Abstract
The intensive labour and time required for conventional methods to identify bacterial fish pathogens have revealed the need to develop alternative methods. Raman spectroscopy has been used in the rapid optical identification of bacterial pathogens in recent years as an alternative method in microbiology. Strains of bacterial fish pathogens (Vibrio anguillarum, Lactococcus garvieae and Yersinia ruckeri) that often cause infectious diseases in fish were here identified and analyzed in terms of their biochemical structures in different media and at different incubation times, and the data were specified by using Raman spectroscopy. The results demonstrated that Raman spectroscopy presents species-specific Raman spectra of each disease-causing bacteria and that it would be more appropriate to choose general microbiological media over selective media for routine studies. Additionally, it was found that species-specific band regions did not differ in 24- and 48-hour cultures, but there could be a difference in peak intensity which may lead to difficult characterization of spectrum. The current study, conducted for the first time with bacterial fish pathogens under different incubation conditions, is believed to provide a basis for the routine use of Raman spectroscopy for quick pathogen identification and the precise determination of the methodology for further research.
Introduction
Pathogen diagnosis with rapid identification methods has emerged as a requirement for the detection of infectious diseases in recent years. Polymerase chain reaction (PCR) has been extensively used; however, typical disadvantages such as the requirements for strain-specific probes, the complexity of interpreting the results, the high costs, and contamination problems have made it difficult to find alternative approaches for bacterial diagnosis.Citation1 Vibrational spectroscopy has been declared a potential alternative method for bacterial identification that offers a unique vibrational spectral pattern of samples with the frequencies of chemical bonds and atoms involved in molecular vibrations.Citation2
Raman spectroscopy allows us to determine the overall molecular constitution of biological samples such as nucleic acids, proteins, carbohydrates, lipids, and inorganic crystalsCitation3 and the results are presented as a phenotypic, fingerprint-like spectrum.Citation2 Because of the unique molecular structure of a biomolecule, Raman spectrum analysis allows for unique spectral fingerprints. Thus, these spectrum markers can be used for disease surveillance, cell and tissue drug effectiveness studies, pathogen identification, and many other biological processes.Citation4–8 The specificity of analytical techniques combined with appropriate chemical processing allows bacteria to be characterized, discriminated, and identified.Citation9–11 Raman spectrums of microorganisms reflect specific biochemical compositions and can be used to identify bacteria species and yeast.Citation3 This method is preferred in microbiological studies by several researchers,Citation12–15 especially for the identification of bacterial agents.Citation2,Citation7,Citation16–18
A number of Gram-positive and Gram-negative bacteria are the causes of various fish diseases worldwide, which are associated with many different clinical manifestations such as erosion, swelling, haemorrhagic septicaemia and ulceration.Citation19 Yersinia ruckeri, Vibrio anguillarum and Lactococcus garvieae are reported to be some of the main disease-causing (Yersiniosis, Vibriosis and Lactococcosis) bacterial pathogens in rainbow trout (Oncorhynchus mykiss Walbaum, 1792),Citation20 and the rapid identification of these pathogens is important to prevent economic loss and maintain efficient prevention strategies.
The conditions of the culture, such as using different media, different incubation times or temperatures, have the potential to affect the discrimination and reproducibility of Raman spectroscopy.Citation21 There are several studies that have shown that changes in Raman spectra have been observed in bacteria that were grown in different culture conditions or media.Citation21–24 However, accurate determination of cell composition in the detection of bacterial cell components by Raman spectroscopy is closely related to culture conditions,Citation25 and in order to be able to identify bacteria using Raman spectroscopy, it is of great importance to know their biochemical contents and to predetermine the basic cell structures under different conditions.Citation26 Parameters such as bacteria preparation, growth phase, growth temperature, and growth time have been reported to affect the spectra obtained by Raman spectroscopy analysis.Citation27
The aim of this study was to determine the effects of different culture conditions on the biochemical structure of bacterial fish pathogens with Raman spectroscopy, which allows rapid identification of bacterial fish pathogens by determining species-specific Raman spectra of each pathogenic bacteria. In addition to determining the effects of different incubation times on the reproductive performance of bacteria, the effects of the use of different media on the biochemical structures of the bacteria were determined. The aim was to provide basic data for the use of Raman spectroscopy as a diagnostic tool in the rapid diagnosis of bacterial diseases, which have an important place in fish farming, to determine the incubation conditions for bacteria in Raman spectroscopy, which provides the most appropriate data, and, following this, to create a database. The findings obtained are biochemical fingerprints of the pathogens studied and include the characterization regions determined by Raman spectroscopy.
Materials and methods
Strains
Lactococcus garvieae, Vibrio anguillarum and Yersinia ruckeri strains were supplied the culture collection of the Fish Disease and Biotechnology Laboratory at the Faculty of Fisheries of Izmir Katip Celebi University. These strains had been isolated from acute cases of diseased rainbow trout and identified at species level by 16s rRNA sequencing. The GenBank accession number of the L. garvieae strains are OP420797 (A1LG22), OP420800 (A3LG22), OP420803 (A6LG22), of the Y. ruckeri strains are OP420805 (B1YR22), OP420860 (B2YR22), OP420869 (B3YR22), and of the V. anguillarum strains are OP422192 (C1VA22), OP422240 (C2VA22) and OP422224 (C5VA22).
Raman sample preparation
Two different experimental setups were established to determine the particular effects of the culture conditions. In the first experiment, selective strain-specific media were used (Thiosulfate Citrate Bile Sucrose Agar (TCBS, Merck, Germany)) for V. anguillarum, Waltman-Shotts MediumCitation28,Citation29 for Y. ruckeri, LG AgarCitation30 for capsuled L. garvieae) while all strains were grown in standard media for microbiology (Tryptic soy agar (TSA, Merck, Germany) and brain heart infusion agar (BHI, Merck, Germany)). They were incubated at 21° C for 24 h and fresh cultures were prepared for further analysis. In the second experiment, L. garvieae, V. anguillarum, and Y. ruckeri strains were grown in standard medium (TSA) at 21° C and then 24 and 48 h cultures were prepared for further analyses to determine the effects of the incubation time.
The colonies were suspended in isotonic sodium chloride and centrifuged for 10 minutes at 6000 rpm (Hermle Labortechnik GmbH, Z206A). After the supernatant was removed, the bacterial pellets were lyophilized at − 40° C, 1 Pa with XO-Instrument Freeze Dry (XO-12B).
Raman measurements
The Raman spectra of each sample as lyophilized powders on substrate were then measured with a Renishaw Raman Spectrometer at the Application and Research Center, Central Research Laboratory, Izmir Katip Celebi University. The source wavelength was 785 nm and X 100 objective was selected for the study. Each sample was measured three times from randomly chosen locations and the averaged spectra were recorded for further analyses. The results were graphitized using SpectraGryph 1.2.8 Spectroscopy Software (Germany). Spectra evaluation and biochemical band detection were performed according to the data reported in the literature.Citation17,Citation31,Citation32 The spectra of each strain were transferred to PCA (Principal Component Analyses) loadings in the Raman Tool Set (BioNEM Lab of the University ‘Magna Graecia’ in Catanzaro, Italy)Citation33 for classification of the data.
Results
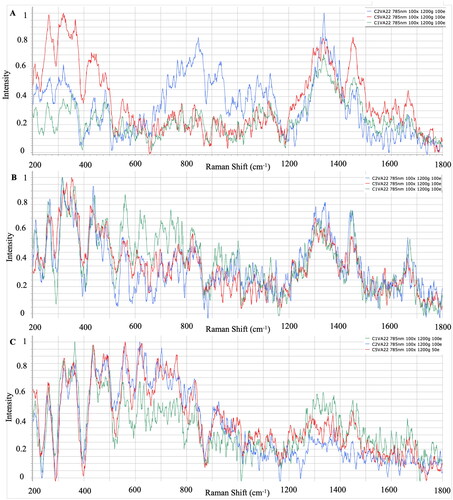
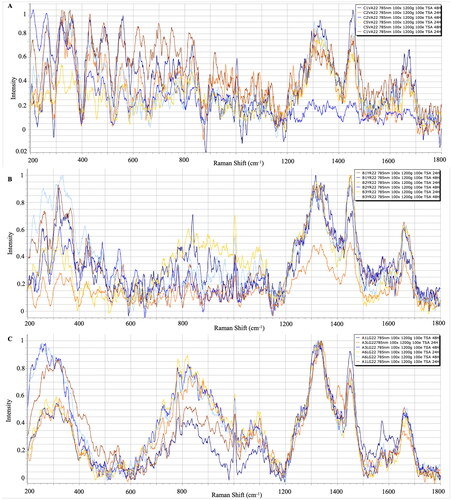
The effects of variable culture conditions on Raman spectroscopy, and the use of different media for different bacterial fish pathogens generated during the first phase of this research, were investigated. shows the average of the processed Raman spectra for each V. anguillarum strain that grew on TCBS, BHI and TSA, respectively. Significant identification peaks in V. anguillarum strains isolated from TCBS agar were similar to TSA and BHI, the regions where polysaccharides are represented were 436–481 cm−1, nucleic acids were represented at 834–846 cm−1, amino acids were represented at 1334–1341 cm-1, and fatty acids were determined in the 1663–1666 cm−1 regions ().
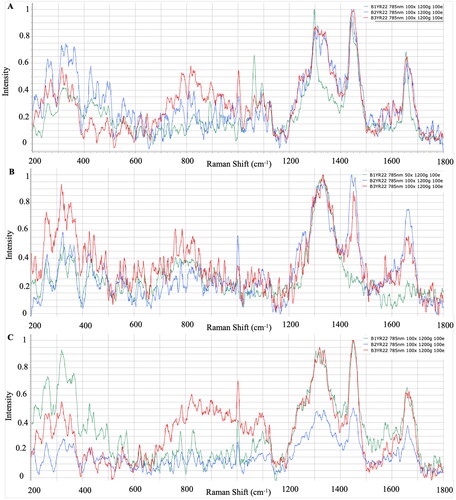
The Raman spectra of three different strains of Y. ruckeri that grew on three different media are presented in . In strains of Y. ruckeri isolated from WS agar, significant identifying peaks were determined in 314–329 cm−1 where polysaccharides were represented, 810–829 cm−1 where nucleic acids were represented, 1063–1092 cm−1 where amino acids were represented, and the 1655–1658 cm−1 regions where fatty acids were represented. The bacteria-specific characteristic peaks were detected in the regions 318–323 cm−1, 1001–1003 cm−1 and 1303–1320 cm−1, 1439–1451 cm−1 and 1655–1661 cm−1 where polysaccharides, amino acids and fatty acids were represented from the strains incubated in TSA and BHI ().
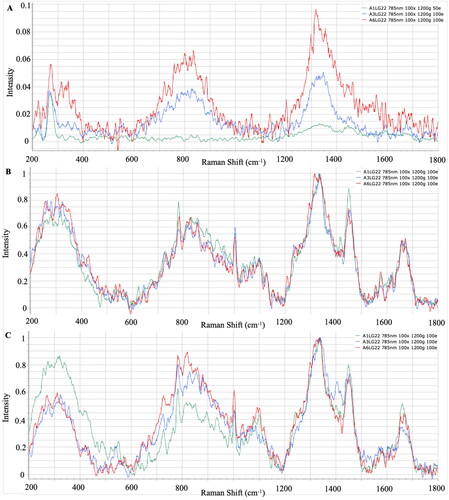
The mean Raman spectra for each L. garvieae strain grown on LG agar, BHI, and TSA are presented in . The typical peaks were centred in 320 cm−1, where polysaccharides were represented; between 781–782 cm−1, where nucleic acids were represented; between 1332–1340 cm−1, where amino acids were represented; and between 1449–1450 cm−1 and 1659–1672 cm−1, where fatty acids were represented from strains isolated from LG agar, TSA and BHI (). However, the intensity of peaks from LG agar strains were detected lower ().
The Raman spectra of V. anguillarum, Y. ruckeri and L. garvieae strains at different incubation period are presented in the figures, and it was determined that the incubation time did not create significant differences in significant characteristic peaks ().
Figure 4. A: 24 and 48 h Raman spectra of V. anguillarum strains (TSA), B: 24 and 48 h Raman spectra of Y. ruckeri strains (TSA), C: 24 and 48-hour Raman spectra of L. garvieae strains (TSA).
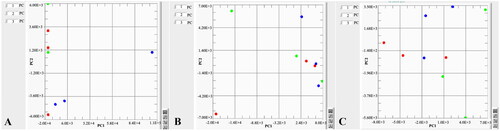
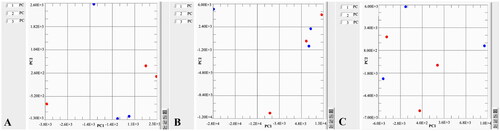
shows the PCA analyses of different mediums used for each bacterial strain. It is clearly indicated that LG agar is distinctly separate from all media used for L. garvieae strains. Individual comparisons of the 24 and 48 h cultures of each species of bacteria were performed by PCA analyses in .
Discussion
This study aimed to determine the changes in the biochemical structure of bacterial fish pathogens caused by different culture conditions using Raman spectroscopy. In the first stage of the study, the Raman spectra of V. anguillarum, L. garvieae and Y. ruckeri pathogens incubated in different media under the same incubation conditions were compared. At this stage of this investigation, the Raman spectra of V. anguillarum, L. garvieae and Y. ruckeri strains were incubated in different microbiological media under the same incubation conditions and the Raman spectrum of each pathogen was compared. In the second stage, the effects of different incubation times on Raman spectra were investigated.
For the identification of microorganisms such techniques as biochemical testing, microscopy, serological methods, mass spectroscopy and molecular methods are commonly used approaches that have advantages and disadvantages.Citation34 Rapid detection of pathogenic bacteria is necessary for effective treatment of bacterial infections and the identification of specific bacteria takes about 48 hours, including cultivation and specific biochemical tests on average.Citation35,Citation36 Raman spectroscopy has been used by many researchers to identify different pathogens isolated from different sources,Citation3,Citation37–40 and it has started to be preferred for routine pathogen identification in microbiology in recent years.[Citation3] In addition to all these advantages, Raman spectroscopy is practicable for biomaterials with low water background and gives accurate and sensitive resultsCitation12 which makes Raman spectroscopy an alternative identification method for disease diagnosis. Harz et al.Citation22 reported in a study on the identification of strains belonging to the Staphylococcus genus under different incubation conditions that it is a very successful method for bacterial identification on a species basis; however, the correct determination of the bacterial cell components is relevant to the culture conditions of bacteria.Citation25 For this reason, it is essential to determine the changes that occur in the biochemical profiles of bacterial fish pathogens through parameters such as different media and incubation conditions in order to create the necessary infrastructure and data banks for the routine identification of fingerprint spectra.
Selective culture media are used to isolate specific species or genera of bacteria, and this has the purpose of eliminating unwanted microbial flora after adding some inhibitors to the culture medium.Citation41 To isolate pathogens from the site of infection, the use of selective media is crucial to make it possible to accurately identify and diagnose the pathogen and initiate treatment; furthermore, it is an important first step in microbiological research on environmental samples.Citation42 Although similar results were obtained in TSA and BHI, which are general microbiological media, peaks were detected in different regions in pathogen-specific selective media. Similarly, Hutsebaut et al.Citation21 reported that TSA and BHI are more suitable for the identification of bacteria, and species-specific spectrum information can be obtained even under different incubation conditions. When the bacterial spectra were examined, it was determined that the peaks of the strains isolated from BHI and TSA gave significant results at similar points. It was observed that the strains of L. garvieae, Y. ruckeri and V. anguillarum gave significant peaks in the selective media LG agar, WS and TCBS at similar points to the general medium TSA and BHI. When the spectrum differences between the species were examined, peaks were commonly observed in the band regions where the basic biochemical components of bacterial cells, polysaccharides, nucleic acids, amino acids and fatty acids were represented, but it was determined that the spectrum intensities differed according to the species. Dinçtürk and TanrıkulCitation18 released the first Raman data of bacterial fish pathogens which were incubated at 21° C in Tryptic Soy Broth (TSB). Despite using different Y. ruckeri, V. anguillarum and L. garvieae strains and different media, the species-specific band regions showed no difference when compared with the present study. The characteristic peaks were detected from both TSB and TSA/BHI cultures. In addition, correlative results were observed from 24 and 48 h cultures of these bacterial pathogens. The similarities proved that the location of significant spectra could be obtained from different media, different incubation times and different strain sources.
Raman spectroscopy has been described as an extremely capable method for bacteria identification, even with different culture conditions.Citation22 Even though Raman spectroscopy is a promising technique for reliable and fast identification of bacterial species, there are some potential limitations of this method; the requirement of a Raman database is crucial for routine pathogen detection.Citation27 However, recommendations for the preparation of samples and data processing guidelines should be introduced, which will significantly promote the use of Raman spectroscopy and convert it into a routine diagnostic method in clinical laboratories,Citation43 and trained personnel and possible automatization is necessary for routine diagnosis and future work.Citation34 Hutsebaut et al.Citation21 determined the effect of culture conditions on three Bacillus species with Raman spectroscopy and concluded that species-specific data can be obtained from Raman spectra, even under different culture parameters. In this current study, it is aimed to obtain a reliable dataset with varying culture conditions for important bacterial fish pathogens, L. garvieae, V. anguillarum, and Y. ruckeri, by Raman spectroscopy.
In conclusion, Raman spectroscopy is an effective method for the identification of some of the important psychrophilic bacteria, V. anguillarum, L. garvieae and Y. ruckeri, and it was determined that species-specific band regions were observed in the general medium even under different culture conditions. The results of the study have determined that Raman spectroscopy can be an important alternative method for the rapid and reliable diagnosis of bacterial fish diseases and it would be more appropriate to use general microbiological media over selective media for routine studies. Furthermore, it was determined that species-specific band regions did not differ in 24 and 48 h cultures and as a result of different incubation conditions, but there could be a difference in peak intensity. It is believed that the current study, conducted for the first time with bacterial fish pathogens under different incubation conditions, provides a basis for the routine use of Raman spectroscopy and the precise determination of the methodology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author, [Dinçtürk, E.], upon reasonable request.
Additional information
Funding
References
- Kastanos E, Kyriakides A, Hadjigeorgiou K, Pitris C. Identification and antibiotic sensitivity of UTI pathogens using Raman spectroscopy. In: Urinary Tract Infections. London: IntechOpen; 2011: 207–226.
- Münchberg U, Rösch P, Bauer M, Popp J. Raman spectroscopic identification of single bacterial cells under antibiotic influence. Anal Bioanal Chem. 2014;406(13):3041–3050.
- Brauchle E, Schenke‐Layland K. Raman spectroscopy in biomedicine–non‐invasive in vitro analysis of cells and extracellular matrix components in tissues. Biotechnol J. 2013;8(3):288–297.
- Moritz TJ, Taylor DS, Krol DM, Fritch J, Chan JW. Detection of doxorubicin-induced apoptosis of leukemic T-lymphocytes by laser tweezers Raman spectroscopy. Biomed Opt Express. 2010;1(4):1138–1147.
- Gautam R, Vanga S, Ariese F, Umapathy S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Techn Instrum. 2015;2(1):38.
- Escoriza MF, VanBriesen JM, Stewart S, Maier J. Raman spectroscopic discrimination of cell response to chemical and physical inactivation. Appl Spectrosc. 2007;61(8):812–823.
- Sil S, Mukherjee R, Kumar NS, Aravind S, Kingston J, Singh UK. Detection and classification of bacteria using Raman spectroscopy combined with multivariate analysis. Def Life Sc Jl. 2017;2(4):435–441.
- Dutta A, Gautam R, Chatterjee S, Ariese F, Sikdar SK, Umapathy S. Ascorbate protects neurons against oxidative stress: a Raman microspectroscopic study. ACS Chem Neurosci. 2015;6(11):1794–1801.
- Maquelin K, Dijkshoorn L, van der Reijden TJ, Puppels GJ. Rapid epidemiological analysis of Acinetobacter strains by Raman spectroscopy. J Microbiol Methods. 2006;64(1):126–131.
- Jarvis RM, Goodacre R. Characterisation and identification of bacteria using SERS. Chem Soc Rev. 2008;37(5):931–936.
- Ashton L, Lau K, Winder CL, Goodacre R. Raman spectroscopy: lighting up the future of microbial identification. Future Microbiol. 2011;6(9):991–997.
- Jung GB, Nam SW, Choi S, Lee GJ, Park HK. Evaluation of antibiotic effects on Pseudomonas aeruginosa biofilm using Raman spectroscopy and multivariate analysis. Biomed Opt Express. 2014;5(9):3238–3251.
- Neugebauer U, Schmid U, Baumann K, et al. Characterization of bacterial growth and the influence of antibiotics by means of UV resonance Raman spectroscopy. Biopolymers: Original Res. Biomolecul. 2006;82(4):306–311.
- Lee KS, Landry Z, Pereira FC, et al. Raman microspectroscopy for microbiology. Nature Rev Methods Primers. 2021;1(1): 80.
- Nakar A, Wagenhaus A, Rösch P, Popp J. Raman spectroscopy for the differentiation of Enterobacteriaceae: a comparison of two methods. Analyst. 2022;147(17):3938–3946.
- Strola SA, Schultz E, Allier CP, DesRoches B, Lemmonier J, Dinten JM. 2013. March). Raman microspectrometer combined with scattering microscopy and lensless imaging for bacteria identification. In Advanced Biomedical and Clinical Diagnostic Systems XI (Vol. 8572, pp. 166–175). SPIE.
- Strola SA, Baritaux JC, Schultz E, et al. Single bacteria identification by Raman spectroscopy. J Biomed Opt. 2014;19(11):111610.
- Dinçtürk E, Tanrıkul TT. First preliminary study on identification of bacterial fish pathogens with Raman spectroscopy. Anim Biotechnol. 2021;34(3):529–537.
- Austin B. Methods for the diagnosis of bacterial fish diseases. Mar Life Sci Technol. 2019;1(1):41–49.
- Toranzo AE. Report about fish bacterial diseases. In: Alvarez-Pellitero P, Barja JL, Basurco B, Berthe F, Toranzo AE, eds. Mediterranean Aquaculture Laboratories, Zaragoza: CIHEAM, 2004: 49–89.
- Hutsebaut D, Maquelin K, De Vos P, Vandenabeele P, Moens L, Puppels GJ. Effect of culture conditions on the achievable taxonomic resolution of Raman spectroscopy disclosed by three Bacillus species. Anal Chem. 2004;76(21):6274–6281.
- Harz M, Rösch P, Peschke K-D, Ronneberger O, Burkhardt H, Popp J. Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst. 2005;130(11):1543–1550.
- De Gelder J, Scheldeman P, Leus K, et al. Raman spectroscopic study of bacterial endospores. Anal Bioanal Chem. 2007a;389(7-8):2143–2151.
- Dettman JR, Goss JM, Ehrhardt CJ, Scott KA, Bannan JD, Robertson JM. Forensic differentiation of Bacillus cereus spores grown using different culture media using Raman spectroscopy. Anal Bioanal Chem. 2015;407(16):4757–4766.
- De Gelder J. 2008. Raman Spectroscopy as a Tool for Studying Bacterial Cell Compounds (Doctoral dissertation). Ghent University.
- Madigan MT, Martinko JM, Parker J. 1997. Brock Biology of Microorganisms. (Vol. 11). Upper Saddle River, NJ: Prentice Hall.
- Pahlow S, Meisel S, Cialla-May D, Weber K, Rösch P, Popp J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv Drug Deliv Rev. 2015;89:105–120.
- Austin B, Austin DA, Munn CB. 2007. Bacterial Fish Pathogens: disease of Farmed and Wild Fish. (Vol. 26, p. 552. Chichester: Springer.
- Waltman WD, Shotts EB. Jr, A medium for the isolation and differentiation of Yersinia ruckeri. Can J Fish Aquat Sci. 1984;41(5):804–806.
- Chang CI, Lee CF, Tsai JM, et al. Development of a selective and differential medium for capsulated L actococcus garvieae. J Fish Dis. 2014;37(8):719–728.
- Escoriza MF, Vanbriesen JM, Stewart S, Maier J. Studying bacterial metabolic states using Raman spectroscopy. Appl Spectrosc. 2006;60(9):971–976.
- De Gelder J, De Gussem K, Vandenabeele P, Moens L. Reference database of Raman spectra of biological molecules. J Raman Spectroscopy. 2007b;38(9):1133–1147.
- Candeloro P, Grande E, Raimondo R, et al. Raman database of amino acids solutions: a critical study of extended multiplicative signal correction. Analyst. 2013;138(24)2013:7331–7340.
- Rebrosova K, Samek O, Kizovsky M, Bernatova S, Hola V, Ruzicka F. Raman spectroscopy—A novel method for identification and characterization of microbes on a single-cell level in clinical settings. Front Cell Infect Microbiol. 2022;12:866463.
- Maurer FP, Christner M, Hentschke M, Rohde H. Advances in rapid identification and susceptibility testing of bacteria in the clinical microbiology laboratory: implications for patient care and antimicrobial stewardship programs. Infect Dis Rep. 2017;9(1):6839.
- Ramesh G, Paul W, Valparambil Puthanveetil V, Raja K, Embekkat Kaviyil J. Raman spectroscopy as a novel technique for the identification of pathogens in a clinical microbiology laboratory. Spectrosc Lett. 2022;55(8):546–551.
- Maquelin K, Kirschner C, Choo-Smith L-P, et al. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J Clin Microbiol. 2003;41(1):324–329.
- Harz M, Kiehntopf M, Stöckel S, et al. Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro‐Raman spectroscopy. J Biophotonics. 2009;2(1-2):70–80.
- Kirschner C, Maquelin K, Pina P, et al. Classification and identification of Enterococci: A comparative phenotypic, genotypic, and vibrational spectroscopic study. J Clin Microbiol. 2001;39(5):1763–1770.
- Maquelin K, Kirschner C, Choo-Smith LP, et al. Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Methods. 2002;51(3):255–271.
- Bonnet M, Lagier JC, Raoult D, Khelaifia S. Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbes New Infect. 2020;34:100622.
- Al-Blooshi SY, Latif MAA, Sabaneh NK, Mgaogao M, Hossain A. Development of a novel selective medium for culture of Gram-negative bacteria. BMC Res Notes. 2021;14(1):211.
- Wang L, Liu W, Tang JW, et al. Applications of Raman spectroscopy in bacterial infections: principles, advantages, and shortcomings. Front Microbiol. 2021;12:683580.