISE), an optimal bandwidth could be found when information of exists. It is not possible to make such an elegant argumentation for pseudometric kernel density estimation. However, an expression, including one-dimensional integrals, for the AMISE of
is derived when f is equal to
.
Theorem 3.13
The asymptotic mean squared error of
and the AMISE of
are
4. Experiments
In this section, a few experiments are presented where various PKDE are compared with the standard KDE and the flexible Bayesian estimator. The experiments involve drawing repetitively an order pair , nrep times. Here, X is a sample of n points that are drawn from a known distribution f and
is a query point that is drawn randomly from an equivalence class
. The sequence
starts at 0 and is uniformly spaced over the relevant domain of f. For every ri, the mean and the quartiles are computed and compared with the known analytic solution. When plotted as a function of ri, the variances and the biases of the estimators are easily seen.
Seven experiments are conducted, where PKDE, KDE and flexible Bayesian are compared on a set of distributions, including the standard normal, the uniform, the one-sided normal and a one-sided normal that is split into two. Variable PKDE and variable KDE are compared only on the standard normal distribution. The parameters are summarised in and .
Table 2. Parameters for all experiments.
Table 3. Parameters for each experiment.
4.1. Results
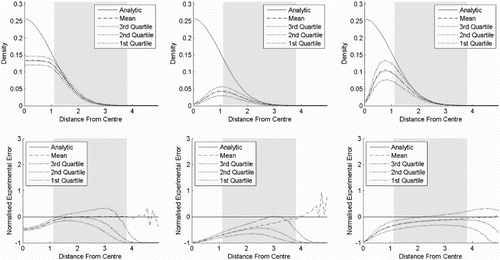
In experiment one (), the estimator is shown in various dimensions while the bandwidth is kept constant. The bandwidth is chosen to 0.5, which is the bandwidth that minimises AMISE (12) in six dimensions. The estimator seems to be unbiased in the tails, while it underestimates near the centre where the level of underestimation increases with the number of dimensions. The tails seem fairly unbiased, but as we move away from the centre, the mean normalised experimental error starts to fluctuate. From the quartile plots, it is seen that the skewness of the normalised experimental error increases with distance from the centre.
Figure 6. Result of experiment one. Kernel density estimates for a standard normal distribution in various number of dimensions, where the bandwidth is 0.5. The top row shows the densities as functions of the distances from the centre, while the bottom row shows the corresponding normalised standard errors. The distance from the centre is chi distributed with order m and the shaded area is where the cumulative distribution function is between 0.025 and 0.975. (a) Two dimensions, (b) four dimensions and (c) six dimensions.
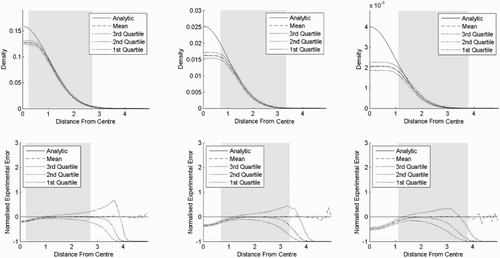
In experiment two (), we see the effect of increasing and decreasing the bandwidth in six dimensions. When the bandwidth is increased, the underestimation near the centre increases while the variance decreases everywhere. When the bandwidth is decreased, the bias is reduced at the cost of a higher variance.
Figure 7. Result of experiment two. Kernel density estimates for a standard normal distribution in six dimensions with various degrees of smoothing. The top row shows the probability density functions as functions of the distances from the centre, while the bottom row shows the corresponding normalised standard errors. The distance from the centre is chi distributed with order six. The shaded area is where the cumulative distribution function is between 0.025 and 0.975. (a) Bandwidth is 0.2 and (b) bandwidth is 0.8.
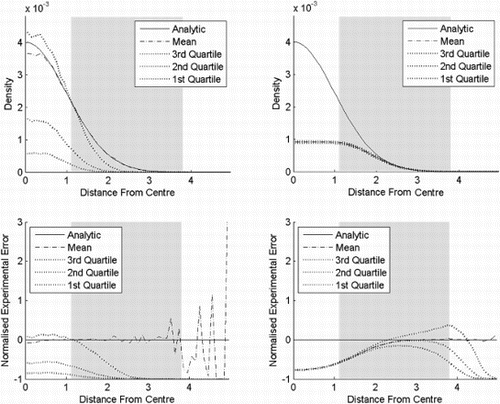
In experiment three (), KDE with fixed bandwidth and flexible Bayesian is shown for comparison with PKDE in experiment one. Since the bandwidth in KDE is the same as for PKDE, the bias and the variance are the same near the centre, while KDE continues to underestimate much further away from the centre. Also, when the analytical graph is passed, KDE starts to overestimate. Obviously, an optimal bandwidth for KDE is smaller than for PKDE, since the bias in the tail is larger. This underestimation and overestimation pattern is also seen in the flexible Bayesian, but to a lower extent. In the case of the flexible Bayesian, we have chosen a bandwidth so that the variance near the centre is approximately the same as the KDE and the PKDE. This means that it is easier to compare the biases.
Figure 8. Result of experiment three. KDE and flexible Bayesian estimates for a standard normal distribution in six dimensions, where the bandwidths are 0.5 and 0.2, respectively. The top row shows the probability density functions as functions of the distances from the centre, while the bottom row shows the corresponding normalised standard errors. The distance from the centre is chi distributed with order six. The shaded area is where the cumulative distribution function is between 0.025 and 0.975. (a) KDE and (b) flexible Bayesian.
In experiment four (), PKDE and KDE with variable bandwidths are shown for comparison. The number of neighbours is chosen so that the variances near the centres are approximately the same as in and , where the bandwidths are fixed. It is interesting to see that the variance of the normalised experimental error is fairly stable and not skewed compared to the fixed bandwidth graphs. The bias in the tails is clear in these plots. Notice also that the PKDE with variable bandwidth has a similar, or smaller, variance and bias than the flexible Bayesian inside the shaded area.
Figure 9. Result of experiment four. Variable PKDE and variable KDE for a standard normal distribution in six dimensions. Two standard deviations of the Gaussian kernel are defined to be the distance (in the pseudometric space) to the 15th nearest neighbour. The top row shows the probability density functions as functions of the distances from the centre, while the bottom row shows the corresponding normalised standard errors. The distance from the centre is chi distributed with order six and the shaded area is where the cumulative distribution function is between 0.025 and 0.975. (a) Variable PKDE and (b) variable KDE.
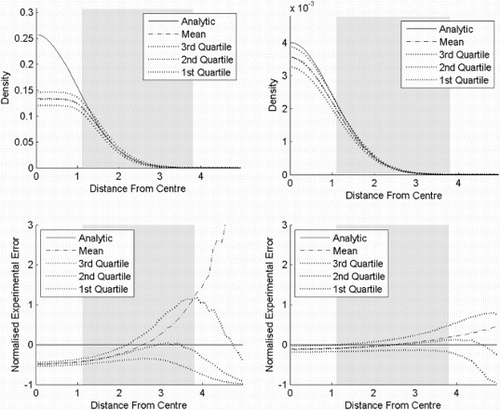
In experiment five (), PKDE, KDE and flexible Bayesian are compared on a uniform population. The bandwidth of the flexible Bayesian is adjusted so the variance near the centre is similar for all estimators. Clearly, PKDE has the sharpest border, while KDE is outperformed by the other two.
Figure 10. Result of experiment five. PKDE, KDE and flexible Bayesian on a six-dimensional uniform distribution on [−0.5, 0.5]. The bandwidth for the flexible Bayesian estimate is 0.04, while it is 0.15 for the other two. The shaded area is where the cumulative distribution function of the (pseudometric) distance to the centre is between 0.025 and 0.975. (a) PKDE, (b) KDE and (c) flexible Bayesian.
![Figure 10. Result of experiment five. PKDE, KDE and flexible Bayesian on a six-dimensional uniform distribution on [−0.5, 0.5]. The bandwidth for the flexible Bayesian estimate is 0.04, while it is 0.15 for the other two. The shaded area is where the cumulative distribution function of the (pseudometric) distance to the centre is between 0.025 and 0.975. (a) PKDE, (b) KDE and (c) flexible Bayesian.](/cms/asset/fc98ba58-f0b0-48ba-ad33-2e024a468809/gnst_a_944524_f0010_b.gif)
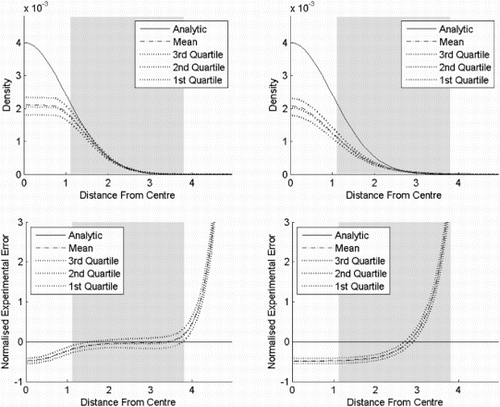
In experiment six (), the estimators are evaluated on a one-sided normal distribution. When the bandwidth is half the bandwidth used in experiment one, PKDE performs identically to PKDE in experiment one. KDE, on the other hand, underestimates far more than in the two sided case in experiment three. This is particularly the case near the centre. The same effect is present for the flexible Bayesian.
Figure 11. Result of experiment six. PKDE, KDE and flexible Bayesian for a one-sided standard normal distribution in six dimensions, where the bandwidths are 0.1 for the flexible Bayesian estimates and 0.25 for the other methods. The top row shows the probability density functions as functions of the distances from the centre, while the bottom row shows the corresponding normalised standard errors. The distance from the centre is chi distributed with order six. The shaded area is where the cumulative distribution function is between 0.025 and 0.975. (a) PKDE, (b) KDE and (c) flexible Bayesian.
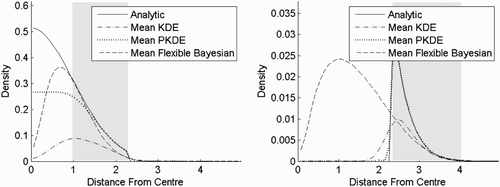
In experiment seven (), the one-sided normal distribution is split into two equally sized populations. The left population contains points where the radius is less than 2.3126 and the right population contains points where the radius is more than 2.3126. The bandwidth settings were the same as in experiment six, which is probably not optimal for a classification. However, it is still clear that PKDE would be favourable in a classification setting. This is because, in the case of PKDE and KDE, there is a trade off in how well the means are fitted on the two populations. The better fit we have on one population the worse the fit will be on the other population. Here, PKDE fits much better on both populations and is therefore favourable. The flexible Bayesian estimator obviously performs very poorly on the right population, because of the severe violation of the independence assumption.
Figure 12. Result of experiment seven. Various kernel density estimates for two populations. The left population has a probability density function that is twice the one-sided standard normal distribution in six dimensions when the radius is less than 2.3126 and 0 elsewhere. The right population has a probability density function that is twice the one-sided standard normal distribution in six dimensions when the radius is more than 2.3126 and 0 elsewhere. The bandwidths are 0.1 for the flexible Bayesian estimates and 0.25 for the other methods. The shaded areas are where the cumulative distribution functions of the (pseudometric) distances to the centre are between 0.025 and 0.975. (a) Left population and (b) right population.
4.2. Discussion of the experiments
KDE and variable KDE are known to be consistent, but in the above experiments the estimators are clearly biased. This is because the sample size is low compared to the number of dimensions. In a practical setting, this is often the case and as a result, nonparametric estimators may suffer from poor statistical power.
In all experiments, the estimators are tested on associated distributions that satisfy the strong condition. In a practical setting, the knowledge about the parameters is less exact, where for instance only the priors are given. In a fair comparison, estimation of parameters should be part of the experiment, but his topic has not been treated in this manuscript.
It is worth noting that PKDE is fairly biased near the centre on the normal distribution. The reason for this is that the radii of the balls with constant volume are larger near the centre. Large contributions from points that are far away (in the pseudometric space) together with a high curvature of the distribution explains this effect. The underestimation is therefore also dependent on the number of dimensions and the choice of bandwidth. The bias near the centre is not seen in the uniform distribution, because the curvature is 0.
In the case of using a variable bandwidth, it seems that the skewness of the normalised experimental error is reduced everywhere at the cost of overestimation in the tails. The reason for the overestimation in the tails is that a query point that is far away from any point in the sample will always have contributions because of the increasing bandwidth. When choosing between variable and fixed PKDE (and also KDE), one should consider the need for accuracy in the tails. In case of classification, it may be desirable to detect very small densities, when the query points are far from any of the points in the population.
In the above experiments, it has also been revealed instances where KDE and flexible Bayesian perform poor. In the case of the uniform population, KDE struggles with the sharp border that is heavily smoothed to decrease variance. This is a great example where PKDE promises great potential over KDE. Notice also that PKDE performs even better than the flexible Bayesian.
In the case of a one-sided population, it is clearly seen that KDE and flexible Bayesian underestimate severely near the centre and continue to underestimate far into the tails. The reason for this is that the estimators are smoothing over orthants where the distribution is 0 by definition. This does not happen in PKDE, because the volume calculation is only done on the relevant orthant.
The one-sided normal distribution that is split in two is added to symbolise a two class classification problem of practical interest. The flexible Bayesian struggles with the right population because of the obvious dependency between explanatory variables. KDE performs poorly on the left population as a result of the effect described in experiment six. It also performs poor on the right population because the boundary is sharp. Clearly, PKDE would perform well if the density estimates were used as inputs to a classification rule.
To understand the full extent of experiment seven, it is important to note that similar results would be evident for a great variety of distributions. Each explanatory variable could be a generalised normal, which could also be either one-sided left, one-sided right or two sided. In addition, the whole distribution could be linearly transformed.
As a concluding remark of the experiments, it is clear that increased statistical power is observed for PKDE with pseudometric of type one, when proper assumptions about the distributions are made. Obviously, if poor assumptions were made, the estimator would suffer from poor robustness. PKDE with a pseudometric of type two has pseudometrics of type one and KDE as special cases. If care is taken when choosing parameters (or priors for parameters), it is therefore possible to increase statistical power at little expense of robustness.
5. Open problems and the use of the new estimators on other problems than density estimation
5.1. Asymptotic argumentation
The literature on kernel density and kernel regression is full of asymptotic argumentation for any variation of estimators. Description of consistency and rates of convergence started with the early works of CitationFix and Hodges (1951), CitationRosenblatt (1956) and CitationParzen (1962) in the one-dimensional case. Optimal rates of convergence for nonparametric estimators were described in CitationStone (1980). It is an interesting question if it is possible to outline an argument for the rates of convergence and AMISE for the estimators described in this paper, provided that the data sample is drawn from an associated distribution. This seems difficult, since the results of the derivations of the bias and AMISE in Section 3.4 contain integrals that seem difficult to treat. However, it is possible that there are approximations that are not seen by the author of this manuscript.
5.2. Parameter selection
In the case of kernel density estimation, the parameters, usually the bandwidth matrix, have usually been selected using either plug-in or cross-validation methods. An introduction can be found in CitationWand and Jones (1994), while more recent articles are given by CitationDuong and Hazelton (2003, Citation2005). In short, the plug-in selector is found by minimising an analytic expression of the AMISE. Since analytical expressions for AMISE do not exist yet, this path for fitting parameters seems difficult. It is far more likely that a cross-validation algorithm is a better route, because it is minimising the mean integrated squared error directly. This is a very important question, but the scope of it is so large that it is left as an open question in this paper.
If we succeed with fitting parameters in the future, the parameters that give the best fit describe the level sets of the estimated density. This is useful for exploration of data and interpreting results. As an example, it was enough to describe the density as a function of distances from the centre in the experiments in Section 4.1.
5.3. Kernel regression
The new nonparametric density estimators can be used on various problems. In terms of regression, most kernel smoothing algorithms can be generalised by using pseudometrics because they are essentially derived from the probability densities (CitationWatson 1964). An overview of kernel smoothing algorithms is given by CitationSimonoff (1996) and CitationWand and Jones (1994). A generalisation of the Nadaraya–Watson kernel-weighted average is proposed as
The model above fits somewhat into the plethora of restricted nonparametric and semiparametric regression methods to reduce the effect of high dimensionality. The additive model has the form of a linear combination of one-dimensional smoothers. This method is strong when the explanatory variables are independent, while dependencies are only dealt with through the use of the back fitting algorithm. In projection pursuit, the additive model is generalised by allowing a projection of the data matrix of explanatory variables before application of the one-dimensional smoothers. The additive models and the projection pursuit models were first defined in CitationFriedman and Stuetzle (1981), with the functional extension given in CitationFerraty, Goia, Salinelli, and Vieu (2013). The projection pursuit regressor has in common with Equation (15) that the visualisation and interpretation of the whole distribution are possible in lower dimensional spaces. However, the regressors will generally have very different forms, since in projection pursuit the regressor is a sum of one-dimensional curves, while Equation (15) has parts of the level sets described by the pseudometric.
In partially linear models the explanatory variables are divided into a linear part and a nonparametric part. The general principles are described in the book of CitationHärdle, Liang, and Gao (2000) and the functional extension is described in CitationAneiros- Péres and Vieu (2006). This method has less in common with Equation (15), since is generally not linearly dependent on any of the explanatory variables.
5.4. Classification
The classification rule can use density estimates for each subpopulation (CitationTaylor 1997). More precisely, the classification rule is
where the πis are the prior probabilities and
s are the density estimates for each subpopulation. Recognise that different pseudometrics can be used for each subpopulation.
5.5. Statistical learning
Statistical learning theory is a framework for machine learning drawing from the fields of statistics and functional analysis as described in CitationMohri, Rostamizadeh, and Talwalkar (2012). The theory deals with the problem of finding a predictive function based on a set of data. The problems that can be solved through this framework are many, including density estimation, classification and regression as discussed above. Another problem that is worth mentioning is data clustering. It is possible that the proposed estimators can be used in density-based clustering (CitationKriegel, Kröger, Sander, and Zimek 2011).
5.6. Some practical considerations related to training parameters for pseudometric kernel density estimation
Clearly, it is not a trivial challenge to find a recipe for training parameters on a wide range of distributions (or problems). Both searching the parameter space and avoiding over fitting are challenges. Limiting the hypothesis space is an important tool to improve computational time and avoid over fitting.
An interesting idea is to use a hypothesis test to find which explanatory variables are dependent on each other and rearrange them so they are grouped together. A semi-naive Bayesian estimator is proposed as
where (oj) is the sequence of the last element in each group and o1 equals 0.
Often, the bottleneck in computational time for kernel density estimation is the training of the parameters. When cross-validation is part of the training, the computational time is dominated by the calculations that are O(n2). In pseudometric kernel density estimation, distance, volume and kernel function calculations are all O(n2). However, all calculations and all coefficients in the volume polynomials can be precomputed before the cross-validation starts. This means that the new estimators can have a computational performance that is comparable to common KDE.
6. Conclusion
An idea for optimising kernel density estimation using pseudometrics is described, where assumptions on the kernel of the distribution can be given. In simulations, the proposed estimators show increased statistical power when proper assumptions are made. As a consequence, this paper describes an approach, where partial knowledge about the distribution can be used effectively. It is also argued that the new estimators may have a potential in statistical learning algorithms such as classification, regression and data clustering.
Acknowledgements
The author thanks Kay Steen, Frode Sørmo, Odd Erik Gundersen, Håkon Lorentzen, Waclaw Kusnierczyk and Helge Langseth and in particular the referees for many helpful comments.
Funding
This work has been partially supported by Verdande Technology and AMIDST. AMIDST is a project that is supported by the European Union's Seventh Framework Programme for research, technological development and demonstration [grant agreement no. 619209].
References
- Aneiros-Péres, G., and Vieu, P. (2006), ‘Semi-functional Partial Linear Regression’, Statistics and Probability Letters, 76, 1102–1110.
- Breiman, L., Meisel, W., and Purcell, E. (1977), ‘Variable Kernel Estimates of Multivariate Densities’, Technometrics, 19, 135–144. doi: 10.1080/00401706.1977.10489521
- Cacoullos, T. (1966), ‘Estimation of a Multivariate Density’, Annals of the Institute of Statistical Mathematics, 18, 179–189. doi: 10.1007/BF02869528
- Delaigle, A., and Hall, P. (2010), ‘Defining Probability Functions for a Distribution of Random Functions’, The Annals of Statistics, 38, 1171–1193. doi: 10.1214/09-AOS741
- Duong, T., and Hazelton, M.L. (2003), ‘Plug-in Bandwidth Matrices for Bivariate Kernel Density Estimation’, Journal of Nonparametric Statistics, 15, 17–30. doi: 10.1080/10485250306039
- Duong, T., and Hazelton, M.L. (2005), ‘Cross-validation Bandwidth Matrices for Multivariate Kernel Density Estimation’, Scandinavian Journal of Statistics, 32, 485–506. doi: 10.1111/j.1467-9469.2005.00445.x
- Epanechnikov, V.K. (1969), ‘Non-parametric Estimation of a Multivariate Probability Density’, Theory of Probability and Its Applications, 14, 153–158. doi: 10.1137/1114019
- Ferraty, F.Y., and Vieu, P. (2006), Nonparametric Functional Data Analysis: Theory and Practice, New York: Springer.
- Ferraty, F., Kudraszow, N., and Vieu, P. (2012), ‘Nonparametric Estimation of a Surrogate Density Function in Infinite-dimensional Spaces’, Journal of Nonparametric Statistics, 24, 447–464. doi: 10.1080/10485252.2012.671943
- Ferraty, F., Goia, A., Salinelli, E., and Vieu, P. (2013), ‘Functional Projection Pursuit Regression’, TEST, 22, 293–320. doi: 10.1007/s11749-012-0306-2
- Fix, E., and Hodges, J.L. (1951), ‘Discriminatory Analysis – Nonparametric Discrimination: Consistency Properties’, Technical Report 4, United States Air Force School of Aviation Medicine, Randolph Field, TX.
- Folland, G.B. (2001), ‘How to Integrate a Polynomial Over a Sphere’, The American Mathematical Monthly, 108, 446–448. doi: 10.2307/2695802
- Friedman, J.H., and Stuetzle, W. (1981), ‘Projection Pursuit Regression’, Journal of the American Statistical Association, 76, 817–823. doi: 10.1080/01621459.1981.10477729
- Friedman, J.H., Stuezle, W., and Schroeder, A. (1984), ‘Projection Pursuit Density Estimation’, Journal of the American Statistical Association, 79, 599–608. doi: 10.1080/01621459.1984.10478086
- Gao, F. (2013), ‘Volumes of Generalized Balls’, The American Mathematical Monthly, 120–130.
- Härdle, W., Liang, H., and Gao, J. (2000), Partially Linear Models, New York: Springer.
- Hoti, F., and Holmström, L. (2003), ‘A Semiparametric Density Estimation Approach to Pattern Recognition’, Pattern Recognition, 37, 409–419. doi: 10.1016/j.patcog.2003.08.004
- Hurvich, C.M., and Simonoff, J.S. (1998), ‘Smoothing Parameter Selection in Nonparametric Regression Using an Improved Akaike Information Criterion’, Journal of the Royal Statistical Society: Series B, 60, 271–293.
- John, G.H., and Langley, P. (1995), ‘Estimating Continuous Distributions in Bayesian Classifiers’, in Proceedings of the Eleventh Annual Conference on Uncertainty in Artificial Intelligence, August, San Mateo, CA: Morgan Kaufmann, pp. 338–345.
- Kriegel, H.P., Kröger, P., Sander, J., and Zimek, A. (2011), ‘Density-Based Clustering’, Wiley Interdisciplinary Reviews Data Mining and Knowledge Discovery, 1, 231–240. doi: 10.1002/widm.30
- Liescher, E. (2005), ‘A Semiparametric Density Estimator Based on Elliptical Distributions’, Journal of Multivariate Analysis, 92, 205–225. doi: 10.1016/j.jmva.2003.09.007
- Loftsgaarden, D.O., and Quesenberry, C.P. (1965), ‘A Nonparametric Estimate of a Multivariate Density Function’, Annals of Mathematical Statistics, 36, 1049–1051. doi: 10.1214/aoms/1177700079
- Mohri, M., Rostamizadeh, A., and Talwalkar, A. (2012), Foundations of Machine Learning, Cambridge: The MIT Press.
- Naito, K. (2004), ‘Semiparametric Density Estimation by Local L2-fitting’, The Annals of Statistics, 32, 1162–1191. doi: 10.1214/009053604000000319
- Parzen, E. (1962), ‘On Estimation of a Probability Density Function and Mode’, The Annals of Mathematical Statistics, 33, 1065–1076. doi: 10.1214/aoms/1177704472
- Rosenblatt, M. (1956), ‘Remarks on Some Nonparametric Estimates of a Density Function’, The Annals on Mathematical Statistics, 27, 832–837. doi: 10.1214/aoms/1177728190
- Simonoff, J.S. (1996), Smoothing Methods in Statistics, New York: Springer Series in Statistics.
- Stone, C.J. (1980), ‘Optimal Rates of Convergence for Nonparametric Estimators’, The Annals of Statistics, 8, 1348–1360. doi: 10.1214/aos/1176345206
- Taylor, C. (1997), ‘Classification and Kernel Density Estimation’, Vistas in Astronomy, 41, 411–417. doi: 10.1016/S0083-6656(97)00046-9
- Terrel, G.R., and Scott, D.W. (1992), ‘Variable Kernel Density Estimation’, The Annals of Statistics, 20, 1236–1265. doi: 10.1214/aos/1176348768
- Wand, M.P., and Jones, M.C. (1994), Kernel Smoothing, Boca Raton: Chapman and Hall/CRC.
- Wang, X. (2005), ‘Volumes of Generalized Unit Balls’, Mathematics Magazine, 78, 390–395. doi: 10.2307/30044198
- Watson, G.S. (1964), ‘Smooth Regression Analysis’, Sankhyā: The Indian Journal of Statistics, 26, 359–372.
- Welling, M., Zemel, R.S., and Hinton, G.E. (2003), ‘Efficient Parametric Projection Pursuit Density Estimation’, in UAI, August, Acapulco, Mexico. San Francisco: Morgan Kaufmann, pp. 575–582.
Appendix 1. Proofs
A.1 Proof of Theorem 3.4
We use that the m-dimensional generalised normal distribution can be expressed as a function g(ε), where
, so
Since
integrates to 1, then the following also holds for g(ε)
Let be a diagonal matrix, where the ith diagonal element
, then we recognise that
which means that
is the generalised unit ball that has been linearly transformed by
. The volume of
is therefore
where p is the harmonic average of the pi’s. This means that Equation (A1) reduces to
Since the integral is Γ(m/p), then
must be
A.2 Proof of Theorem 3.5
Let be a diagonal matrix, where the ith diagonal element is
, then we recognise that
which means that
is the generalised unit ball that has been linearly transformed by
. The volume of
is therefore
A.3 Proof of Theorem 3.7
We recognise that
Each equivalence class in
is therefore uniquely determined by u, thus
. Moreover,
must integrate to 1, which means that the following is valid for g(u)
The strong condition is satisfied, because
A.4 Proof of Theorem 3.8
When
When
A.5 Proof of Theorem 3.10
We recognise that
which means that each equivalence class in
is uniquely determined by u and
, thus
.
Since integrates to 1, then the following is valid for
A.6 Proof of Theorem 3.11
By a simple change of variables, we recognise that
and also that
can be found without loss of generality, by choosing
. Therefore
Now let and
, then the inner integral is
If we substitute for β, we see that the inner integral is a function U of α
where s is defined in Theorem 3.11. The full integral is
and if we let
be the volume of the Lq ball with radius α in m2-dimensions, then we can integrate over α, so that
Substituting and reorganising gives
If we change the variables in the integrals with
, we see that the integral parts can be expressed with incomplete beta functions. The two integrals are
respectively, so
A.7 Proof of Theorem 3.12
In the case when , s is zero and in the case when
,
as
. From Theorem 3.11, we see that
The proof is completed by noting that,
and that all the gamma functions either cancel out or go to 1 as
.
A.8 Proof of Theorem 3.13
The expected value of is
where
is 1/n at the
’s and 0 elsewhere. This is seen, because
. Moreover, the variance of
is
because
is equal to 1/n times the first part plus (n−1)/n times the second part in the square brackets. Equations (13) and (14) are derived by changing variables to αy. The bottom equation in Equation (12) is obtained by integrating
over
and changing variables to αx.