Abstract
Litter size in sheep is a complex trait controlled by micro-effective polygenes. APAF1, CLSTN2, CTH, PLCB1, PLCB4, and CHST11 are all involved in mammalian reproduction. However, the effects of these genes on litter size in sheep are still unclear. Therefore, in this study, we used Sequenom MassARRAY® SNP assay technology to type the single nucleotide polymorphisms (SNPs) loci of six genes in five sheep breeds. The results showed that most sheep breeds contain three genotypes at each locus. Then, we conducted population genetic analysis on the SNPs of six genes and found that the polymorphic information content in all sheep breeds ranged from 0 to 0.37, and most sheep breeds were in Hardy-Weinberg equilibrium (p > 0.05). In addition, association analysis in Small Tail Han sheep indicated that the rs399534524 locus in CLSTN2 was highly associated with first parity litter size, and litter size in ewes with CT genotype was higher than that in ewes with CC genotype or TT genotype. Furthermore, the rs407142552 locus in CTH was highly associated with second parity litter size in Small Tail Han sheep, and litter size in ewes with CT genotype was higher than that in ewes with TT genotype. Finally, we predicted the CTH and CLSTN2 protein interaction network and found that HTR1E, NOM1, CCDC174 and ALPK3 interact with CLSTN2 and have been reported as candidate genes related to litter size in sheep. These results suggest that they may be useful genetic markers for increasing litter size in sheep.
Introduction
Reproductive traits of sheep are one of the most important economic traits that directly determine the production costs and economic benefits of sheep production. Ewes that produce more lambs per litter can provide a large number of lambs, thus increasing economic efficiency. Therefore, it is important to identify molecular markers that influence litter size in sheep. Marker-assisted selection significantly shortens the breeding cycle and improves breeding success compared to traditional methods. The recent discovery of SNPs has been recognized as one of the most useful types of genetic markers.Citation1,Citation2 In particular, point mutations in genes such as bone morphogenetic protein receptor, type 1B (BMPR1B),Citation3 growth differentiation factor 9 (GDF9),Citation4 and bone morphogenetic protein 15 (BMP15)Citation5 have been implicated in sheep reproduction and result in litter size differences. We compiled a number of genes that may affect fertility in sheep and found all the SNPs in the region where each gene is located based on previous resequencing data. The population differentiation index (FST) was then calculated for each SNPs locus. The above obtained SNPs annotation results and FST were integrated and screened according to the following conditions: ① non-synonymous mutations with FST > 0.05; ② SNPs in introns with FST > 0.15. We identified SNPs in six genes that were suitable for screening and could be potential genetic markers influencing litter size in sheep.
The protease apoptosis activating factor-1 (APAF1) gene induces programmed cell death and is a critical factor in the mitochondrial-dependent death pathway. And apoptosis has been reported to be involved in oogenesis, folliculogenesis, oocyte loss/selection and atresia.Citation6,Citation7 APAF1 has been found to be abundant in the granulosa cells of early ovarian antral follicles.Citation8 In addition, APAF1 is one of the apoptotic signaling pathways mediated by porcine granulosa cells.Citation9 And APAF1 mutations can lead to a decrease in reproductive efficiency in Holstein dairy cattle.Citation10 Calsyntenin-2 (CLSTN2), also known as alcadein-γ, plays a role in a variety of biological processes, including cell growth, differentiation, cell death and tumorigenesis.Citation11–13 A previous study found that CLSTN2 is expressed in the uterus of rats.Citation14 Moreover, this gene is a candidate gene associated with litter size in sheep.Citation15 Studies have shown that the indel in CLSTN2 is a candidate gene affecting goat fertility.Citation13 Cystathionine-γ-lyase (CTH) is a widely distributed enzyme that catalyzes the formation and transformation of hydrogen sulfide (H2S).Citation16 The production of endogenous H2S is involved in the preovulatory cascade reaction and follicular rupture during ovulation in mice. Research has shown that CTH is expressed in mouse and human granulosa cells and increases during ovulation. Furthermore, inhibition of CTH activity can reduce the ovulation rate in mice.Citation17 And CTH mRNA has also been reported in porcine cumulus cells and oocytes.Citation18 Moreover, a decrease in CTH activity may also lead to placental abnormalities in pre-eclampsia and fetal growth restriction.Citation19,Citation20 Phospholipase C beta 1 (PLCB1) and phospholipase C beta 4 (PLCB4) are members of the phospholipase C beta family. The PLCB1 protein is a downstream effector of the G protein-coupled GnRH receptor. G protein-coupled receptor (GPCR) plays a central role in mediating fertility because some hormones such as GnRH, FSH, LH, and oxytocin signal through GPCR.Citation21 In addition, PLCB1 has been identified as a candidate gene for human primary ovarian failure and ovarian dysfunction.Citation22 PLCB4 regulates PLCB1 and has a function in calcium signaling,Citation23 which plays an important role in proper uterine quiescence and smooth muscle contraction during pregnancy.Citation24 Notably, a meta-analysis of GWAS found that PLCB1 and PLCB4 were associated with litter size in sheep.Citation25 Carbohydrate sulfotransferase 11 (CHST11) is a target gene of transforming growth factor β (TGFβ)-like growth factor,Citation26 which may regulate litter size in sheep through TGFβ and BMP signaling.Citation27 Furthermore, CHST11 was found to regulate proliferation and apoptosis of mouse embryonic chondrocytes.Citation28
All of the above genes play a role in animal reproduction, so they may be involved in sheep reproduction and lead to differences in litter size and may be important genetic markers for sheep breeding. Therefore, this study aims to investigate the association between SNPs of six genes and litter size. This information has not been previously elucidated, but is expected to provide valuable genetic markers for sheep breeding.
Materials and methods
Animal preparation and sample collection
The Science Research Department (in charge of animal welfare issues) of the Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS-CAAS; Beijing, P. R. China) has approved all procedures involving laboratory animals. Ethics approval was also granted by the animal ethics committee of IAS-CAAS (No. IASCAAS-AE-03; December 12th, 2016).
The five sheep breeds used in this experiment included a total of 768 ewes, of which three were polytocous breeds (Small Tail Han sheep, n = 384; Cele Black sheep, n = 96; and Hu sheep, n = 96) and two were monotocous breeds (Sunite sheep, n = 96; and Bamei Mutton sheep, n = 96). The details of the five sheep breeds are displayed in . Blood samples were collected from all sheep via the jugular vein, anticoagulated with EDTA, and stored at −20 °C. Simultaneously record parity and litter size of the Small Tail Han sheep.
Table 1. Basic information on ewes used in this study.
Blood DNA extraction
DNA was extracted using a blood DNA extraction kit (Tiangen Biotechnology Co., Ltd., Beijing, China). The extracted DNA samples were tested for concentration using NanoDrop 2000 (Thermo Fisher, USA) and quality was assessed by 1.5% agarose gel electrophoresis, and qualified samples were selected for subsequent experimental analysis.
Genotyping
The rs399534524 locus in CLSTN2, the rs407142552 locus in CTH, the rs414162829 locus in PLCB1, the chr13_1651705 locus in PLCB4 (Genomic version: Oar_v3.1), the rs1092455701 locus in CHST11, and the rs416244410 and rs426974615 locus in APAF1 use Sequenom MassARRAY® SNP technology for genotyping in polytocous and monotocous breeds of sheep. The primers for sheep genes used in this study were designed based on their sequence in GenBank (). Then, these primers were synthesized by Beijing Compass Biotechnology Co., Ltd. (Beijing, China).
Table 2. Primers used in the present study.
Protein interaction networks predicted by STRING database
We predicted the protein interaction networks involving CTH and CLSTN2 through the STRING database v.11.0Citation29 (https://string-db.org), which is a useful tool for predicting protein interactions.
Statistical analysis
The allele and genotype frequency, polymorphism information content (PIC), heterozygosity (HE), effective allele number (NE), and p-value (Chi-square test) were calculated using the genotyping data. It was considered that the ewe population with p > 0.05 (Chi-square test) was in Hardy-Weinberg equilibrium. To determine the association between genotypes and litter size, the adjusted linear model, yijn = μ + Pi+ Gj + IPG + eijn was applied, in which yijn is the phenotypic value (litter size); μ represents the population mean; Pi indicates the fixed effect of the ith parity (i = 1, 2, or 3); Gj represents the effect of the jth genotypes (j = 1, 2, or 3); IPG is the interactive effect of parity and genotype; and eijn indicates the random error.
Results
Genotyping and population genetic analysis of SNPs in six genes
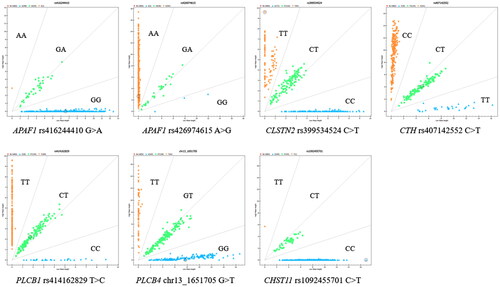
Two SNPs were detected in APAF1 (rs416244410 and rs426974615) and one SNP in five other genes (CLSTN2: rs399534524, CTH: rs407142552, PLCB1: rs414162829, PLCB4: chr13_1651705, CHST11: rs1092455701), respectively (). The loci rs416244410, rs399534524, rs407142552 and chr13_1651705 are in the intron region, whereas the loci rs426974615, rs414162829 and rs1092455701 are in the exon region. Among them, the rs416244410 and rs426974615 loci in APAF1 had low polymorphisms (PIC < 0.25) in all five sheep breeds and were in Hardy-Weinberg equilibrium (p > 0.05). The rs399534524 locus in CLSTN2 was moderately polymorphic (0.25 < PIC < 0.5) in all five sheep breeds. In addition, this locus was in Hardy-Weinberg equilibrium (p > 0.05) in Cele Black sheep, while it was not in Hardy-Weinberg equilibrium (p < 0.05) in other breeds. The rs407142552 locus in CTH was low polymorphism (PIC < 0.25) in Bamei Mutton sheep, while it was moderately polymorphism (0.25 < PIC < 0.5) in Small Tail Han sheep, Hu sheep, Cele Black sheep and Sunite sheep. Furthermore, this locus was in Hardy-Weinberg equilibrium (p > 0.05) in all five sheep breeds. The rs414162829 locus in PLCB1 and the chr13_1651705 locus in PLCB4 showed moderate polymorphism (0.25 < PIC < 0.5) in all five sheep breeds, and all were in Hardy-Weinberg equilibrium (p > 0.05). The rs1092455701 locus in CHST11 was low polymorphism (PIC < 0.25) and under Hardy-Weinberg equilibrium (p > 0.05) in all five sheep breeds ().
Table 3. Population genetic analysis of SNPs with different genes in five sheep breeds.
Genotype and allele frequencies of seven SNPs in the six genes in monotocous and polytocous sheep
We classified the five breeds into two categories based on litter size characteristics: polytocous (Small Tail Han sheep, Cele Black sheep, and Hu sheep) and monotocous (Sunite sheep and Bamei Mutton sheep). Genotyping of seven SNPs in five sheep breeds showed that the allele and genotype frequencies of the rs416244410 locus in APAF1, the rs399534524 locus in CLSTN2, the rs407142552 locus in CTH, and the rs1092455701 locus in CHST11 were significantly different (p < 0.05) between polytocous and monotocous sheep breeds (). The allele frequency of the rs426974615 locus in APAF1 was significantly different (p < 0.05) between polytocous and monotocous sheep breeds, but the genotype frequency was not significantly different (p > 0.05). In addition, the allele and genotype frequencies of the rs414162829 locus in PLCB1 and the chr13_1651705 locus in PLCB4 were not significantly different (p > 0.05) in polytocous and monotocous breeds.
Table 4. Genotype and allele frequencies in sheep populations of different fecundity.
Associations between seven loci in six genes with litter size in Small Tail Han sheep
Association analysis () revealed that the rs399534524 locus in CLSTN2 was significantly associated with the first parity litter size in Small Tail Han sheep (p < 0.05), and the litter size of ewes with CT genotype was higher than that of ewes with CC and TT genotypes in all parities. The rs407142552 locus in CTH was significantly associated with the second parity litter size in Small Tail Han sheep (p < 0.05), and the litter size of ewes with CT genotype was higher than that of ewes with TT genotype in all parities. However, locus rs426974615 and rs416244410 in APAF1, the rs414162829 in PLCB1, the chr13_1651705 in PLCB4, and the rs1092455701 in CHST11 have no significant correlation with litter size in Small Tail Han sheep (p > 0.05).
Table 5. Least squares mean and standard error of litter size in Small Tail Han sheep with different genotypes.
Predicted protein interaction networks involving CTH and CLSTN2
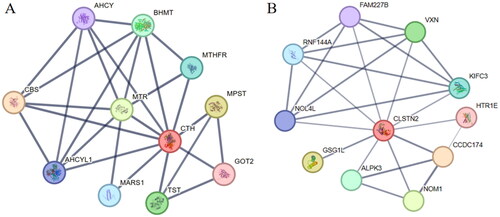
To further explore the functions of CTH and CLSTN2 in sheep reproductive traits, proteins associated with them were predicted using the STRING website. The results () showed that CTH and CLSTN2 were predicted to interact with 10 proteins, respectively. Among them, HTR1E,Citation30 NOM1, CCDC174 and ALPK3, which interact with CLSTN2, have been reported as candidate genes related to sheep litter size that may be involved in the reproductive process.Citation15 Therefore, it is hypothesized that CLSTN2 is important for the development of reproductive traits in sheep.
Discussion
CLSTN2 is a calcium-related gene involved in cell proliferation, differentiation, cell death, tumorigenesis and follicle expression.Citation13,Citation31 Previous reports have shown that CLSTN2 is expressed in the uterus of rats, followed by an increase in 17-β-estradiol during the estrous cycle.Citation14 In another study, the CLSTN2 gene was expressed in the ovary and fallopian tube of goat tissue.Citation13 The ovary produces follicles containing eggs, which has a significant impact on mammalian fertility. Therefore, gene expression in the ovary is likely to be related to litter size. Additionally, a genome-wide association study revealed that CLSTN2 may be a candidate gene associated with litter size in Pelibuey sheep.Citation15 And research has shown that the indel within the CLSTN2 gene is a candidate gene that affects goat fertility and can be used for marker assisted selection in goats.Citation13 In addition, the gene coordinates the secretion of follicle-stimulating hormone from the hypothalamus,Citation32 resulting in elevated estrogen levels, prolonged estrous cycle, and prolonged luteal phase, which inhibits ovulation and affects litter size.Citation14 In this study, we found that the rs399534524 locus in CLSTN2 contained three genotypes in five sheep breeds, and the PIC values showed a moderate level of polymorphism, suggesting that it can be used for selection in sheep breeding; moreover, the locus was not in Hardy-Weinberg equilibrium in four breeds except Cele Black sheep, which may be the result of natural and artificial selection. We also found that the rs399534524 locus in CLSTN2 differed significantly in genotype frequency and allele frequency between polytocous and monotocous breeds, implying that this locus is significantly associated with lambing traits. Furthermore, we determined that the rs399534524 locus in CLSTN2 is highly correlated with first parity litter size in Small Tail Han sheep, and the litter size of ewes with CT genotype is higher than that of ewes with CC or TT genotype in all parities. This is consistent with previous studies suggesting that it may affect reproductive traits in sheep. Most physiological processes are supported by multiple proteins; therefore, building networks of protein interactions can enhance our understanding of complex traits such as reproduction. We predicted the protein interaction network involving CLSTN2, which interacts with 10 proteins, among which HTR1E,Citation30 NOM1, CCDC174 and ALPK3 have been reported as candidate genes related to sheep litter size and may be involved in the reproductive process.Citation15 The HTR1E gene was enriched in the cAMP signaling pathway, where persistent cAMP signals from internalized LH receptors contribute to the transduction of LH effects within follicle cells and the oocyte.Citation15,Citation33 The NOM1 gene is a member of the MIF4G/MA3 family, which is involved in processes that affect translation.Citation34 It is also an essential eukaryotic serine/threonine phosphatase required for many cellular processes, including cell division, signal transduction, and metabolism.Citation35 The CCDC174 gene is essential for neuronal differentiation.Citation36 Expression levels of CCDC174 as a potential marker for assessing semen quality and fertility in bulls.Citation37 The ALPK3 gene has been implicated in a variety of cellular processes, including protein translation, Mg2+ homeostasis, intracellular trafficking, cell migration, adhesion and proliferation.Citation38 These genes have not been reported in ewe reproduction and lambing and require further investigation. Therefore, we believe that the rs399534524 locus in CLSTN2 can be used as a potential molecular marker for litter size traits in Small Tail Han sheep.
CTH is one of the major enzymes involved in the production of the gasotransmitter H2S in the body, using cystathionine and cysteine as substrates.Citation39 The production of free radicals such as H2S has been linked to the regulation of ovarian function, including ovulation. Research has found that the H2S generation system plays an important role in the propagation of preovulatory cascade reactions and follicular rupture.Citation17 As a vasodilator and angiogenesis molecule, H2S can regulate uterine vasodilation and stimulate placental angiogenesis, which affects the ovarian cycle and pregnancy.Citation40,Citation41 And endogenous H2S is one of the key factors in maintaining uterine quiescence during pregnancy.Citation42 Additionally, research has found that CTH can be expressed in ovarian granulosa cells and affect ovulation rate in mice.Citation17 Moreover, the H2S-releasing enzyme CTH is present in porcine oocytes and produces endogenous H2S, which is involved in the regulation of oocyte maturation and oocyte mound expansion.Citation18 The CTH gene affects animal reproduction, but its effect on litter size in sheep has not been studied. In this study, we found that the rs407142552 locus in CTH contained three genotypes in sheep breeds except Bamei Mutton sheep, which may be due to low polymorphism, and three genotypes may be detected if we increase the number of ewes tested. The PIC values of the remaining four breeds showed moderate polymorphism. In addition, the locus was in Hardy-Weinberg equilibrium in five breeds. We also found that the rs407142552 locus showed a significant difference in genotype frequency and allele frequency between polytocous and monotocous breeds, suggesting that this locus is significantly associated with lambing traits. The present study also determined the relationship between rs407142552 and litter size in Small Tail Han sheep. It is worth noting that our results indicate a significant correlation between the rs407142552 locus in CTH and second parity litter size in Small Tail Han sheep. The ewes with CT genotype were significantly higher than the ewes with TT genotype, and it is also similar in the first and third parity. The results suggest that the rs407142552 locus may influence litter size in sheep. In addition, a protein interaction network was predicted in which CTH interacts with 10 proteins. CBS is expressed in oocytes and granulosa cellsCitation18,Citation43 and interacts with CTH to produce H2S, which plays a role in the ovulation process.Citation17 AHCYL1 is a novel estrogen-stimulated gene expressed in chicken oviduct epithelial cells and may affect growth and development and calcium metabolism in mature hens oviducts through the estrogen-mediated ERK1/2 MAPK cell signaling pathway.Citation44 Choline and folate intake in healthy pregnant women may be associated with polymorphisms in MTHFR, BHMT.Citation45 However, these genes have been less studied in reproduction, especially sheep reproduction has not been reported and needs further research. Therefore, we think that the rs407142552 locus in CTH can be used as a potential molecular marker for litter size traits in Small Tail Han sheep.
Apoptosis has been shown to play a critical role in maintaining ovarian homeostasis in mammals.Citation7,Citation46 In mammalian ovaries, more than 99% of follicles undergo a degenerative process known as ‘atresia’, and only a few follicles ovulate during ovarian follicular development. Granulosa cell apoptosis plays a critical role in this process, follicular atresia.Citation47 APAF1 induces apoptosis by binding to cytochrome c and caspase-9, and the resulting complex accumulates in the cytoplasm where caspase-9 subsequently activates the caspase cascade leading to cell death.Citation7 In the ovary, APAF1 was found to be abundant in granulosa cells of early antral follicles and the APAF1 and caspase-9 mediated mitochondrial signaling pathway plays a critical role in determining granulosa cell fate in porcine ovarian atresia. PLCB1 and PLCB4 are members of the phospholipase C beta family.Citation25 PLCB1 has been implicated in ovarian function, particularly in the regulation of oocyte meiotic timing.Citation48,Citation49 Female mice with homozygous deletion mutations in the PLCB1 gene have altered reproductive behavior, ovulation, and pre-implantation embryo development and exhibit impaired implantation.Citation49–51 In addition, PLCB1 is a downstream effector of the GPCR signaling pathway, which plays a central role in mediating reproductive physiology; many reproductive hormones, particularly GnRH, FSH, LH, and oxytocin, are signaled by the GPCR. PLCB1 appears to play an important role in mediating GPCR signaling in reproductive physiology, as PLCB1 null mice are sterile and have pleiotropic reproductive defects. PLCB1 is essential for uterine implantation preparation and defects in PLCB1 mediated signaling during implantation are associated with aberrant ovarian steroid signaling and endogenous cannabinoid metabolism.Citation21 PLCB4 acts as a master regulator that modulates PLCB1 and plays a role in calcium signaling, which is essential for proper uterine quiescence and smooth muscle contraction during gestation. In addition, PLCB4 plays a role in the embryonic development of the internal and external structures of the face, and when development is impaired, disruption of development can lead to fetal death.Citation24 Notably, a meta-analysis of global genomic studies found that both PLCB1 and PLCB4 were associated with litter size in sheep.Citation25 CHST11 is a target gene for TGFβ-like growth factors (such as TGFβ, BMP2, and activin).Citation26 Study finds that CHST11 regulates proliferation and apoptosis in mouse embryo chondrocytes.Citation28 CHST11 mRNA levels increase when BMP4 interacts with the phosphorylated Smad3 binding site in the CHST11 promoter.Citation52 Furthermore, CHST11 has been associated with ovarian cancer, endometrial carcinomas and porcine farrowing interval.Citation53–55 These results suggest that CHST11 may regulate litter size in sheep through TGFβ and BMP signaling. We detected and analyzed SNPs and population genetic analysis, and found that the polymorphism information content of all sheep breeds ranged from 0 to 0.37, with most sheep breeds in Hardy-Weinberg equilibrium. In addition, the allele and genotype frequencies of the rs416244410 locus in APAF1 and the rs1092455701 locus in CHST11 differed significantly between polytocous and monotocous breeds. This suggests that SNP loci may be significantly associated with lambing traits. However, no significant correlation was found between these SNP loci and litter size in Small Tail Han sheep. Further investigation of the mechanisms is required.
Conclusions
The present study revealed that the allele and genotype frequencies of SNP for APAF1 (rs416244410), CLSTN2 (rs399534524), CTH (rs407142552), and CHST11 (rs1092455701) were significantly different between polytocous and monotocous breeds, and it was speculated that there might be a correlation between these loci and lambing traits. In addition, SNP rs399534524 in CLSTN2 and rs407142552 in CTH were significantly correlated with litter size in STH sheep, suggesting that SNPs rs399534524 and rs407142552 could be used as potential genetic markers in sheep breeding. Increased sample populations and additional function analyses of these two SNPs are needed to help verify our results.
Authors’ contributions
Kai Liu performed the experiments, analyzed the data, and wrote the first draft. Yufang Liu participated in tissue and serum collection and sample pretreatment. Mingxing Chu participated in the experimental design and revision of the manuscript.
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Zhang X, Yuan R, Bai Y, et al. A deletion mutation within the goat AKAP13 gene is significantly associated with litter size. Anim Biotechnol. 2023;34(2):1–11.
- Abd EL-Hack ME, Abdelnour SA, Swelum AA, et al. The application of gene marker-assisted selection and proteomics for the best meat quality criteria and body measurements in Qinchuan cattle breed. Mol Biol Rep. 2018;45(5):1445–1456.
- Akhatayeva Z, Bi Y, He Y, et al. Survey of the relationship between polymorphisms within the BMPR1B gene and sheep reproductive traits. Anim Biotechnol. 2023;34(3):718–727.
- Chu MX, Yang J, Feng T, et al. GDF9 as a candidate gene for prolificacy of Small Tail Han sheep. Mol Biol Rep. 2011;38(8):5199–5204.
- Calvo JH, Chantepie L, Serrano M, et al. A new allele in the BMP15 gene (FecX(RA)) that affects prolificacy co-segregates with FecX(R) and FecX(GR) in Rasa Aragonesa sheep. Theriogenology. 2020;144:107–111.
- Kaman T, Karasakal ÖF, Özkan Oktay E, et al. In silico approach to the analysis of SNPs in the human APAF1 gene. Turk J Biol. 2019;43(6):371–381.
- Hussein MR. Apoptosis in the ovary: molecular mechanisms. Hum Reprod Update. 2005;11(2):162–177.
- Deveraux QL, Reed JC. IAP family proteins–suppressors of apoptosis. Genes Dev. 1999;13(3):239–252.
- Manabe N, Goto Y, Matsuda-Minehata F, et al. Regulation mechanism of selective atresia in porcine follicles: regulation of granulosa cell apoptosis during atresia. J Reprod Dev. 2004;50(5):493–514.
- Adams HA, Sonstegard TS, Vanraden PM, et al. Identification of a nonsense mutation in APAF1 that is likely causal for a decrease in reproductive efficiency in Holstein dairy cattle. J Dairy Sci. 2016;99(8):6693–6701.
- Aravind P, Bulbule SR, Hemalatha N, et al. Elevation of gene expression of calcineurin, calmodulin and calsyntenin in oxidative stress induced PC12 cells. Genes Dis. 2021;8(1):87–93.
- Rindler MJ, Xu CF, Gumper I, et al. Calsyntenins are secretory granule proteins in anterior pituitary gland and pancreatic islet alpha cells. J Histochem Cytochem. 2008;56(4):381–388.
- Wijayanti D, Bai Y, Hanif Q, et al. Goat CLSTN2 gene: tissue expression profile, genetic variation, and its associations with litter size. Anim Biotechnol. 2022;34(7):2674–2683.
- Hong EJ, Park SH, Choi KC, et al. Identification of estrogen-regulated genes by microarray analysis of the uterus of immature rats exposed to endocrine disrupting chemicals. Reprod Biol Endocrinol. 2006;4(1):49.
- Hernández-Montiel W, Martínez-Núñez MA, Ramón-Ugalde JP, et al. Genome-wide association study reveals candidate genes for litter size traits in Pelibuey sheep. Animals. 2020;10(3):434.
- Jurkowska H, Kaczor-Kamińska M, Bronowicka-Adamska P, et al. Cystathionine γ-lyase. Postepy Hig Med Dosw. 2014;68:1–9.
- Estienne A, Portela VM, Choi Y, et al. The endogenous hydrogen sulfide generating system regulates ovulation. Free Radic Biol Med. 2019;138:43–52.
- Nevoral J, Žalmanová T, Zámostná K, et al. Endogenously produced hydrogen sulfide is involved in porcine oocyte maturation in vitro. Nitric Oxide. 2015;51:24–35.
- Wang K, Ahmad S, Cai M, et al. Dysregulation of hydrogen sulfide producing enzyme cystathionine γ-lyase contributes to maternal hypertension and placental abnormalities in preeclampsia. Circulation. 2013;127(25):2514–2522.
- Lu L, Kingdom J, Burton GJ, et al. Placental stem villus arterial remodeling associated with reduced hydrogen sulfide synthesis contributes to human fetal growth restriction. Am J Pathol. 2017;187(4):908–920.
- Filis P, Kind PC, Spears N. Implantation failure in mice with a disruption in phospholipase C beta 1 gene: lack of embryonic attachment, aberrant steroid hormone signalling and defective endocannabinoid metabolism. Mol Hum Reprod. 2013;19(5):290–301.
- Zorrilla M, Yatsenko AN. The genetics of infertility: current status of the field. Curr Genet Med Rep. 2013;1(4):247–260.
- Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80(4):1291–1335.
- Oliver KF, Wahl AM, Dick M, et al. Genomic analysis of spontaneous abortion in holstein heifers and primiparous cows. Genes (Basel). 2019;10(12):954.
- Gholizadeh M, Esmaeili-Fard SM. Meta-analysis of genome-wide association studies for litter size in sheep. Theriogenology. 2022;180:103–112.
- KLüPPEL M, Vallis KA, Wrana JL. A high-throughput induction gene trap approach defines C4ST as a target of BMP signaling. Mech Dev. 2002;118(1–2):77–89.
- Tao L, He X, Jiang Y, et al. Genome-wide analyses reveal genetic convergence of prolificacy between goats and sheep. Genes (Basel). 2021;12(4):480.
- KLüPPEL M, Wight TN, Chan C, et al. Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development. 2005;132(17):3989–4003.
- VON Mering C, Jensen LJ, Snel B, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433–7.
- Tao L, He XY, Wang FY, et al. Identification of genes associated with litter size combining genomic approaches in Luzhong mutton sheep. Anim Genet. 2021;52(4):545–549.
- Cai D, Wang Z, Zhou Z, et al. Integration of transcriptome sequencing and whole genome resequencing reveal candidate genes in egg production of upright and pendulous-comb chickens. Poult Sci. 2023;102(4):102504.
- Choi SG, Wang Q, Jia J, et al. Growth differentiation factor 9 (GDF9) forms an incoherent feed-forward loop modulating follicle-stimulating hormone β-subunit (FSHβ) gene expression. J Biol Chem. 2014;289(23):16164–16175.
- Lyga S, Volpe S, Werthmann RC, et al. Persistent cAMP signaling by internalized LH receptors in ovarian follicles. Endocrinology. 2016;157(4):1613–1621.
- Simmons HM, Ruis BL, Kapoor M, et al. Identification of NOM1, a nucleolar, eIF4A binding protein encoded within the chromosome 7q36 breakpoint region targeted in cases of pediatric acute myeloid leukemia. Gene. 2005;347(1):137–145.
- Gunawardena SR, Ruis BL, Meyer JA, et al. NOM1 targets protein phosphatase I to the nucleolus. J Biol Chem. 2008;283(1):398–404.
- Volodarsky M, Lichtig H, Leibson T, et al. CDC174, a novel component of the exon junction complex whose mutation underlies a syndrome of hypotonia and psychomotor developmental delay. Hum Mol Genet. 2015;24(22):6485–6491.
- Selvaraju S, Swathi D, Ramya L, et al. Orchestrating the expression levels of sperm mRNAs reveals CCDC174 as an important determinant of semen quality and bull fertility. Syst Biol Reprod Med. 2021;67(1):89–101.
- Middelbeek J, Clark K, Venselaar H, et al. The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell Mol Life Sci. 2010;67(6):875–890.
- Peleli M, Antoniadou I, Rodrigues-Junior DM, et al. Cystathionine gamma-lyase (CTH) inhibition attenuates glioblastoma formation. Redox Biol. 2023;64:102773.
- Lechuga TJ, Qi QR, Magness RR, et al. Ovine uterine artery hydrogen sulfide biosynthesis in vivo: effects of ovarian cycle and pregnancy. Biol Reprod. 2019;100(6):1630–1636.
- Chen DB, Feng L, Hodges JK, et al. Human trophoblast-derived hydrogen sulfide stimulates placental artery endothelial cell angiogenesis. Biol Reprod. 2017;97(3):478–489.
- You X, Chen Z, Zhao H, et al. Endogenous hydrogen sulfide contributes to uterine quiescence during pregnancy. Reproduction. 2017;153(5):535–543.
- Liang R, Yu WD, Du JB, et al. Localization of cystathionine beta synthase in mice ovaries and its expression profile during follicular development. Chin Med J (Engl). 2006;119(22):1877–1883.
- Jeong W, Kim J, Ahn SE, et al. AHCYL1 is mediated by estrogen-induced ERK1/2 MAPK cell signaling and microRNA regulation to effect functional aspects of the avian oviduct. PLoS One. 2012;7(11):e49204.
- Chmurzynska A, Seremak-Mrozikiewicz A, Malinowska AM, et al. Associations between folate and choline intake, homocysteine metabolism, and genetic polymorphism of MTHFR, BHMT and PEMT in healthy pregnant polish women. Nutr Diet. 2020;77(3):368–372.
- Manabe N, Matsuda-Minehata F, Goto Y, et al. Role of cell death ligand and receptor system on regulation of follicular atresia in pig ovaries. Reprod Domest Anim. 2008;43 Suppl 2(s2):268–272.
- Inoue N, Matsuda F, Goto Y, et al. Role of cell-death ligand-receptor system of granulosa cells in selective follicular atresia in porcine ovary. J Reprod Dev. 2011;57(2):169–175.
- Ledig S, RöPKE A, Wieacker P. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev. 2010;4(4-5):225–232.
- Filis P, Lannagan T, Thomson A, et al. Phospholipase C-beta1 signaling affects reproductive behavior, ovulation, and implantation. Endocrinology. 2009;150(7):3259–3266.
- Böhm D, Schwegler H, Kotthaus L, et al. Disruption of PLC-beta 1-mediated signal transduction in mutant mice causes age-dependent hippocampal mossy fiber sprouting and neurodegeneration. Molecular Cellular Neurosci. 2002;21(4):584–601.
- Ballester M, Molist J, Lopez-Bejar M, et al. Disruption of the mouse phospholipase C-beta1 gene in a beta-lactoglobulin transgenic line affects viability, growth, and fertility in mice. Gene. 2004;341:279–289.
- Bhattacharyya S, Feferman L, Tobacman JK. Regulation of chondroitin-4-sulfotransferase (CHST11) expression by opposing effects of arylsulfatase B on BMP4 and Wnt9A. Biochim Biophys Acta. 2015;1849(3):342–352.
- Oliveira-Ferrer L, Heßling A, Trillsch F, et al. Prognostic impact of chondroitin-4-sulfotransferase CHST11 in ovarian cancer. Tumour Biol. 2015;36(11):9023–9030.
- Farkas SA, Sorbe BG, Nilsson TK. Epigenetic changes as prognostic predictors in endometrial carcinomas. Epigenetics. 2017;12(1):19–26.
- Wu P, Wang K, Zhou J, et al. GWAS on imputed whole-genome resequencing from genotyping-by-sequencing data for farrowing interval of different parities in pigs. Front Genet. 2019;10:1012.