 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The present study was designed with the aim to study morphometric characterization as well as phylogeny and diversity of the local Surguli goat at their breeding tract district Kohat through mitochondrial DNA region, i.e., Cytochrome C Oxidase Subunit One (CO1) gene. Morphometric data and blood samples were collected from thirty (30) pure goats. Morphometric analysis showed that sex had significant effect (p < 0.05) on body weight, body length, hearth girth and horn length while no significant effect (p > 0.05) was observed for other characteristics. The results also indicated that age had significant effect (p < 0.05) on height at rump, ear length, horn length and tail length while no significant effect (p > 0.05) was observed for other characteristics. The phylogenetic analysis through CO1 nucleotide sequences within nucleotide range 1–767 showed nine polymorphic sites segregating into eight haplotypes. The mean intraspecific diversity and mean interspecific diversity were calculated as 0.23 and 2.36%, respectively. Phylogenetic tree revealed that Capra Ibex and native Surguli goat have common ancestors. The morphometric and molecular results obtained from the present study can be exploited as a selection tool for breeding and overall improvement.
Introduction
Asia comes first having 597 million of goat population, which is 59% of the total world population. Asian countries’ present share of goat products, such as milk, meat and skin in the world are estimated to be 58.3, 70.7 and 76.5 percent, respectively.Citation1
Small ruminants, particularly native breed kinds, play a significant role to the livelihoods of a considerable part of human population in the tropics from socio-economic aspects.Citation2–4 Thus, combined trials with emphasis on administration and genetic progress to improve animal outputs are of decisive significance.Citation5,Citation6 Economical and biological efficiency of production enterprises generally improves by increasing productivity and reproductive performance of small ruminants.Citation7–11
Pakistani goats are originated from two wild species Markhor (Capra falconeri) and Ibex (Capra ibex).Citation12 Pakistan, as the world’s leading goat producer, contributes 80.3 million goats to the total livestock population of the country. The annual contribution of goat production to the national economy was calculated to be 991 thousand tons of milk and 0.3 million of skin. Furthermore, goat meat is estimated to be 275 tons.Citation13
There have been 36 goats breeds identified so far from different geographical zones of Pakistan.Citation12 Other breeds can be added to this list because of within breed diversity which is not very well presented and a number of unregistered breeds, especially in Khyber Pakhtunkhwa province. It has a variety of documented native goat breeds and undocumented goats. Surguli goat is one of the undocumented goats belonging to Kohat division and the surrounding areas and can also be found in other southern region i.e., Lakki Marwat and D.I Khan.Citation14 The base line survey is not available for Surguli goat; however, research study has been conducted on morphometric characterization in comparison with other goat breeds of the province.Citation14
The phenotypic and genetic characteristics of animals are used to characterize them. Coat color, hair or skin, horns (shape and age), adult size in terms of live weight and body measurements such as wither height are often used in phenotypic characterization of a breed Mahammi et al.,Citation15 Meghen et al.,Citation16 Benhamadi et al.,Citation17 Labbaci et al.,Citation18 Mediouni et al.,Citation19 Djaout et al.,Citation20 Benyarou et al.,Citation21 Belantar et al.,Citation22 Benyarou et al.,Citation21 Meghelli et al.Citation23 and Mogharbi et al.Citation24
Besides from morphometric characterization of breeds, the mitochondrial genome has been regarded nowadays as a strong tool for identifying and determining genetic markers and diversities to explore the phylogenetic associations amongst populations or species. This novel method is used globally due to quick evolving, easiness in sampling and radially obtainable from any tissue.Citation25–29 Every eukaryotic cell is enriched with mitochondrial organelle located inside the cytoplasm.
Mitochondria has small sized maternally transmitted genome which is lacking genetic recombination and repair mechanism. Mitochondrial DNA is organized as a circular double stranded DNA molecule in most eukaryotes.Citation30 The strands are categorized into H-strand (Heavy strand) and L-strand (Light strand). H-strand is guanine rich as compared to L-strand, which is cytosine rich. The length of mitochondrial DNA varies between species to species (15,000–17,000 bp).
Cytochrome c oxidase subunit one gene (CO1) is one of 37 genes of mitochondrial DNA which codes for protein and is located from nucleotide 5904–7444 on H-strand.Citation31 The CO1 gene is pronounced globally for the evaluation of molecular diversity as it proved a powerful toolkit for identity of closely related species.Citation32 Research studies have confirmed that CO1 mitochondrial gene is extremely conservative across species.Citation33,Citation34 The CO1 gene has successfully differentiated various ruminant species into different taxa and has been proven to be a reliable tool for ruminant phylogeny and taxonomy.Citation35 Through CO1 gene sequences, the domestic Pakistani goat breeds can be differentiated from the foreign breeds.Citation36 Conservation management and specie monitoring need utilization of the statistics attained through genetic methods like phylogenetics, genetic information and evolution.Citation37 In recent years, studies have revealed the relationships between Pakistani goat breeds and species,Citation12 therefore the present study was aimed to characterize the Surguli goat based on morphometric characterization as well as molecular characterization through CO1. This will be the first study of its kind on phylogeny and identification of Khyber Pakhtunkhwa goat breeds through CO1 gene. The key goal of the present study was to examine the origin of Surguli goat and level of the genetic diversity within this goat for future documentation and considering Surguli an independent goat breed.
Materials and methods
Sample collection and DNA extraction
The breeding track of Surguli goat district Kohat located at the Southern belt of Khyber Pakhtunkhwa province was visited in April 2021 and true representative goats were selected for morphometric and molecular characterization. The blood samples and morphometric data of Surguli goats were collected at Arid Zone Small Ruminant Research Institute, Ghulam Banda, Kohat, Pakistan including registered farmers of the area. A team of experts from Arid Zone SRRI Ghulam Banda, Kohat (33.5889° N, 71.4429° E) was accompanied to select a typical Surguli goats as shown in the . Moreover, history-based pedigree was recorded from the owner of each animal to ensure pure breed. A total of thirty Surguli goats including fifteen(3 bucks and 12 does) from research farm and fifteen (6 bucks and 9 does) from registered farmers of different age groups i.e., 2–5 years of age having no common ancestors were randomly sampled for morphometric data and blood collection.
An electronic weighing balance (JMS Machineries Pvt. LTD.PAK) and a measuring tape (cm) (JMS Machineries Pvt. LTD.PAK) were used to measure the body weight (kg) and other morphometric characteristics, respectively.
Morphometric parameters of the Surguli goats that were collected included body weight, body length, hearth girth, horn length, ear length, head length, neck length, height of animal at wither and rump and tail length.
Animal handling, restraining and collection of data were performed in accordance with the descriptions lists provided by FAO.Citation38 Morphometric data were collected on semi-structured data collection sheets (Annexure 1) along with visual judgment and measurement. It was made sure that animals were standing on a plan surface during their body measurements.
For molecular characterization blood sample were collected from the animals, which were characterized morphometrically. The surface of jugular vein was disinfected with 70% ethanol and punctured with sterile and disposable syringe for collection of 3 mL blood, which were gently transferred to EDTA tubes (REF-XLGA-E3K3, Xinle®, China). DNA was extracted through non-enzymatic salting out method from blood samples as described by Suguna et al.Citation39 The research work was performed at Histopathology Lab, College of Veterinary Sciences FAH & VS, The University of Agriculture Peshawar, Khyber Pakhtunkhwa, Pakistan (34.0206° N, 71.4814° E).
PCR amplification of CO1
The specific region of 767-bp of CO1 gene was amplified using: F-5′-ACAGGACTTGGTAAAAAGAGG-3′ and R-3′-ATACTTCAGGGTGTCCAAAG-5′ primers (accession no. KP662714.1). The designed primers were sent to Macrogen®, Korea, for synthesis. A total of 25-µL PCR reaction was prepared for each sample in a PCR tube by adding 5 µL of master mix (Cat. No.SM213-0250, GeneDireX, Inc.), 1.5 µl of reverse primer and 1.5 µL of forward primer, 12 µl of PCR water and 5 µL of template DNA. The reaction was carried out in thermal cycler BIORAD® using the following protocol: 94 °C for 5 min and 34 cycles of 94 °C for 30 s, 55.1 °C and 59 °C for 30 s, 72 °C for 1 min and 72 °C for 10 min.
Sequencing of CO1 gene
The targeted region on isolated DNA was amplified by PCR techniques using specific primers of CO1 gene. The sequencing reactions of mitochondrial CO1 gene were performed through Sanger sequencing method, as described by Sanger et al.Citation40 The targeted regions were subjected to chain termination PCR and million to billion copies were terminated at random lengths by 5′-ddNTPs. Then, terminated oligonucleotides were separated in gel electrophoresis via supply of electric current. Finally, gel was analyzed, and DNA sequence was determined by fluorescence tags through automated Sanger sequencing and results were generated by computer in AB1 format.
Data analysis
Morphometric analysis
The morphometric data was organized in Microsoft excel sheet and analyzed using statistical package SAS, version 6.9. Body weight, which is economic trait were taken as response variable. Keeping in view the importance of body weight as an important growth parameter, the same was also predicted from body length and hearth girth as class variables. Following regression model was used:
Where:
Y = The dependent variable (body weight)
β0= Intercept
β1=Independent variables (Body length and hearth girth)
ε= random residuals
To find the statistical differences among the sample goat populations, a general linear model approach (PROC GLM) was applied. Sex and age groups of the goats were fitted as fixed variables whereas body weight and other morphometric parameters as response variables, least square means with their corresponding standard errors were found out for each parameter over sex and age to test statistically difference by Duncan Multiple Range test. The following general linear model was used.
Where;
Yijkl = each dependent variable
µ = Population mean
Ai = ith effect of age (2, 3, 4 and 5 years)
Sj = jth effect of sex (Male and Female)
eijk = random residuals error
Molecular analysis
The reference sequence of COI gene (accession no. KP662714.1) was downloaded from NCBI GenBank (www.ncbi.nlm.nih.gov). The sequencing results of COI of the current study were compared with its reference sequences, in which single-nucleotide polymorphism (SNP) positions were detected. Every identical sequence was considered as single haplotype. Multiple sequence alignment was conducted through ClustalW method in MEGAX software.Citation41 A phylogenetic tree was constructed from haplotypes of CO1 gene via maximum likelihood method, The Kimura-2 parameter method was used to estimate genetic distance within CO1 gene of current study, with other related species using the same software.
Results and discussion
Effect of sex and age on body weight and morphometric measurements
The effect of sex on body weight (kg) and morphometric measurements (cm) in the study is depicted in . Body weight was observed significantly (p < 0.05) different between the sexes. The body weight of goats has been observed to be sexually dimorphic.Citation22,Citation42–44 In the present study, measurements were generally higher in males than females (). This is in consonance with Hamayun et al.Citation45 Adeyinka and Mohammed,Citation42 Nazeer and ShahCitation14 and Akpa et al.Citation46 On the other handCitation47 and Adamu et al.Citation48 worked on West African Dwarf goats, Red Sokoto and Sahel goats and reported that females were superior in body weight and other morphometric parameters across all age groups, which is contradicting with the present study findings. The effect of sex on body weight and several morphometric features shows that there is a typical difference between sexes caused by hormonal action, with males developing quicker than females.Citation49,Citation50 The results also showed that sex had significant (p˂0.05) effect on the body weight, heart girth, body length and horn length and no significant (p > 0.05) effect on heights of wither and rump, head length, ear length, neck length and tail length in Surguli goats which is in consonance with the work of Aliyu et al.Citation51 Effect of sex on body measurements has been reported in previous studies.Citation52,Citation53 The effect of age on body weight (kg) and morphometric measurements (cm) in the study is depicted in . Results of the present study showed that age had significant (p < 0.05) effect on height at rump, ear length, horn length and tail length and no significant (p > 0.05) effect on body weight, body length, heart girth, height at wither, head length and neck length (). The results are in agreement with the reports of Aliyu et al.Citation51 Results of the present study revealed that body weight increased with the advancement of age and then declined after attaining optimum growth at the ages of 4 and 5 years respectively. These variations are in accordance with the early reports of Hamayun et al.Citation45 and Fajemilehin and Salako.Citation47 Similarly, Getahun et al.Citation54 also reported that all body measurements increased as age group increased from 1.5 to 2 years and 3.5 to 4 years of age groups followed by a persistent decline.
Table 1. Effect of sex on the body weight (kg) and morphometric measurements (cm) of Surguli goats.
Table 2. Effect of age on the body weight (kg) and morphometric measurements (cm) of Surguli goats (mean ± SE).
All measurements for different body parameters were included in model and stepwise elimination process was assumed to exclude the unfit body measurements. Results of the presents study showed that body weight of Surguli goat was predicted more precisely using all the body measurements in the present study, however, due to higher adjusted R2 value of 92%, suggested that the employed model was best fitted (). Many researchers found the influential role on the regression model of body weight to all body measurements at all ages and sex.Citation55 The results of the current study are also supported by the study of Iqbal et al.Citation56 who worked on prediction of body weight through body measurements in Beetal goats.
Table 3. Equations for predicting live weight of Surguli goat from body parameters.
Polymorphism and genetic diversity in the Surguli goat population
Analysis of amplified region (767-bp) of CO1 gene identified polymorphic sites at the positions 5, 8, 10, 12, 13, 33, 260, 350 and 756, which might be used as genetic markers amongst individual animals within population of the Surguli goat as shown in .
Table 4. SNPs positions in nucleotides sequence of CO1 gene with reference sequence Capra hircus mitochondrial genome accession no (KP662714.1).
Eight haplotypes were identified based on polymorphic sites found in nucleotide sequences of the CO1 gene when compared to the mitochondrial genome of the Capra hircus. Every identical polymorphic site in CO1 gene sequence was assumed to be the same haplotype. The largest haplotype comprised of two individuals whereas the remaining 7 haplotypes were represented by single sequence as data files obtained through DnaSP6 Software. The dots indicating identical base pairs with the reference mitochondrial genome of the Capra hircus as presented in . Both transversion and transition cause changes in nucleotide composition, which could be employed as genetic markers to study natural populations.Citation29,Citation57,Citation58 Nucleotide substitutions caused by transversion, or transition can display a detailed picture of a population’s evolutionary and geographic history.Citation57,Citation58 The nine polymorphic locations were determined by comparing the COI gene sequences of Surguli goat samples with GenBank sequences of other goats, which revealed that the CO1 gene has low polymorphism. In a previous study on CO1, similar low polymorphism was reported in Indonesian Lakor goat sequences having four polymorphic positions.Citation59 When compared to previous investigations on other mitochondrial regions, the CO1 gene showed low polymorphism asCitation60 have obtained only four polymorphic sites in CO1 sequences of Azi-kheli River buffaloes as compared to mt D-loop region where 28 polymorphic sites were identified. Similarly, Sultana et al.Citation61 also reported 129 polymorphic sites in Pakistani domestic goats by using mtDNA D-loop and cytochrome b gene sequences.
The mean intra-specific diversity of nucleotide sequences of CO1 within Surguli goats as well as the mean inter-specific diversity of present study with closely related goats’ species were calculated by Kimura-2 parameter method in MEGAX softwareCitation41 as shown in . The published nucleotide sequences of CO1 related goat breeds were assembled from NCBI data base in the current study included Capra Ibex COX1 gene (NC_020623.1); Meigu goat (C. hircus) (KM244714.1); Chinese Tibetan goat (C. hircus) (KJ940969.1); Qaidam Cashmere goat (C. hircus) (MG603753.1). The nucleotide sequence of the CO1 gene may be used to successfully identify species, for which the sequence variation should be sufficiently low within species while the variation among closely related species needs to be sufficiently high that a clear threshold between interspecific and intraspecific genetic variations can be demarcated.Citation62 The results of present study showed intraspecific diversity of 0.23% within Surguli goats and interspecific diversity of 2.36% with other related species. Based on previous studies, 2.5% sequence difference is recommended in the CO1 gene sequence for species identification,Citation63 while another study suggested that higher than 2 percent CO1 nucleotide sequence variation is enough to distinguish animal species.Citation64 The outcomes of present study for distinction of Surguli goat with closely related goats species is in consonance with previously reported studies.
Table 5. Intraspecific diversity within CO1 sequences of the samples (M33-M42) and interspecific diversity with CO1 sequences of closely related species with specific accession numbers assembled from NCBI.
Molecular phylogeny of the Surguli goat through COI sequences
Phylogenetic relationship of the Surguli goat samples within nucleotide range 1–767-bp of the COI and other published C. hircus species sequences retrieved from GenBank was performed. Phylogenetic tree was built from the haplotypes of CO1 nucleotides sequence with one reference haplotypes of C. hircus mitochondrion (KP662714.1) along with already reported haplotypes of closely related capra species including C. Ibex COX1 gene (NC_020623.1); C. Falconeri COX1 gene (NC_020622.1); C. Sibirica COX1 gene (NC_020626.1); Meigu goat (C. hircus) (KM244714.1); Chinese Tibetan goat (C. hircus) (KJ940969.1); Qaidam Cashmere goat (C. hircus) (MG603753.1); Capra nubiana COX1 gene (NC_020624.1); C. Aegagrus mitochondrion (KT290893.1); Capra hircus Isolate LS16 (J920218.1), through Neighbor-Joining and Tamura-Nei model toolkits in MEGAX software. The evolutionary paradigm displayed that the CO1 gene may clearly distinguish the intra and inter-species levels linked to phylogenetic tree. The phylogram branches built on the COI gene sequences exhibited that Surguli goat is grouping with others individuals that were used to design this phylogenetic tree as shown in the . The present study phylogenetic tree revealed that evolutionarily, Surguli goat built-up a close relationship with wild goats’ species; C. Ibex COX1 gene (NC_020623.1), C. Aegagrus mitochondrion (KT290893.1), Qaidam Cashmere goat (C. hircus) (MG603753.1), C. Sibirica COX1 gene (NC_020626.1) and with two Chinese goat breeds; The Meigu goat (KM 244714.1), Chinese Tibetan goat (C. hircus) (KJ940969.1).
Figure 2. Phylogenetic tree constructed by N–J method and Tamura-Nei model in MEGAX software from CO1 haplotypes found in the present study and previously published haplotypes of other related species assembled from NCBI.
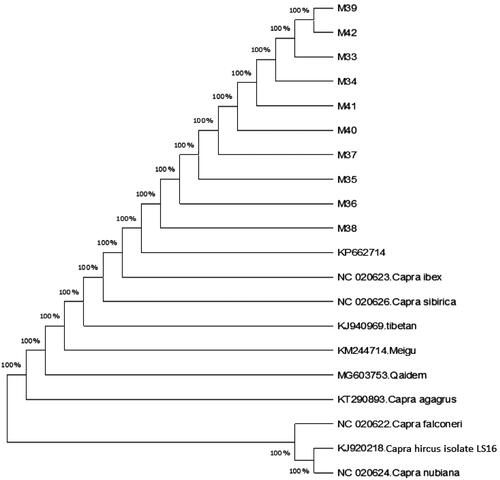
A well-resolved phylogenetic tree can help to define the species’ taxonomy, population history and evolutionary processes.Citation59 The COI gene has successfully proven to distinguish between various ruminant species and could be consider as an efficient tool for the ruminants’ identification and their classification into different taxonomic categories.Citation65 Ali et al.Citation36 also suggested that COI gene sequences can be used as a possible tool to differentiate local Pakistani goat breeds from the exotic ones.
The phylogenetic tree revealed that the Surguli goats shared ancestry tree with C. ibex COX1 gene (NC_020623.1), C. aegagrus mitochondrion (KT290893.1), Qaidam Cashmere goat (C. hircus) (MG603753.1), C. sibirica COX1 gene (NC_020626.1) and with two Chinese goat breeds: The Meigu goat (KM 244714.1), Chinese Tibetan goat (C. hircus) (KJ940969.1). MasonCitation66 has reported C. aegagrus (bezoar) as the direct ancestor of the domesticated goat and C. falconeri (Markhor) as the contributor to mostly Asian goats. According to previous studies C. aegagrus was considered as a single ancestor to Asian domestic goats.Citation67,Citation68 The alternative hypothesis by HarrisCitation69 proposed that at least two other species of wild goats, the C. falconeri (Markhor) and the Ibex (C. ibex) are possible candidates as ancestors for the domestic goats. In addition, Chen et al.Citation70 also reported that the Asian domestic goats are originated and evolved from two wild types of goats (C. aegagrus and C. ibex) of Middle East, which are still existing in Tibet and Inner Mongolia. The goats are possibly domesticated about 10,500 years ago in high, rocky mountainous areas spreading from the Taurus mountains of Turkey into PakistanCitation71 and then spread swiftly because of human migration and trade patterns.Citation72,Citation73 This hypothesis supports our results, which showed that C. ibex might be the patriarchic descendant of the Surguli goat studied in this research, which has gone through adaptation in various environment at different periods.
The Neighbor-Joining tree topology () clearly showed that Surguli goat and other goats from GenBank have close evolutionary relationship. According to Rumanta et al.Citation59 the three wild species of goat; Ibex (Capra ibex), Bezoars (Capra aegagrus) and Markhors (Capra falconeri) have close relationship to the present-day domestic goat (C. hircus). However, the two wild species were ruled out, because the present study results obtained by phylogenetic analysis of CO1 clearly showed that these species are distantly related to Surguli goat. Moreover, the present study results are also supported by Khan et al.Citation12 who reported, Markhor (Capra falconeri) and Ibex (Capra ibex) as the wild relatives of the Pakistani goats. Similarly, Sultana et al.Citation61 worked on D-loop and Cyt b regions sequences in Pakistani goats and proposed Sindh Ibex (C.a. bylyti) as a possible ancestor of domestic goats.
Conclusions
Morphometry of Sex is a significant indicator of body weight, body length, heart girth and horn length while age of the animals could be used as best predictor of rump height, ear length, horn length and tail length. This could be exploited as a selection tool for breeding and overall improvement. The molecular characterization indicated that Surguli goat have diverse population structure having high genetic diversity which can be used for strategic selection and further multiplication. The Neighbor Joining phylogenetic tree revealed that Capra Ibex and native Surguli goat have common ancestors.
Supplemental Material
Download MS Word (16 KB)Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through a large group research project under a grant number (R.G.P.2/66/44).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Skapetas B, Bampidis V. Goat production in the WORLD: present situation and trends. Livest Res Rural Dev. 2016;28(11):200.
- Ahsani MR, Mohammadabadi MR, Shamsaddini MB. Clostridium perfringens isolate typing by multiplex PCR. J Venom Anim Toxins Incl Trop Dis. 2010;16(4):573–578.
- Mohammadabadi MR. Tissue-specific mRNA expression profile of ESR2 gene in goat. Agric Biotechnol J. 2021;12(4):169–184.
- Mousavizadeh A, Mohammadabadi MR, Torabi A, Nasiri MR, Ghiasi H. Genetic polymorphism at the growth hormone locus in Iranian Talli goats by polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP). Iranian J Biotech. 2009;7(1):51–53.
- Masoudzadeh SH, Mohammadabadi MR, Khezri A, et al. Dlk1 gene expression in different tissues of lamb. Iran J Appl Anim Sci. 2020;10:669–677.
- Shahsavari M, Mohammadabadi M, Khezri A, et al. Correlation between insulin-like growth factor 1 gene expression and fennel (Foeniculum vulgare) seed powder consumption in muscle of sheep. Anim Biotechnol. 2021;34(4):882–892.
- Amiri Roudbar M, Mohammadabadi MR, Mehrgardi AA, Abdollahi-Arpanahi A. Estimates of variance components due to parent-of-origin effects for body weight in Iran-Black sheep. Small Rumin Res. 2017;149:1–5.
- Amiri Roudbar M, Abdollahi-Arpanahi R, Ayatollahi Mehrgardi A, Mohammadabadi M, Taheri Yeganeh A, Rosa GJM. Estimation of the variance due to parent-of-origin effects for productive and reproductive traits in Lori-Bakhtiari sheep. Small Rumin Res. 2018;160:95–102.
- Mohamadipoor L, Mohammadabadi M, Amiri Z, et al. Signature selection analysis reveals candidate genes associated with production traits in Iranian sheep breeds. BMC Vet Res. 2021;17(1):369.
- Mohammadabadi MR. Inter-simple sequence repeat loci associations with predicted breeding values of body weight in Kermani sheep. Genet 3rd Millen. 2016;14(4):4383–4390.
- Mohammadabadi MR, Tohidinejad F. Charachteristics determination of Rheb gene and protein in Raini Cashmere goat. Iran J Appl Anim Sci. 2017;7(2):289–295.
- Khan MS, Khan MA, Mahmood S. Genetic resources and diversity in Pakistani goats. Int J Agric Biol. 2008;10(2):227–231.
- GOP. Pakistan Economic Survey, 2020–2021. 2020–2021. Partners available at: http://www.finance.gov.pk/survey_ 1921.html.
- Nazeer M, Shah SH. Morphological characterization of indigenous goats breeds of Khyber Pakhtunkhwa, Pakistan. Sarhad J Agric. 2018;34(2):258–267.
- Mahammi FZ, Gaouar SBS, Tabet-Aoul N, Tixier-Boichard M, Saidi-Mehtar N. Morpho-biometric characteristics and breeding systems of local chickens in the Oranie region (West Algeria). Cahiers Agric. 2014;23(6):382–392.
- Meghen C, MacHugh DE, Bradley DG. Genetic characterization and West African cattle. World Anim Rev. 1994;78(1):59–66.
- Benhamadi M, Mezouar K, Benyarou M, Bouandas S, Gaouar BS. Morphometric characterization of the equine barbe breed in northwest of Algeria. Genet Biodivers J. 2017;1(2):48–65.
- Labbaci MA, Djaout M, Benyarou AA, Ameur S, Gaouar BS. Morphometric characterization and typology of donkey farming (Equus asinus) in the Wilaya of Tlemcen. Genet Biodivers J. 2018;2(1):56–68.
- Mediouni MR, Said S, Ameur AA, Gaouar SBS. Phenotypic and morphometric diversity of Indigenous Turkey (Meleagris Gallopavo) from Wilaya of Tlemcen, Northwest of Algeria. Genet Biodivers J. 2020;4(3):1–10.
- Djaout A, Belharfi FZ, Kourichi A, Larabi N, Benidir M, Gaouar SBS. Morphometric assessment and physico-chemical description of the milk of Arbia goat breed in province of Tlemcen. Genet Biodiv J. 2022;6(2):127–141.
- Benyarou M, Ameur AA, Gaouar SBS. Contribution to the study of two local bovine breeds in Wilaya of Tlemcen: morphometric characterization and DNA biobank. Genet Biodivers J. 2018;2(1):44–55.
- Belantar I, Tefiel H, Gaouar SBS. Phenotypic characterization of local goat population in western Algeria (Wilaya of Relizane) with morphometric measurements and milk analysis. Genet Biodivers J. 2018;2(2):55–66.
- Meghelli I, Kaouadji Z, Yilmaz O, Cemal I, Karaca O, Gaouar SBS. Morphometric characterization and estimating body weight of two Algerian camel breeds using morphometric measurements. Trop Anim Health Prod. 2020;52(5):2505–2512.
- Mogharbi A, RM, Mediouni A, A, Ameur N, Azzi, SBS, Gaouar. Morphometric characterization of domestic rabbits (Oryctolagus cuniculus domesticus L.) in western Algeria. Genet Biodivers J. 2021;5(3):72–79.
- Al-Jumaili AS, Boudali SF, Kebede A, et al. The maternal origin of indigenous domestic chicken from the Middle East, the north and the horn of Africa. BMC Genet. 2020;21(1):30.
- Boudali SF, Al-Jumaili AS, Bouandas A, et al. Maternal origin and genetic diversity of Algerian domestic chicken (Gallus gallus domesticus) from North-Western Africa based on mitochondrial DNA analysis. Anim Biotechnol. 2022;33(3):457–467.
- Ghernouti N, Bodinier M, Ranebi D, Maftah A, Petit D, Gaouar SBS. Control region of mtDNA identifies three migration events of sheep breeds in Algeria. Small Rumin. Res. 2017;155(1):66–71.
- İbiş O, Kılıç M, Özcan S, Tez C. Genetic characterization of the Turkish gray hamster (Cricetulus migratorius) based on mitochondrial cytochrome b and 12S rRNA sequences. Mitochondrial. DNA A DNA Mapp Seq Anal. 2018;29(6):819–830.
- Kunda RM, Handayani NSN, Wijayanto H, Widayanti R. Study of genetic marker of Cuscuses (Marsupialia: Phalangeridae) from Maluku and Papua based on cytochrome b gene sequences. Pak J Biol Sci. 2016;19(3):122–135.
- Andrews R, Kubacka MI, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147–147.
- Zong NC, Li H, Li H, et al. Integration of cardiac proteome biology and medicine by a specialized knowledgebase. Circ Res. 2013;113(9):1043–1053.
- Hebert PD, Cywinska A, Ball SL, Dewaard JR. Biological identifications through DNA barcodes. Proc R Soc Lond B. 2003;270(1512):313–321.
- Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, Passamonti M. A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 2014;30(12):555–564.
- Waugh J. DNA barcoding in animal species: progress, potential and pitfalls. Bioessays. 2007;29(2):188–197.
- Nagarajan M, Nimisha K, Kumar S. Mitochondrial DNA variability of domestic river buffalo (Bubalus bubalis) populations: genetic evidence for domestication of river buffalo in Indian subcontinent. Genome Biol Evol. 2015;7(5):1252–1259.
- Ali A, Rehman A, William K. Phylogenetic analysis of Capra hircus commonly found goat breeds of Pakistan using DNA barcode. J Bioresour Manag. 2016;3(1):1.
- Schwartz MK, Luikart G, Waples RS. Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol. 2007;22(1):25–33.
- FAO. Status of animal genetic resources. In: Rischkowsky B, Pilling D, Scherf B, eds. The State of the World’s Animal Genetic Resources for Food and Agriculture. 1st ed. Rome, Italy: Commission on genetic resources for food and agriculture, Food and Agriculture Organization of the United Nations; 2007: 23–48.
- Suguna S, Nandal D, Kamble S, Bharatha A, Kunkulol R. Genomic DNA isolation from human whole blood samples by non enzymatic salting out method. Int J Pharm Sci. 2014;6(6):198–199.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463–5467.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549.
- Adeyinka I, Mohammed ID. Relationship of live weight and linear body measurement in two breeds of goat of Northern Nigeria. J Anim Vet Adv. 2006;11(5):891–893.
- Benyoub KQ, Ameur Ameur A, Gaouar SBS. Phenotypic characterization of local goats populations in western Algerian: Morphometric measurements and milk quality. Genet Biodivers J. 2018;2(1):69–76.
- Fantazi K, Tolone M, Amato B, et al. Characterization of morphological traits in Algerian indigenous goats by multivariate analysis. Genet Biodivers J. 2017;1(2):20–30.
- Hamayun K, Muhammad F, Ahmad R, Nawaz G, Zubair M. Relationship of body weight with linear body measurements in goats. J Anim Vet Adv. 2006;5(6):452–455.
- Akpa GN, Duru S, Amos TT. Influence of strain and sex on estimation of within-age-group body weight of Nigerian Maradi goats from their linear body measurements. Trop Agric. 1998;75(4):462–467.
- Fajemilehin OS, Salako AE. Body measurement characteristics of the West African dwarf (WAD) goat in deciduous forest zone of Southwestern Nigeria. Afr J Biotechnol. 2008;7(14):2521–2526.
- Adamu H, Ma’aruf BS, Shuaibu A, Umar HA, Maigado AI. A study on some morphometric traits of red Sokoto and Sahel goats in Maigatari local government area of Jigawa state. NJAP. 2020;47(4):15–23.
- Kakuma Y, Ichimaru T, Yonezawa T, et al. Androgen induces production of male effect pheromone in female goats. J Reprod Dev. 2007;53(4):829–834.
- Seifemichael M, Kefelegn K, Negassi A, Banerjee AK. Variability in linear body measurements and their application in predicting body weight of Afar goats in Ethiopia. Int J Interdiscip Multidiscip Stud. 2014;1(4):17–25.
- Aliyu AM, Rotimi EA, Galadima NM. Effect of age and sex on morphometric measurements of Sahelian goats in Faskari local government area of Katsina state. FJS. 2021;5(1):11–15.
- Akpa GN, Abubakar MY, Nwagu BI, Alphonsus C. Effect of dam mating weight, month of kidding and sex of kids on growth characteristics of Red Sokoto kids. Niger Agric J. 2010;41(1):57–61.
- Bazzi H, Ghazaghi M. Effects of environmental factors on body weight of Sistani goat at different ages. J. Anim. Vet. Adv. 2011;10(21):2819–2823.
- Getahun S, Ahmed S, Zemene W. Morphometric characterization of indigenous goats in East Gojjam Zone, Amhara Region, Ethiopia. Int J Adv Res Biol Sci. 2020;7(2):47–62.
- Dakhlan A, Qisthon A, Hamdani MDI. Predicting body weight based on body measurements at different ages and sex in Saburai goat. Adv Anim Vet Sci. 2021;9(11):1791–1799.
- Iqbal M, Javed K, Ahmad N. Prediction of body weight through body measurements in Beetal goats. Pak J Sci. 2013;65(4):458.
- Widayanti R, Agustianti T, Suprayogi RM, Kunda, S, Pakpahan. Phylogenetic relationship of cuscuses (Marsupialia: Phalangeridae) from Papua and Maluku based on mitochondrial sequences of NADH dehydrogenase sub-unit 1 gene. J Biotechnol. 2016;15(1–2):17–25.
- Widayanti R, Wijayanto H, Wendo WD, Kunda RM. Identification of genetic diversity 12S ribosomal RNA genes as genetic markers for species determining species cuscuses. J. Vet. 2015;16(2):227–235.
- Rumanta M, Kunda RM, Volkandari SD, Indriawati I, Kakisin P. Genetic characterization and phylogenetic study of Lakor goat from Southwest Maluku Regency based on mitochondrial COI gene. Vet World. 2020;13(6):1209–1220.
- Ali F, Ahmad I, Ali MI, et al. Mitochondrial phylogenetic and diversity analysis in Azi-Kheli buffalo. Trop Anim Health Prod. 2021;53(5):512.
- Sultana S, Mannen H, Tsuji S. Mitochondrial DNA diversity of Pakistani goats. Anim Genet. 2003;34(6):417–421. 10.1046/j.0268-9146.2003.01040.x.
- Luo J-y, Yan D, Song J-y, et al. A strategy for trade monitoring and substitution of the organs of threatened animals. Sci Rep. 2013;3(1):3108.
- Tobe SS, Kitchener AC, Linacre AM. Reconstructing mammalian phylogenies: a detailed comparison of the cytochrome b and cytochrome oxidase subunit I mitochondrial genes. PLOS One. 2010;5(11):e14156.
- Yan D, Luo JY, Han YM, et al. Forensic DNA barcoding and bio-response studies of animal horn products used in traditional medicine. PLOS One. 2013;8(2):e55854.
- Nagarajan M, Nimisha K, Thomas S. Molecular characterisation of ruminant mammals using DNA barcodes. In: Trivedi S, Rehman H, Saggu S, Panneerselvam C, Ghosh S, eds. DNA Barcoding and Molecular Phylogeny. Cham: Springer; 2020: 203–216.
- Mason IL. Evolution of Domesticated Animals. New York, USA: Prentice Hall Press; 1984: 468.
- Liu RY, Yang GS, Lei CZ. The genetic diversity of mtDNA D-loop and the origin of Chinese goats. Yi Chuan Xue Bao. 2006;33(5):420–428.
- Wang J, Chen YL, Wang XL, Yang XZ. The genetic diversity of seven indigenous Chinese goat breeds. Small Rumin Res. 2008;74(1–3):231–237.
- Harris DR. The distribution and ancestry of the domestic goat. Proc Linn Soc Lond. 1962;173(2):79–91.
- Chen SY, Su YH, Wu SF, Sha T, Zhang YP. Mitochondrial diversity and phylogeographic structure of Chinese domestic goats. Mol Phylogenet Evol. 2005;37(3):804–814.
- Zeder MA, Hesse B. The initial domestication of goats (Capra hircus) in the Zagros Mountains 10,000 years ago. Science. 2000;287(5461):2254–2257.
- Fernández H, Hughes S, Vigne J-D, et al. Divergent mtDNA lineages of goats in an early Neolithic site, far from the initial domestication areas. Proc Natl Acad Sci USA. 2006;103(42):15375–15379.
- Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P. Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc Natl Acad Sci U S A. 2001;98(10):5927–5932.

