Abstract
Fibroblast growth factor 7 (FGF7) is involved in lipid metabolism, which is considered as a candidate gene with close relation with muscle development by eGWAs and RNA-Seq analyses. To date, limited research has been conducted on the relationship between FGF7 gene and growth traits. The main objective of this work was to further investigate the association between novel InDel within FGF7 gene and growth traits in goat. Herein, FGF7 mRNA expression levels were investigated in various Fuqing goat tissues. We found that FGF7 gene was expressed in six adult goat tissues with the highest mRNA levels in adipose tissue. This result suggested that FGF7 gene might play a critical role in fat deposition. We also detected potential polymorphisms in Fuqing, Nubian and Jianyang Daer breeds. A 22-bp InDel polymorphism in FGF7 gene was detected in 396 goats and the three genotypes were designated as II, ID, and DD. Correlation analysis revealed that InDel polymorphism was significantly associated with growth traits (P < 0.05). Goats with genotypes ID and/or II had superior growth traits compared to those with genotype DD. In summary, our findings suggested that the 22-bp InDel within FGF7 gene could act as a molecular marker to improve the growth traits of goats in breeding programs.
1. Introduction
Fibroblast growth factors (FGFs) play critical roles in multiple biological processes including cellular proliferation, differentiation, and survival fibroblast[Citation1,Citation2]. Approximately, 23 secreted and intracellular FGF families have been identified in vertebrates. Their functions differ based on FGF family and the type of cell from which they were derived[Citation3], such as, embryogenesis, tissues regeneration[Citation4], carcinogenesis[Citation5], adipose development[Citation6] and bone homeostasis[Citation7].
Fibroblast growth factor 7 (FGF7) is a member of fibroblast growth factor family. It is located on chromosome 10 of goats and contains 4 exons and 3 introns. Complete coding region is 585 bp in length and coding protein contains 194 amino acids. FGF7 is closely associated with the regulation of multiple biological processes, including cell survival, differentiation, proliferation, migration[Citation2,Citation8] and follicular development[Citation9,Citation10]. FGF7 is especially involved in insulin secretion of pancreatic b-cells[Citation11]. Recombinant FGF7 derived from tobacco plants was found to promote wound healing in diabetic rats[Citation12]. FGF7 is also produced in osteogenically differentiated cells and improves bone marrow engraftment through a regulatory-T-cell-dependent mechanism[Citation13]. Research has shown that the addition of exogenous FGF7 facilitated osteogenic differentiation of embryonic stem cells by activating extracellular-signal-regulated kinase (ERK)/runt-related transcription factor 2 (Runx2) signaling pathway[Citation14,Citation15]. Overexpression of FGF7 correlates with inflammatory bowel disease[Citation16], while FGF7-deficiency impairs inhibitory synapse formation[Citation17] and liver regeneration[Citation18]. FGF7 is essential to growth and development in mammalians.
FGF7 gene was considered as a candidate regulator by eGWAs and RNA-Seq analyses[Citation19]. For example, FGF7 was closely related to muscle development[Citation20], lipid metabolism[Citation19] and bone formation[Citation21]. GWAS results showed that FGF7 gene was highly related with abdominal fat weight in chicken, indicating that FGF7 gene had a positive regulatory effect on abdominal fat deposition[Citation22]. Previous studies have shown that FGF7 was associated with wool growth in sheep[Citation23]. By detecting mRNA expression of FGF7 gene in cow ovaries at different times of GnRH administration, it was found that FGF7 was involved differently in follicle maturation and CL formation processes[Citation24]. Some InDel loci in the ATBF1, STAT5A, CPT1a and CFAP43 genes of goat have been shown to significantly influence growth traits in our previous studies[Citation25–28]. While research on mutant polymorphisms within the FGF7 gene has only been carried out in cancer[Citation16] or in some other diseases[Citation29]. Hitherto, FGF7 gene is assumed as a potential target for improving growth traits in livestock. Until now, however, goat FGF7 gene and its role in growth traits have never been reported. In this study, a total of 396 samples, containing three goat breeds, were employed for detecting novel FGF7 indel variants. The main objective was to evaluate the association between the genetic variations of FGF7 gene and growth traits in goats, which could benefit the selection and breeding of goat through marker-assisted selection (MAS).
2. Materials and methods
Experimental animals and procedures performed in this research were approved by International Animal Care and Use Committee of Fujian Academy of Agricultural Sciences (FAAS), Fujian province, China. The care and use of experimental animals was fully consistent with local animal welfare laws, guidelines, and policies.
2.1. Samples and data collection
Genomic DNA was extracted from the blood samples of all 396 mature female goats in three breeds of Fuqing goat (FQ, n = 131), Nubian goat (NB, n = 144), and Jianyang Daer Goat (JY, n = 121), The selected goats were approximately 2 years old and had been kept under the same diet and environmental conditions. The zoometric data of all animals were recorded, which included body weight (BW), body height (BH), body length (BL), chest circumference (ChC), chest depth (ChD), chest width (ChW), hucklebone width (HhW), and cannon circumference (CaC). These traits were measured based on the method introduced by Gilbert et al. (1993)[Citation30]. Trunk index (TI), body length index (BLI), chest circumference index (ChCI), cannon circumference index (CaCI), chest width index (CWI), and huckle bone width index (HuWI) were also calculated as advised by Wu et al. (2014)[Citation26].
In addition, 4 Fuqing adult wethers were fed and slaughtered in FAAS goat farm. 9 tissue types, including heart, liver, spleen, lung, kidney, leg muscle, longissimus muscle, perirenal fat, and brain, were collected for reverse-transcription quantitative real-time polymerase chain reaction (qRT-PCRs). All samples were frozen in liquid nitrogen and stored at −80 °C.
2.2. DNA isolation and genomic DNA pools construction
Genomic DNA was extracted from the blood samples of all 396 animals based on phenol-chloroform method. The obtained DNA sample was diluted to 50 ng/μL and stored at −80 °C for genomic DNA pool construction and polymorphism detection. To establish genomic DNA pools, a total of 50 DNA samples were randomly selected from each breed. These DNA pools were employed for screening InDel variations in FGF7 gene by PCR and sequencing techniques.
2.3. Primer design, amplifications, and genotyping
NCBI database (GenBank accession number NC_030836.1), Primer 5.0 was applied to design primers for the amplification of FGF7 gene in goat. The locus ins31499 was genotyped using P7 primer pair (). Each PCR reaction was performed in a 25 μL reaction mixture containing 50 ng genomic DNA, 0.5 µM of each primer, 1× buffer (including 1.5 mM MgCl2), 200 μM dNTPs (dATP, dTTP, dCTP, and dGTP), and 0.625 units of Taq DNA polymerase (TaKaRa, Dalian, China). Cycling protocol was 5 min at 95 °C for pre-denaturation, followed by 40 cycles at 94 °C for 30 s, annealing for 30 s, 72 °C for 10 s, and a final extension at 72 °C for 10 min. PCR products were then analyzed by 3.5% agarose gel electrophoresis.
Table 1. the primer information.
2.4. Total RNA isolation and strand cDNA synthesis
Total RNA was isolated from samples using RNAiso Plus reagent (TaKaRa, Dalian, China). Genomic DNA was removed by RNase-free DNase I treatment. The concentration of isolated RNA was measured with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA) and 1% agarose gel electrophoresis. Reverse-transcription PCRs (RT-PCRs) were conducted for the synthesis of complementary DNA (cDNA) using a PrimeScript™ RT reagent Kit (TaKaRa). 0.5 μg total RNA was mixed with 5 × PrimeScriptTM buffer, 0.5 μL Random 6 mers primer (100 μM), 0.5 μL PrimeScriptTM RT Enzyme Mix I, and RNase-free water to a total volume of 10 μL. The mixture was incubated at 37 °C for 15 min and the reaction was stopped by exposure to 85 °C for 5 s.
2.5. Bioinformatics analysis
The sequences of DNA and amino acid were aligned using MEGA 5.1 (http://www.megasoftware.net/) and BioXM 2.6 (Nanjing Agricultural University, Nanjing, China). Phylogenetic trees were constructed using the neighbor-joining method implemented in MEGA 5.0 and NCBI pairwise alignments (http://www.ncbi.nlm.nih.gov/blast).
2.6 qRT-PCR
The primers P8 and P9 for mRNA expression analysis were designed based on the sequence of goat FGF7 gene (NCBI: NC_030817.1) (). To strengthen mRNA specificity in amplification products, gene-specific primers for exons 14 and 15 were designed to decrease genomic DNA interference (). qRT-PCRs were performed with an Eastep® qRT-PCR Master Mix Kit (Promega) in an Eppendorf Mastercycler ep Realplex 4 Real-Time PCR system. Reaction system and qRT-PCR were implemented as previously described[Citation31].
2.7. Statistical analyses
Genotypic and allelic frequencies of goat InDels in FGF7 gene were directly calculated and polymorphism information content (PIC) was examined using GDIcall online calculator (http://www.msrcall.com/Gdicall.aspx). χ2 test was applied to determine the presence of polymorphism in Hardy-Weinberg equilibrium (HWE). Analysis of variance (ANOVA) was applied to calculate the association of genotypes and growth traits in tested goat populations. SPSS statistical software (version 18.0) (IBM Corp., Armonk, NY, USA) was used for calculations. A general linear model: Yijk =μ + αi+βj+εijk was designed to investigate the association of the 22-bp InDel with growth traits, where Yijk was the observation of the growth trait (body height, etc.) evaluated on the ith level of the fixed factor age (αi) and the jth level of the fixed factor genotype (βj), μ was the overall mean for each growth trait and εijk was the random error for the ijkth individual[Citation32].
2−ΔΔCq method was applied to calculate the expression levels of FGF7 gene with GAPDH as reference gene[Citation33]. No-template control (NTC) was included which contained ddH2O instead of cDNA template. Relative expression levels of FGF7 gene in two groups were analyzed by Student’s t-test. Values in the tables and figures with upper case letters indicated statistical significance at P < 0.01 and those with lower case letters indicated statistical significance at P < 0.05.
3. Result
3.1. Genotypic and genetic parameters of InDel within goat FGF7 gene
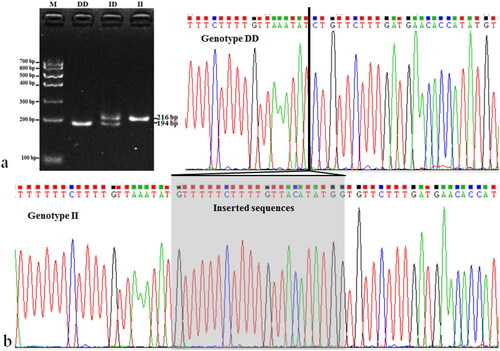
A novel 22-bp insertion polymorphism in intron 2 region (NC_030817.1: g.31499_31521 ins GTTTTTCTTTTGTTACATATGG, ins31499) was detected in FGF7 gene by DNA pool sequencing. Three genotypes were identified in test populations by direct 3.5% agarose gel electrophoresis. There was one band (216 bp) for genotype II, one band (194 bp) for genotype DD, and two bands (194/216 bp) for genotype ID ().
3.2. Bioinformatics analysis of the FGF7
The goat FGF7 transcript encoded 194 amino acids. We observed the coding sequence of goat FGF7 to share 96%, 100%, 100%, 100%, 99%, 100%, 94%, 94%, 99%, and 97% similarity Ovis aries (ACC93619.1), Bos taurus (XP_005211918.1), Bos indicus (XP_019824423.1), Bos grunniens (UQE85735.1), Bubalus bubalis (AFH66797.1), Camelus dromedarius (XP_010996823.1), Mus musculus (AAH52847.1), Rattus rattus (XP_032760022.1), Sus scrofa (XP_020929430.1), Pan troglodytes (XP_001167319.1) (). We performed phylogenetic reconstruction based on the FGF7 sequence, which observed that the goat FGF7 was most closely related to Bos taurus FGF7 and most distantly related to the clade comprising sequences from R. rattus and M. musculus ().
Figure 2. Phylogenetic analysis of FGF7 in 11 Species. Note: The tree was constructed from the amino acid sequences by the neighbor-joining method using MEGA5.1. The numbers on the joints were values of bootstrap test, and the branch length represents the evolutionary time.
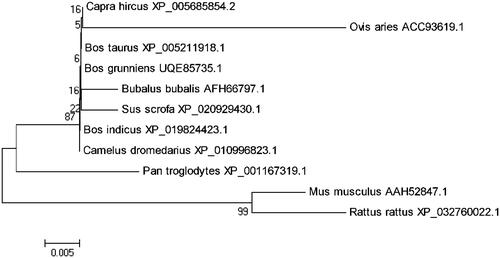
Table 2. Accession numbers and description of the amino acid sequences used in tree building.
3.3. Genetic diversity of 22-bp InDel in three goat populations
Genotypic and allelic frequencies as well as polymorphic indicators including gene homozygosity (Ho), heterozygosity (He), effective allele numbers (Ne), and polymorphism information content (PIC) were calculated on based on genotype results (). χ2 test was applied to explore the existence of polymorphisms in Hardy-Weinberg equilibrium (HWE). The obtained results showed that the frequency of II genotype was the highest in FQ group while that of II genotype was the highest in NB and JY breeds. The results also suggested that the frequency of “I” allele was higher than that of “D” in FQ breed, unlike those in NB and JY breeds. χ2 test results (P > 0.05) and that of JY was not in HWE (P < 0.05). 22-dp InDel was of moderate genetic diversity (0.25 < PIC < 0.5) in all populations ().
Table 3. Genetic polymorphism of InDel within FGF7 gene.
3.4. Association between InDel variant and growth traits in goat
Association analysis between InDel locus and growth traits showed that 22-bp InDel genotype was strongly associated with several goat growth traits (P < 0.05) (). The obtained results showed that BL with II genotype was extremely larger than that with ID genotype (P = 0.007) in NB breed. 22-bp InDel was highly associated with growth traits, including BW (P = 0.032), BH (P = 0.03), ChC (P = 0.046), ChW (P = 0.042), ChD (P = 0.035), and CaC (P = 0.05) in FQ goats, BH (P = 0.03), ChW (P = 0.021), CaC (P = 0.017) of NB breed, and ChCI (P = 0.036) and CaCI (P = 0.013) in JY population. More importantly, individuals with II genotype had superior growth traits than those with DD genotype.
Table 4. Relationship between the InDel of the FGF7 gene and growth traits in goat.
3.5. Relative expressions of FGF7 gene in goat
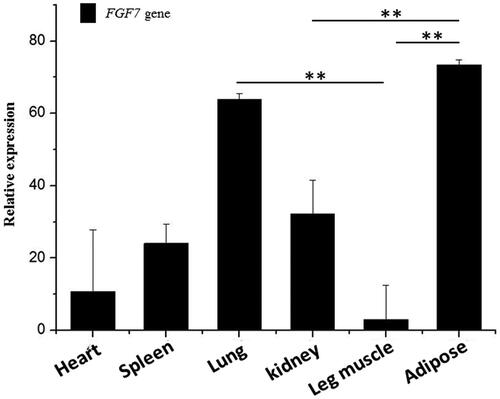
The expression of FGF7 gene was detected in 9 tissues collected by qRT-PCR. FGF7 was found to be expressed in 6 tissues with the highest mRNA levels in lung, adipose tissues, kidney, spleen and heart. Leg muscles showed low expression of this gene ().
4. Discussion
Body weight and growth performance are important economic traits, which have become crucial factors in molecular breeding. InDel molecular markers have been extensively applied to improve economic traits in livestock and poultry because of their simplicity and convenience characteristics. The result of phylogenetic analysis showed that the FGF7 gene was very conserved in ruminants (). In this study, a predicted InDel (NC_030817.1: g.31499_31521 ins GTTTTTCTTTTGTTACATATGG) of FGF7 gene was identified in one indigenous goat breed (FQ) and two introduced breeds (NB and JY). The sequence of 22-bp InDel was different than the predicted sequences of FGF7 gene in European Variation Archive database (http://www.ebi.ac.uk/eva/, variant ID: rs671485500) due to the presence of different breeds. The predicted sequence of which was based on African dwarf goats and environment.
FGF7 gene was found to be closely related to fat deposition[Citation34] and was considered as a candidate regulator for meat performance by eGWAs and RNA-Seq analyses[Citation35]. In this study, qRT-PCR results showed that FGF7 gene was expressed in heart, spleen, lung, kidney, leg muscle, and fat tissues. Noticeably, this gene was abundantly expressed in adipose; similar results have been reported for mice[Citation36], human[Citation37], and chicken[Citation21]. Previous studies have demonstrated that FGF7 increased fatty acid mobilization and hepatic triglyceride secretion[Citation38]. Adipose is a diversified tissue organ with different depots secreting different hormones. It could also contribute to FGF7 generation. Alternatively, FGF7 might promote the proliferation and differentiation of preadipocytes via activating a family of receptor tyrosine kinases[Citation39]. FGF7 gene could play a role in lipid deposition and transport in adult goats.
The allelic frequency of genes could reflect genetic diversity among different groups, indicating that new mutations were introduced to some extent. In this study, 22-bp InDel genotype frequency was different between Fuqing, Nubian and Jianyang Daer goats. “I” allele was the predominant allele in Fuqing breed, but Nubian and Jianyang Daer goats showed contrary results. Fuqing goat is a Chinese domestic breed that grows on the east coast of China while Nubian and Jianyang Daer goats are introduced breeds. Interestingly, Jianyang Daer goat was bred from Jianyang indigenous goats and Nubian goats[Citation40] and therefore, NB and JY showed similar genotypic frequencies. The results suggested that they might undergo different selection pressures during evolutionary processes. Moreover, the 22-bp InDel variant could be a diagnostic DNA marker in goat.
In animal evolution, stable inheritance and a certain degree of genetic variation occur simultaneously[Citation41]. As a special form of genetic variation, InDel has advantages of easy detection and high accuracy. In recent years, it has been demonstrated that the abundance of InDels loci was important in animal growth and development, and has been applied in animal molecular breeding[Citation42,Citation43]. We described the correlation between the 22-bp InDel polymorphism of FGF7 gene and growth traits in goats. The InDel locus was strongly associated with growth traits including BW, BL, BH, ChC, ChW, ChD, CaC, ChCI, and CaCI in collected groups (). The effect of the InDel locus is greater in Fuqing group compared with Nubian and Jianyang Daer groups, the difference of which might be from breeding pressure. Fuqing goat is a relatively primitive Chinese native breed. In contrast, Nubian and Jianyang Daer goat are highly selected breeding breeds. Importantly, goats with genotypes ID and/or II had superior growth traits compared to those with genotype DD. This suggested that “I” allele conferred positive effects on growth traits compared with “D” allele. So far, few studies have analyzed the association between mutations in the FGF7 gene and productive traits, but there are some studies that have shown similar results in FGF homologous genes such as FGF2[Citation44] and FGF5[Citation45]. Our previous studies have shown that InDel polymorphisms were highly related to production traits in goat[Citation26,Citation46]. Some of InDels, such as the 11-bp deletion of CSN1S1 gene was located in introns 9, which significantly affected milk performance and body measurement traits in goats[Citation47,Citation48]. The 22 bp InDel of the FGF7 gene might affect gene expression levels, thereby regulating lipid deposition and osteogenic differentiation in goats. Van Laere et al. (2003)[Citation49] reported that intronic variation could affect gene expression level and increasing porcine meat production. The SNP (IGF2-intron3-G3072A) was mapped to the QTL region. The mutation was found to occur in an evolutionarily conserved CpG island that was hypomethylated in skeletal muscle. The obtained results demonstrated that intronic mutations are important for gene expression and control of phenotypic variation. Therefore, we considered that the novel 22-bp InDel within FGF7 gene influencing growth traits might also have similar regulatory functions. Besides, weight gain is benefits to the quality of meat flavor, texture parameters. The report showed that the intramuscular fat content was found to increase with higher slaughter weight[Citation50]. Increasing carcass weight is associated with increased fat and moisture content, lower cooking loss and higher water holding capacity of the pork meat[Citation51]. Hence, the novel 22-bp InDel within FGF7 gene could be a valid genetic marker in relation to growth trait in goat breeding.
In summary, the expression of the goat FGF7 gene was detected in heart, spleen, lung, kidney, leg muscle and adipose tissue, and was abundantly expressed in fat. By identifying a new 22-bp InDel of the FGF7 gene in intron 2 region and analyzing the association of this site with growth traits of Fuqing, Nubian and Jianyang Daer goats. We found significant associations of InDel with goat growth traits. Importantly, goats with genotypes ID and/or II had superior growth traits compared to those with genotype DD. These results will help us understand the regulatory mechanisms of FGF7 in goats and suggest a molecular marker that may help improve genetic selection.
| Abbreviations | ||
| FGF | = | Fibroblast growth factor |
| FGF7 | = | Fibroblast growth factor 7 |
| InDel | = | insertion/deletion |
| GWAS | = | Genome Wide Association Study |
| RNA-Seq | = | Transcriptome sequencing technology |
| II | = | insertion/insertion |
| ID | = | insertion/deletion |
| DD | = | deletion/deletion |
| SNP | = | single nucleotide polymorphism |
| MAS | = | marker-assisted selection |
| SPSS | = | statistical product and service solutions |
| bp | = | base pair |
| INS | = | intron |
| EX | = | exon |
| HWE | = | Hardy-Weinberg equilibrium |
| Ho | = | homozygosity |
| He | = | heterozygosity |
| Ne | = | effective allele numbers |
| PIC | = | polymorphism information content |
| PCR | = | polymerase chain reaction |
| qRT-PCR | = | reverse-transcription quantitative real-time polymerase chain reaction |
| Cq | = | quantification cycle |
| LSM ± SE | = | Least square mean ± standard error |
| FQ | = | Fuqing goat |
| NB | = | Nubian goat |
| JY | = | Jianyang Daer goat |
| BW | = | body weight |
| BH | = | body height |
| BL | = | body length |
| ChC | = | chest circumference |
| ChD | = | chest depth |
| ChW | = | chest width |
| HuW | = | hucklebone width |
| CaC | = | cannon circumference |
| TI | = | trunk index |
| BLI | = | body length index |
| ChCI | = | chest circumference index |
| CaCI | = | cannon circumference index |
| CWI | = | chest width index |
| HuWI | = | hucklebone width index. |
Acknowledgments
This work was supported by This work was supported by the Natural Science Foundation of Fujian Province (2022J01815), the Public Research Project of Fujian province (2022R10260015, 2023R1024004), Innovation Team Project of Fujian Academy of Agricultural Sciences (CXTD2021006-2).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Barrientos S, Stojadinovic O, Golinko MS, et al. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16(5):585–601.
- D'Andrea LD, Del GA, De Rosa L, et al. Peptides targeting angiogenesis related growth factor receptors. Curr Pharm Des. 2009;15(21):2414–2429.
- Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215–266.
- Trowbridge JM, Rudisill JA, Ron D, et al. Dermatan Sulfate Binds and Potentiates Activity of Keratinocyte Growth Factor (FGF-7). J Biol Chem. 2002;277(45):42815–42820.
- Birnbaum D, deLapeyriere O, Adnane J, et al. Role of FGFs and FGF receptors in human carcinogenesis. Ann N Y Acad Sci. 1991;638(1):409–411.
- Mejhert N, Galitzky J, Pettersson AT, et al. Mapping of the Fibroblast Growth Factors in Human White Adipose Tissue. J Clin Endocrinol Metab. 2010;95(5):2451–2457.
- Su N, Jin M, Chen L. Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res. 2014;2(1):14003.
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–129.
- Berisha B, Sinowatz F, Schams D. Expression and localization of fibroblast growth factor (FGF) family members during the final growth of bovine ovarian follicles. Mol Reprod Dev. 2004;67(2):162–171.
- Zhu MS, Kong DQ, Tian RY, et al. Platelet sonicates activate hair follicle stem cells and mediate enhanced hair follicle regeneration. J Cell Mol Med. 2020;24(2):1786–1794.
- Van Haeften TW, Twickler TB. Insulin‐like growth factors and pancreas beta cells. Eur J Clin Invest. 2004;34(4):249–255.
- Feng ZG, Pang SF, Guo DJ, et al. Recombinant Keratinocyte Growth Factor 1 in Tobacco Potentially Promotes Wound Healing in Diabetic Rats. Biomed Res Int. 2014;2014(1):579632–579639.
- Bruinsma M, van Soest PL, Leenen PJ, et al. Keratinocyte growth factor improves allogeneic bone marrow engraftment through a CD4+ Foxp3+ regulatory T cell-dependent mechanism. J Immunol. 2009;182(12):7364–7369.
- Jeon Y, Kook S, Rho S, et al. Fibroblast growth factor-7 facilitates osteogenic differentiation of embryonic stem cells through the activation of ERK/Runx2 signaling. Mol Cell Biochem. 2013;382(1-2):37–45.
- Kook S, Jeon Y, Park S, et al. Periodontal fibroblasts modulate proliferation and osteogenic differentiation of embryonic stem cells through production of fibroblast growth factors. J Periodontol. 2014;85(4):645–654.
- Yang L, Zhou F, Zheng D, et al. FGF/FGFR signaling: From lung development to respiratory diseases. Cytokine Growth Factor Rev. 2021;62(1):94–104.
- Takase HM, Itoh T, Ino S, et al. FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration. Genes Dev. 2013;27(2):169–181.
- Lee CH, Javed D, Althaus AL, et al. Neurogenesis is enhanced and mossy fiber sprouting arises in FGF7-deficient mice during development. Mol Cell Neurosci. 2012;51(3-4):61–67.
- Gerhard GS, Styer AM, Strodel WE, et al. Gene expression profiling in subcutaneous, visceral and epigastric adipose tissues of patients with extreme obesity. Int J Obes (Lond). 2014;38(3):371–378.
- Du YF, Ding QL, Li Y, et al. Identification of Differentially Expressed Genes and Pathways for Myofiber Characteristics in Soleus Muscles between Chicken Breeds Differing in Meat Quality. Anim Biotechnol. 2017;28(2):83–93.
- Soto-Pedre E, Siddiqui MK, Mordi I, et al. Evidence of a Causal Relationship between Serum Thyroid-Stimulating Hormone and Osteoporotic Bone Fractures. Eur Thyroid J. 2021;10(6):439–446.
- Zhang H, Shen LY, Xu ZC, et al. Haplotype-based genome-wide association studies for carcass and growth traits in chicken. Poult Sci. 2020;99(5):2349–2361.
- Lee CY, Yang CY, Lin CC, et al. Hair growth is promoted by BeauTop via expression of EGF and FGF-7. Mol Med Rep. 2018;17(6):8047–8052.
- Berisha B, Welter H, Shimizu T, et al. Expression of fibroblast growth factor 1 (FGF1) and FGF7 in mature follicles during the periovulatory period after GnRH in the cow. J Reprod Dev. 2006;52(2):307–313.
- Zhang XY, Wu XF, Jia WC, et al. Novel nucleotide variations, haplotypes structure and associations with growth related traits of goat AT motif-binding factor (ATBF1) gene. Asian-Australas J Anim Sci. 2015;28(10):1394–1406.
- Wu XF, Jia WC, Zhang JJ, et al. Determination of the novel genetic variants of goat STAT5A gene and their effects on body measurement traits in two Chinese native breeds. Small Ruminant Res. 2014;121(2-3):232–243.
- Li WY, Liu Y, Gao CF, et al. A novel duplicated insertion/deletion (InDel) of the CPT1a gene and its effects on growth traits in goat. Anim Biotechnol. 2021;32(3):343–351.
- Mi F, Wu XF, Wang Z, et al. Relationships between the Mini-InDel variants within the goat CFAP43 gene and body traits. Animals. 2022;12(24):3447.
- Huang Y, Wang H, Yang Y. Expression of fibroblast growth factor 5 (FGF5) and its influence on survival of breast cancer patients. Med Sci Monit. 2018;24(1):3524–3530.
- Gilbert RP, Bailey DR, Shannon NH. Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets. J Anim Sci. 1993;71(7):1712–1720.
- Wu XF, Liu Y, Gao CF, et al. Novel alternative splicing variants of ACOX1 and their differential expression patterns in goats. Arch Anim Breed. 2018;61(1):59–70.
- Bi Y, He LB, Feng B, et al. A 5-bp mutation within MSTN/GDF8 gene was significantly associated with growth traits in Inner Mongolia White Cashmere goats. Anim Biotechnol. 2021;32(5):610–615.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408.
- Elghazi L, Cras-Méneur C, Czernichow P, et al. Role for FGFR2IIIb-mediated signals in controlling pancreatic endocrine progenitor cell proliferation. Proc Natl Acad Sci U S A. 2002;99(6):3884–3889.
- Zhang YY, Xue XL, Liu Y, et al. Genome-wide comparative analyses reveal selection signatures underlying adaptation and production in Tibetan and Poll Dorset sheep. Sci Rep. 2021;11(1):2466.
- He ZY, Yu JW, Zhou CY, et al. MiR-143 is not essential for adipose development as revealed by in vivo antisense targeting. Biotechnol Lett. 2013;35(4):499–507.
- Tabuso M, Adya R, Stark R, et al. Fibrotic phenotype of peritumour mesenteric adipose tissue in human colon cancer: a potential hallmark of metastatic properties. Int J Mol Sci. 2021;22(5):2430.
- Steiling H, Wüstefeld T, Bugnon P, et al. Fibroblast growth factor receptor signalling is crucial for liver homeostasis and regeneration. Oncogene. 2003;22(28):4380–4388.
- Patel NG, Kumar S, Eggo MC. Essential role of fibroblast growth factor signaling in preadipoctye differentiation. J Clin Endocrinol Metab. 2005;90(2):1226–1232.
- Hu JT, Zhong T, Wang LJ, et al. Complete sequence and characterization of mitochondrial genome of Jianyang Daer goat (Capra hircus). Mitochondrial DNA A. 2016;27(5):2104–2105.
- Triantaphyllopoulos KA, Ikonomopoulos I, Bannister AJ. Epigenetics and inheritance of phenotype variation in livestock. Epigenet Chromatin. 2016;9(1):1–18.
- Wei CJ, Niu YF, Chen BJ, et al. Genetic effect of an InDel in the promoter region of the NUDT15 and its effect on myoblast proliferation in chickens. BMC Genomics. 2022;23(1):138.
- Wang SH, Zhao HD, Wu ML, et al. Exploring of InDel in bovine PSAP gene and their association with growth traits in different development stages. Anim Biotechnol. 2022;33(1):1–12.
- Ghaffarilaleh V, Javanmard A, Saberivand A, et al. Variation and frequency of supernumerary teats, litter size, histological features and the fibroblast growth factor 2 (FGF-2) gene expression pattern in goats. Theriogenology. 2022;179(1):141–148.
- Li YF, Song S, Zhang ZK, et al. A deletion variant within the FGF5 gene in goats is associated with gene expression levels and cashmere growth. Anim Genet. 2022;53(5):657–664.
- Zhou Q, Hu HN, Yang YT, et al. Insertion/deletion (Indel) variant of the goat RORA gene is associated with growth traits. Anim Biotechnol. 2022;34(7):2175–2182.
- Zhang YH, Wang K, Liu JW, et al. An 11-bp indel polymorphism within the CSN1S1 gene is associated with milk performance and body measurement traits in Chinese goats. Animals. 2019;9(12):1114.
- Wang K, Yan HL, Xu H, et al. A novel indel within goat casein alpha S1 gene is significantly associated with litter size. Gene. 2018;671(1):161–169.
- Van Laere A, Nguyen M, Braunschweig M, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425(6960):832–836.
- Weatherup RN, Beattie VE, Moss BW, et al. The effect of increasing slaughter weight on the production performance and meat quality of finishing pigs. Anim Sci. 1998;67(3):591–600.
- Ba HV, Seo HW, Seong PN, et al. Live weights at slaughter significantly affect the meat quality and flavor components of pork meat. Anim Sci J. 2019;90(5):667–679.