Abstract
Different antibiotics are used to treat mastitis in dairy cows that is caused by Escherichia coli (E. coli). Antimicrobial resistance in food-producing animals in China has been monitored since 2000. Surveillance data have shown that the prevalence of multiresistant E. coli in animals has increased significantly. This study aimed to investigate the occurrence and molecular characteristics of resistance determinants in E. coli strains (n = 105) obtained from lactating cows with clinical bovine mastitis (CBM) in China. A total of 220 cows with clinical mastitis, which has swollen mammary udder with reduced and red or gangrenous milk, were selected from 5000 cows. The results showed 94.3% of the isolates were recognized as multidrug resistant. The isolates (30.5%) were positive for the class I integrase gene along with seven gene cassettes that were accountable for resistance to trimethoprim resistance (dfrA17, dfr2d and dfrA1), aminoglycosides resistance (aadA1 and aadA5) and chloramphenicol resistance (catB3 and catB2), respectively. The blaTEM gene was present in all the isolates, and these carried the blaCTX gene. A double mutation in gyrA (i.e., Ser83Leu and Asp87Asn) was observed in all fluoroquinolone-resistant isolates. In total, nine fluoroquinolone-resistant E. coli isolates were identified with five different types of mutations in parC. In four (44.4%) isolates, Ser458Ala was present in parE, and in all nine (9/9) fluoroquinolone-resistant isolates, Pro385Ala was present in gyrB. Meanwhile, fluoroquinolone was observed as highly resistant, especially in isolates with gyrA and parC mutations. In summary, the findings of this research recognize the fluoroquinolone resistance mechanism and disclose integron prevalence and ESBLs in E. coli isolates from lactating cattle with CBM.
Introduction
In the twenty-first century, antibiotic resistance is a serious health issue and causing a huge problem for humans, animals and the environment and threatening life on the globe. The phenomenon occurs when pathogenic microbes are nonresponsive to the killing or inhibitory property of standard doses of antimicrobials. The cow and buffalo take a significant share of the antibiotic resistance for being reservoirs of the resistant strains. Different bacteria cause various bacterial diseases in animals such as mastitis. In China, dairy cows, buffaloes, goats and sheep play a crucial role in the economy of the country and also fulfil the requirements of the local herdsman. These animals are reared for the production of milk, meat, wool and hair. In China, many dairy cows and goat farms are available and produce millions of tons of milk annually. Due to the high production of milk, these animals are easily susceptible to viral, bacterial and parasitic infections which alter the health of the animals as well as reduce the milk production and alternatively affect the economy of the country. Dairy cattle are more susceptible to mastitis than other species such as buffalo, goat and sheep. Bovine mastitis (BM) is the main health problem in high-producing cows in dairy farms. The common use of antimicrobial agents in disease treatment can potentially lead to the development of antimicrobial-resistant bacteria.Citation1
BM is a prevalent disease with an occurrence rate between 23% and 40% in dairy cow farms.Citation2 BM causes many economic losses in most dairy farms, including treatment, discarded milk, labour, fatality costs, repeated cases of mastitis, decreased milk yield and milk quality changes.Citation3,Citation4 Numerous microorganisms associated with cases of BM have been isolated. The most frequently isolated pathogens that resulted in clinical BM (CBM) in China are Escherichia coli, Klebsiella spp., coagulase-negative staphylococci, Streptococcus dysgalactiae and Staphylococcus aureus.Citation5,Citation6 CBM caused by E. coli ranges from a moderate disease with a short duration to serious, per acute and life-threatening diseases. Appropriate antimicrobial therapy can effectively alleviate the symptoms and reduce the risk of complications of CBM. To date, some antimicrobial agents, i.e., beta-lactams, tetracyclines, macrolides, aminoglycosides and fluoroquinolones, have been commonly used to treat CBM in China.Citation7 Among these drugs, the most widely used antibiotics with convincing evidence of their effectiveness in treating E. coli mastitis are fluoroquinolones and cephalosporins, particularly 3rd and 4th generation drugs.Citation8 With the widespread use of antimicrobials, multidrug resistant E. coli isolates have emerged in many dairy farms.Citation1,Citation9
Integrons are found in the isolates of E. coli that are multidrug resistant from lactating cattle. As natural genetic elements, Integrons can capture, integrate and mobilize antibiotic-resistant gene cassettes. The majority of antibiotic-resistant genes in clinical isolates of Gram-positive bacteria are Class 1 integrons.Citation10,Citation11 There are already more than 130 gene cassettes known, and the encoded products would make bacteria resistant to practically all antibiotics.Citation12 According to previous research, class I integrons were prevalent in clinical E. coli isolates, as well as contributing to the development of antimicrobial resistance in BM isolates.Citation13 A latent pool of antimicrobial resistance genes exists in E. coli isolates from animals carrying class I integrons, while integrons can be found on plasmids or in transposons, facilitating the transfer of antibiotic resistance genes among bacteria.Citation14 In recent years, extended-spectrum ß-lactamases (ESBLs) have been increasingly detected among Gram-negative bacteria isolated from animals. They are mediated by the blaSHV, blaTEM and blaCTX-M genes in Gram-negative bacteria.Citation15 Based on studies from different hospitals, the CHINET national bacterial surveillance project found a 56.2% detection rate of ESBL-producing E. coli in 2009.Citation16 ESBLs and class 1 and class 2 integrons are regularly recovered from domestic livestock, demonstrating that animals can serve as reservoirs for resistance genes.Citation1
The antibiotic class known as fluoroquinolones is extremely effective and has several benefits, such as high oral absorption, wide volume of distribution and broad-spectrum antibacterial action,Citation17 and is frequently used for dealing with both Gram-negative and positive bacterial infections in clinics. Fluoroquinolones are considered a critically important antibiotic by the World Health Organization (WHO) due to their clinical importance in the field of human and animal medicine.Citation18 A significant public health risk arises from the consistent use of fluoroquinolones to treat livestock.Citation19 Fluoroquinolone resistance can be caused by various phenomena, of which numerous point mutations in the quinolone resistance-determining region (QRDR) of the DNA gyrase (gyrA and gyrB) and topoisomerase IV enzymes (parC and parE) are more important.Citation20 Chromosomal mutations in QRDRs resulted in amino acid replacement, structurally altering the target protein and later altering the binding affinity of the drug.Citation21 In addition, the mechanism of fluoroquinolone resistance also included plasmid-mediated quinolone resistance (PMQR). A major facilitator superfamily-type quinolone efflux pump qepA gene is an encoded gene, an aminoglycoside acetyltransferase, aac(6′)-Ib-cr gene, which confers reduced susceptibility to ciprofloxacin, and a QNR gene, which protects DNA gyrase and topoisomerase IV from quinolone inhabitation.Citation22 Although PMQR genes only provide moderate fluoroquinolone resistance, they can help select mutation in topoisomerase and gyrase genes, leading to greater levels of fluoroquinolone resistance.Citation23
Our study elucidate the molecular characteristics and prevalence of antibiotic resistance in E. coli strains from CBM in order to produce baseline data that will be used to assess future antimicrobial resistance risk. In addition, our study investigates why fluoroquinolone-resistant E. coli are found in CBM cases following over 20 years of fluoroquinolone use in Chinese veterinary medicine.
Materials and methods
Selection of clinical mastitis cases in dairy cows and sample collection
The methods and all the protocols used in this research were permitted by the related rules and regulations of the scientific research academic ethics committee, Inner Mongolia Agricultural University ([2020]086) that approved the study. All authors complied with the above guidelines and regulations. The clinical mastitis cases were selected in five dairy farming bases around Hohhot, China’s ‘milk capital’, of China. A total of 220 cows with clinical mastitis and swollen mammary glands with reduced and red or gangrenous milk were selected from 5000 cows. The owners of the five dairy farms agreed to carry out the study, and the milk of cows suffering from clinical mastitis was collected after obtaining the owner’s permission. Some milk samples were collected from cows with clinical mastitis at postpartum. The other samples were collected during the whole lactation. From the samples, there were 143 mild mastitis cases, 55 moderate mastitis cases and 22 severe mastitis cases. Mild mastitis cases (Grade 1): The breast is not abnormal on palpation, and the milk has visible changes, thinning of milk, somatic cell count of more than 500,000/mL and strong positive CMT test. Moderate mastitis cases (Grade 2): there are more serious pathological changes in the breast tissue, and the affected udders quarters are red, swollen, hot and painful. The milk has visible changes, turning grey or watery, with clots. However, the dairy cows with moderate mastitis have no systemic symptoms. Severe mastitis (Grade 3): there are more serious pathological changes in the breast tissue, and the affected udder quarters are red, swollen, hot and painful. The milk has visible changes, turning grey or watery. There are clots in the milk, and dairy cows with severe mastitis have obviously systemic symptoms. The cows’ udders and nipples were washed with warm water, and the nipples were sterilized with iodophor. An aliquot of 10 mL of milk was collected from each udder quarter of the dairy cows with clinical mastitis and stored in sterilized test tubes. Samples of milk were brought to the laboratory in ice and analysed within 12 h of collection. About 252 milk samples were collected between 2015 and 2020.
Bacterial isolation and confirmation
Eosin methylene blue agar was used and 10 µL of milk sample was streaked and incubated for 18–24 h at 37 °C. Presumptive E. coli colonies with the purple-dark green metallic luster were further confirmed with gram staining as per instruction of the manufacturer. Vitek system (bioMerieux) was used. Then, uida gene was identified by polymerase chain reaction (PCR) to confirm E. coli and uida gene is a specific gene of E. coli to confirm the Identification. PCR was used to further biochemically verify the E. coli isolate according to the protocol mentioned earlier.Citation1
Antibacterial susceptibility determination
A total of 22 antibiotic agents were bought from the CIVDC China and were used in current research. The categories of antimicrobials are as follows: aminoglycosides (kanamycin, amikacin, streptomycin and gentamycin), penicillins (ampicillin and amoxicillin), cephalosporins (ceftiofur, cefalotin and cefazolin), tetracyclines (oxytetracycline, doxycycline and oxytetrcycline), colistin, phenylpropanol (chloramphenicol and florfenicol), folic acid antagonists (sulphadiazine, sulphamethoxydiazine and trimethoprim), fluoroquinolones (norfloxacin, ciprofloxacin, lomefloxacin and enrofloxacin). Penicillins and cephalosporins both belong to β- Lactamides. Multidrug resistance (MDR) was defined as acquired nonsusceptibility to at least one agent in three or more antimicrobial categories.Citation24 According to the rules and regulations of the Clinical and Laboratory Standard Institutes 2015 performance Standards for susceptibility testing, the antibiotic drugs were dissolved and diluted.Citation25 Every day, new dilutions of all substances were made.
On a 96-well polystyrene plate with a ‘U’ type bottom, Minimal inhibitory concentrations (MICs) for E. coli isolates were calculated by the microdilution protocol 2015 clinical and laboratory standards were used to interpret the findings.Citation25 The quality control strain for bacterial isolation and drug susceptibility testing was standard strain ATCC25922.
Molecular epidemiological analysis of E. coli ESBLs genes and class I, II and III integrons
The ESBLs genes were detected by multiplex PCR. First, the DNA templates including genomic and plasmid DNA from E. coli isolates were prepared using Bacteric DNA Kit (Promega, Madison, WI) and MiniBest Plasmid Purification Kit. The major members of the β-lactamase gene family (i.e., the blaTEM, blaSHV and blaCTX genes) were amplified by multiplex PCR. PCR products were analyzed by electrophoresis on 1% agarose gels stained with ethidium bromide.Citation26
PCR was used to detect the integrons in all of the isolates. The PCR settings employed to find the gene cassettes and class I integrons have previously been disclosed. The PCR amplification consisted of initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 30 s, class II integron annealing at 52 °C for 30 s, class III integron annealing at 50 °C for 30 s and extension at 72 °C for 40 s, followed by a final extension at 72 °C for 5 min.Citation12 Through electrophoresis in 1.0% agarose gels, the amplification products were separated, and they were then visible in ultraviolet light. Primer sets covering integron-variable areas were used to create the amplifications, which were then gel-purified and cloned into the pGEM-T Easy vector (Promega) using conventional techniques. All reactions were repeated three times. To determine whether different isolates carried identical integrons, the amplicons of similar sizes were compared by restriction fragment length polymorphism (RFLP) analysis using the enzyme EcoRI and standard methods,Citation1 If the amplicons from two strains yielded the same RFLP pattern, the integrons were considered identical. If the PCR product contained a different RFLP pattern, the new product was also sequenced.
QRDR gene mutation analysis
The QRDRs of gyrA, parC, gyrB and parE genes were amplified from nine strains of fluoroquinolone-resistant E. coli by PCR. Using a Wizard Genomic DNA Purification Kit from Promega, genomic DNA was extracted from quinolone-resistant E. coli isolates. Employing previously defined PCR primers and amplification conditions, the QRDRs of the genes gyrA, parC, gyrB and parE were amplified. The PCR protocol was done as mentioned in our earlier study Zhao et al. (2014)Citation1 and primer sequences and PCR product sizes of genes for antimicrobial resistance are mentioned in . The amplification results have been isolated by electrophoresis on 1.0% agarose gel and seen in ultraviolet light.
Table 1. Primers sequences and PCR product sizes of genes for antimicrobial resistance.
PMQR genes detection by PCR
The availability of the PMQR genes qnrA, qnrS, qnrB, qnrC, qnrD, qepA and aac(6′)-Ib-cr was identified according to the previously mentioned.Citation27
Sequencing of the DNA and data analysis
Nucleotide sequences were sequenced with ABI PRISM Big Dye Primer Cycle Sequencing Ready Reaction Kit and ABI 377 DNA auto-sequencing machine by using T7 and SP6 sequence primers (Perkin-Elmer Company, Waltham, MA). For analysis of the data, the software DNASTAR (DNASTAR, Inc., St. Madison, WI) and the programme NCBI-BLAST (www. ncbi.nlm.nih.gov) were used. The nucleotide sequence data generated in this study have been submitted to GenBank under accession numbers X06373, AE000447, M58408, M58409, EF488368, EF488369 and EF488370.
Results
Antimicrobial susceptibility
In , the results of the antimicrobial susceptibility of 105 E. coli isolates with CBM to 22 antimicrobial agents are presented. As shown in , greater resistant incidence rates were shown for sulphamethoxydiazine (100%), sulphadiazine (99.0%), tetracycline (76.2%), oxytetracycline (71.4%), ampicillin (69.5%), cefazolin (62.9%) and amoxicillin (60.0%). However, most of the isolates were susceptible to fland amoxicilli. The isolates’ resistance rates of the different antibiotics such as ciprofloxacin, lemofloxicin, norfloxacin and norfloxacin were 28.6%, 27.6%, 27.6% and 27.6%, respectively. The high susceptibility rate of E. coli strains to ceftiofur was 89.5%, followed by colistin (74.3), amikacin (69.5), kanamycin (65.7%), streptomycin (63.8%), gentamicin (63.8%), chloramphenicol (55.2%) and florfenicol (54.3%).
Table 2. In vitro susceptibilities of 105 E. coli strains isolated from CBM to 22 antimicrobial agents.
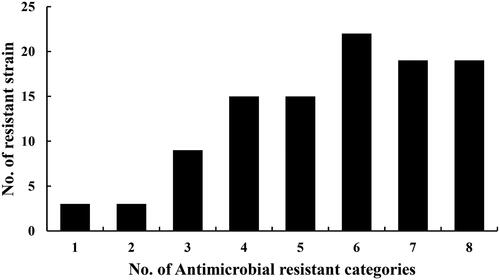
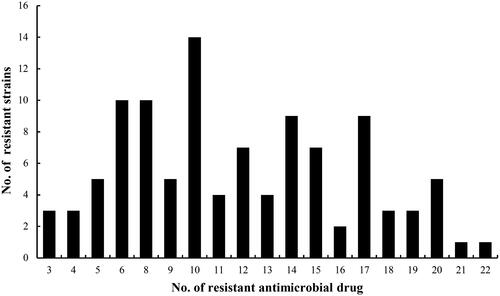
As shown in , the results showed 94.3% (99/105) of the isolates were recognized as multidrug-resistant and only 6 isolates were resistant to one or two antimicrobial categories. The resistant phenotypes of the different multidrug resistant isolates were different. But most of them were resistant to trimethoprim, sulphamethoxydiazine, sulphadiazine, tetracycline, oxytetracycline, ampicillin, amoxicillin and cefazolin. A total of six strains of E. coli were found to have MDR three or more antimicrobial categories when 105 isolates were tested against 22 antimicrobial drugs. Seventy-five isolates were resistant to more than five antibiotic categories of drugs mentioned earlier, accounting for 71.4%. From , there were 69 isolates resistant to more than 10 antibiotic drugs used in the experiments, and account for 65.7%. The results of susceptibility of antimicrobial displayed that those 15 isolates were simultaneously resistant to ciprofloxacin, enrofloxacin, norfloxacin and lomefloxacin, confirmed as fluoroquinolone-resistant E. coli isolates.
Class 1 integrons and ESBLs genes characterization
A total of 30.5% (32/105) of the isolates were noticed for the existence of intI1 (class I integrase gene), which was the indicator for class I integrons. None of the isolates harboured class II integrons and class III integrons. Thus, the integron-borne gene cassettes were cloned and sequenced. As shown in , the integrons contained one to three gene cassettes and the combinations of these gene cassettes.
Table 3. Antibiotic resistance phenotypes and resistance-determining regions in E. coli isolates from dairy cattle with CBM.
Five distinct kinds of gene cassette arrays were characterized. These were aadA1, dfr2d-catB3-aadA1, dfrA1-catB2-aadA1, dfrA1-aadA1 and dfrA17-aadA5, respectively. Of them, dfrA17–aadA5 (43.8%) was found to be the most prevalent gene cassettes among these isolates, followed by aadA1 (34.4%), dfr2d-catB3-aadA1 (21.9%), dfrA1-aadA1(15.6%), dfrA1-catB2-aadA1(12.5%). E. coli isolates (105) from CBM were nominated to determine ß-lactamase genes by PCR protocol. All of E. coli isolates were found to carry the blaTEM gene (100%), but not the blaSHV gene. There were five isolates carrying the blaCTX gene. The isolates that carried the blaTEM and blaCTX genes were resilient to penicillin, aminoglycosides, sulphonamides, trimethoprim, tetracyclines, chloramphenicol and fluoroquinolones.
Mutations in QRDR
The genes such as parC, parE, gyrA and gyrB, mutation were distinguished by sequencing in 15 fluoroquinolone-resistant E. coli isolates. As shown in , Ser83Leu, Asp87Asn, Ser83Leu and Asp87Asn substitutions were observed in GyrA. In parC, five dissimilar swaps were identified in all fluoroquinolone-resistant isolates. Among them, the most common point substitution in the strains was Ser58Ile (80%), followed by Arg138Val (33.3%), Asn141Lys (26.7%), Arg138Asp (13.3%) and Pro140Thr (13.3%). Six isolates were found to have one substitution in ParE (i.e., Ser458Ala), and all of the fluoroquinolone-resistant isolates had the substitution Pro385→Ala in GyrB. Then, the relationship between gene mutations and drug-resistance phenotypes in fluoroquinolone-resistant strains was analyzed. A certain correlation was revealed between the number of mutations mostly identified in the QRDRs region of the gyrA, parC and parE genes and the rising MICs of fluoroquinolones (). Four isolates with a ciprofloxacin MIC of 4–8 µg/mL had no one or one point mutation in the QRDRs region of the gyrA, and parC genes, such as the isolates #80, #82, #90 and #105. Two isolates (#1 and #40) had a ciprofloxacin MIC of 16 µg/mL, harbouring one- or two-point mutations in these genes. Five isolates (#27, #62, #69, #73 and #86) with a ciprofloxacin MIC of approximately 32 µg/mL had three and five mutations in the gyrA, parC and parE genes. All isolates with high-level ciprofloxacin resistance (≥64 µg/mL) carried five or six mutations in these genes. Therefore, there was a correlation between the number of mutations in QRDR and increased fluoroquinolone MICs. Fluoroquinolone resistance at a high degree was observed, especially in isolates with both gyrA and parC mutations. However, the gyrB gene’s QRDR mutation was present in all of the fluoroquinolone-resistant isolates.
Table 4. Fluoroquinolone susceptibility and substitutions in GyrA, GyrB, ParC and ParE QRDRs in E. coli isolates from dairy cattle with CBM.
Plasmid-mediated quinolone resistance
The 15 fluoroquinolone-resistant isolates from CBM were determined for the availability of PMQR mechanisms. No PMQR genes were found in these fluoroquinolone-resistant E. coli isolates from CBM.
Discussion
BM is a crucial problem for dairy farmers which affects the production performance of the dairy cows and also affects the health and economy.Citation29 The cornerstone of this disease’s treatment is antibiotic therapy, but its therapeutic efficacy is constrained by the rising prevalence of infections resistant to antibiotics.Citation30 In our research, most isolates were susceptible to amikacin, kanamycin, streptomycin, gentamicin, fluoroquinolones, ceftiofur and colistin. Instead, greater resistant incidence rates were unveiled for amoxicillin, ampicillin, cefazolin, oxytetracycline, tetracycline, sulphadiazine, sulphamethoxydiazine and trimethoprim, which may be derived from the various, long-term and widespread use of these antimicrobials in the studied dairy farms. About 65.7% of E. coli isolates from CBM were resistant to more than 10 antimicrobial drugs. It is consistent with the findings that E. coli isolates from animals have been resistant to many antimicrobial agents used in clinics.Citation30,Citation31 Fluoroquinolones are potent antimicrobial agents for the treatment of various infections caused by E. coli in veterinary medicine. Unfortunately, fluoroquinolone-resistant E. coli in food-producing animals has been increasing in China because of the extensive usage of quinolones.Citation32 We reported a similar occurrence (27.6%) of quinolone-resistant E. coli from BM cases, as compared with previous studies about dairy cattle suffering mastitis (22.9%) and endometritis (25.0%) in China.Citation1 However, our result was much higher than data from developed countries, where the reported prevalence of quinolone-resistant E. coli in BM cases was 0% for both Canada and the United States.Citation33,Citation34 It may indicate that the widespread use of this class of antibiotics in dairy cows has contributed to a greater prevalence of quinolone resistance. Because the frequent occurrence of resistant isolates from food-producing animals would seriously threaten public health through the food chain, Chinese governmental agencies have taken measures to supervise antibiotic usage critically. Moreover, the use of quinolones in food-producing animals such as dairy cows had already been banned in China in 2015.Citation35 Antimicrobial resistance in food-producing animals in China has been monitored since 2000. The resistance level of E. coli isolates from poultry to fluoroquinolones has increased by 50%.Citation36 Previous studies on the resistance of E. coli to fluoroquinolones have been mainly based on data from swine and poultry.Citation37 In 2008, Wang et al.Citation38 reported that most E .coli isolates from BM were susceptible to fluroquinolones (70.69–77.59%). The resistance rates of these isolates for enrofloxacin, norfloxacin, lomefloxacin and ciprofloxacin were about 30%. In this study, the resistance rates of E. coli isolates from CBM to ciprofloxacin, lemofloxicin, norfloxacin and norfloxacin were 28.6%, 27.6%, 27.6% and 27.6%, respectively. The above results showed that the resistance of E. coli to fluoroquinolones did not increase in cows with mastitis. Although fluoroquinolones have been used for the treatment of different diseases in bovine and previously its prevalence was recorded between 20% and 30% in the past decade. The resistance of E. coli isolated from BM to fluoroquinolones drugs has always kept at a low level, which is closely related to rigorous measures taken by Chinese governmental agencies that all antimicrobials had been banned for animal growth promotion.
Generally, there are three major sources of MDR in BM such as; virulence factors, biofilm and some efflux pump activities.Citation39 Some virulence factors such as haemolysins are responsible for the lysis of some immune cells of the host to invade and colonize into the host udder.Citation40 In addition to this, the biofilm formation in the pathogenic organism acts as a protective layer against host immune response and antibiotic therapy.Citation41 Finally, some efflux pump act as antibiotic resistance such as ESBLs, plasmid-mediated AmpC β-lactamases, carbapenemases and generalized efflux pump activity, but the RND-based tripartite efflux pump AcrAB-TolC is considered as most important factor for the antibacterial drug resistance.Citation42
Fluoroquinolone resistance in E. coli is mostly linked to specific mutations in the QRDRs of the gyrA and parC genes of DNA topoisomerase II (DNA gyrase) and topoisomerase IV.Citation43 These alterations may change how hydrogen bonds are formed, and it appears that interactions between quinolones and DNA are influenced by the amino acids’ negative charges complicate DNA-gyrase.Citation44 The results of the study showed that these amino-acid substitutions Ser83Leu and Asp87Asn in the QRDR of gyrA were detected in 60.0% of E. coli isolates from CBM in China, which were in line with previous studies that reported point mutations in quinolone-resistant E. coli occurred more frequently in S83L/D87N in gyrA.Citation45,Citation46 The results of the study conducted by BANSAL also demonstrated utmost substitutions related to quinolone resistance ensue in the QRDR at the serine (Ser83) and aspartate (Asp87) gyrA protein’s residues in E. coli isolates.Citation47 In our earlier research findings, we primarily noticed the substitutions in terms of amino acid positions Ser83Leu, Asp87Asn, Ser83Leu and Asp87Asn substitutions in gyrA, which are positioned within the QRDR.
In our research trial, in the QRDR of the parC protein different five amino-acid substitutions were found in 15 fluoroquinolone-resistant E. coli isolates from CBM, such as Arg138Asp, Ser58Ile, Pro140Thr, Arg138Val and Asn141Lys. The results indicated fluoroquinolone-resistance arises initially through mutations of gyrA, and additional mutation of parC leads to highly resistant E. coli isolates from cattle mastitis cases in China.
This was in line with past research findings that mutations targeting the DNA gyrase (topoisomerase II) were the main cause of fluoroquinolone resistance, with mutations in the topoisomerase IV causing higher levels of resistance.Citation48 As reported by Sandhya Bansal,Citation48 DNA gyrase (GyrA and GyrB) and topoisomerase IV (ParC and ParE) are the two essential type II topoisomerases. These enzymes act via inhibition of DNA replication. Alteration of a single amino acid at Ser-83 in GyrA is sufficient to generate decreased susceptibility to ciprofloxacin. Moreover, the accumulation of amino acid changes in GyrA and the simultaneous presence of ParC alterations contribute to high-level resistance to ciprofloxacin.
So, further, parC mutations are supposed to play a crucial role in highly resistant strain formation.Citation43 Sáenz et al.Citation49 previously stated that in GyrA a single amino acid exchange was deficient to confer a greater fluoroquinolone resistance phenotype in E. coli isolates. It was the same as our study’s conclusion that gyrA and parC mutations may contribute to the greater level of quinolone resistance in E. coli isolates from CBM in China. Moreover, in our study, all fluoroquinolone-resistant isolates with greater levels of ciprofloxacin resistance (≥32 µg/mL) carried double mutations in gyrA combined with one to three mutations in parC. These findings were in line with earlier findings that in the QRDR total point of mutations have been related to upsurge levels of fluoroquinolone-resistance.Citation22,Citation47
In this research trial, no PMQR determinants were noticed in all fluoroquinolone-resistant isolates. A similar report regarding PMQR genes had been published in a previous study.Citation1 However, the results of the study done by Yang et al. displayed that the dominant PMQR genes among fluoroquinolone-resistant strains were aac(6′)-Ib-cr (44.0%) and oqxA/B (24.0%). In contrast, qepA, qnrB and qnrS were detected with lower frequencies.Citation9 The results of this study revealed that, despite the growing interest in and reporting of PMQR mechanisms, the reduced susceptibility to quinolones in all E. coli isolates from CBM in China was caused by chromosomal mutations in the QRDR region rather than by plasmid-mediated quinolone resistance genes.
In this study, the prevalence of ESBLs in quinolone-resistant E. coli isolates was 100%. Our results showed that all quinolone-resistant E. coli isolates were recognized as β-lactamase producers. The predominant β-lactamase gene in the fluoroquinolone-resistant isolates was blaTEM, followed by blaCTX, and the blaSHV was not detected. Our results did not align with previous studies that confirmed blaCTX-M was the utmost common β-lactams in E. coli from dairy cattle mastitis.Citation50,Citation51 In this study, the positive-integron incidence rate in E. coli isolates from CBM is 30.50%, which is lower than the data for the Beijing area of China, which was reported as 56.9%.Citation38 Different studies reported that the ESBLs and integron prevalence are linked to various antibiotic pressures imposed by the intensive use of antibiotics in different environments. The results of this research showed that β-Lactamase genes blaTEM and blaCTX are frequently found together with integrons in fluoroquinolone-resistant isolates.Citation1,Citation29 The fact that ESBL genes could be acquired by strains harbouring particular integrons may increase the possibilities of selection of these isolates by a variety of different antimicrobials. Moreover, ESBL genes can be located on integrons, which may facilitate the spread of such genetic elements.Citation52
Conclusion
The results of the study showed a total of 220 cows with clinical mastitis were selected from 5000 cows. The incidence rate of CBM was 4.4% in current research. The isolates of E. coli from CBM were resistant to sulphonamides, tetracyclines, trimethoprim and penicillin, but most of them were susceptible to colistin, amikacin, ceftiofur and fluoroquinolones. Mutations in chromosomal genes encoding gyrase and topoisomerase IV are the main mechanisms responsible for high-level fluoroquinolone resistance and plasmid-mediated quinolone resistance (PMQR) have not been identified. Lastly, the findings of this study identified the prevalence of class I integrons and β-Lactamase genes in E. coli isolates from dairy cattle with CBM in different farms of the around Hohhot Inner Mongolia China. The isolates were positive for the class I integrase gene along with seven gene cassettes that were accountable for resistance to trimethoprim, aminoglycosides and chloramphenicol, respectively. The isolates that carried the blaTEM and blaCTX genes were resistant to penicillin, aminoglycosides, sulphonamides, trimethoprim, tetracyclines, chloramphenicol and fluoroquinolones. Since MDR and potential pathogenicity of E. coli is a serious problem among dairy cows in China, we predict that our findings will beneficial for relevant discussions to guide the urgent development of surveillance approaches to help control this phenomenon.
Ethics approval and consent to participate
The methods and all the protocols used in this research were permitted by the related rules and regulations of the scientific research academic ethics committee, Inner Mongolia Agricultural University ([2020]086) that approved the study.
Consent for publication
Not applicable
Authors contribution
Data curation, Chen Song and Weiguang Zhou; Formal analysis, Hailan Ma, Hongliang Fan and Jinshan Cao; Funding acquisition, Jinshan Cao; Investigation, Shuting Fan and Weiguang Zhou; Methodology, Hongxia Zhao; Software, Hailan Ma and Hongliang Fan; Visualization, Shuting Fan; Writing – original draft, Hongxia Zhao; Writing – review and editing, Chen Song.
Acknowledgement
The authors are grateful to the members of the Veterinary Pharmacology and Toxicology Laboratory of Inner Mongolia Agricultural University. The authors thank the Veterinary Pharmacology and Toxicology Laboratory of China Agricultural University for donating the standard strain of E. coli.
Disclosure statement
No conflict of interest in this article exists.
Data availability statement
Upon reasonable request, the corresponding author will provide all documents, data, and protocols that support the study’s findings. The links and accession numbers of the target genes are as follows: gyrA (https://www.ncbi.nlm.nih.gov/nuccore/X06373.1/;X06373), parC (https://www.ncbi.nlm.nih.gov/nuccore/M58408.1/;M58408), GyrB (https://www.ncbi.nlm.nih.gov/nuccore/AE000447.1/; AE000447), ParE (https://www.ncbi.nlm.nih.gov/nuccore/M58409.1/; M58409).
Additional information
Funding
References
- Zhao H-X, Zhao J-L, Shen J-Z, et al. Prevalence and molecular characterization of fluoroquinolone resistance in Escherichia coli isolates from dairy cattle with endometritis in China. Microb Drug Resist. 2014;20(2):1–13.
- Ruegg PL. A 100-year review: mastitis detection, management, and prevention. J Dairy Sci. 2017;100(12):10381–10397.
- Zhang Z, Li X, Yang F, et al. Influences of season, parity, lactation, udder area, milk yield, and clinical symptoms on intramammary infection in dairy cows. J Dairy Sci. 2016;99(8):6484–6493.
- Bobos S, Radinovic M, Vidic B, Pajic M, Vidic V, Galfi A. Mastitis therapy: direct and indirect costs. Bio Anim Husb. 2013;29(2):269–275.
- Krömker V, Leimbach S. Mastitis treatment—reduction in antibiotic usage in dairy cows. Reprod Domest Anim. 2017;52(S3):21–29.
- Gao J, Barkema HW, Zhang L, et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci. 2017;100(6):4797–4806.
- Gao X, Fan C, Zhang Z, et al. Enterococcal isolates from bovine subclinical and clinical mastitis: antimicrobial resistance and integron-gene cassette distribution. Microb Pathog. 2019;129:82–87.
- Poutrel B, Stegemann MR, Roy O, Pothier F, Tilt N, Payne-Johnson M. Evaluation of the efficacy of systemic danofloxacin in the treatment of induced acute Escherichia coli bovine mastitis. J Dairy Res. 2008;75(3):310–318.
- Yang F, Zhang S, Shang X, Wang L, Li H, Wang X. Characteristics of quinolone-resistant Escherichia coli isolated from bovine mastitis in China. J Dairy Sci. 2018;101(7):6244–6252.
- Betteridge T, Partridge SR, Iredell JR, Stokes H. Genetic context and structural diversity of class 1 integrons from human commensal bacteria in a hospital intensive care unit. Antimicrob Agents Chemother. 2011;55(8):3939–3943.
- Labbate M, Case RJ, Stokes HW. The integron/gene cassette system: an active player in bacterial adaptation. Horizontal Gene Transfer: Genomes in Flux. Totowa, NJ: Humana Press; 2 009:103–125.
- Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33(4):757–784.
- Hooper DC. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis. 2001;7(2):337–341.
- Onyango DM, Kakai R, Ny WE, Ghebremedhin B, Konig W, Kong B. Integron-plasmid mediated antibiotic resistance and virulence factors in clinical Salmonella enterica serovars in rural Western Kenya. Afr J Pharm Pharmacol. 2010;4(7):490–497.
- Copăcianu B, Tuchiluş C, Poiata A, Iancu LS. Research regarding extended-spectrum beta-lactamases produced by enterobacteria strains. Rev Med Chir Soc Med Nat Iasi. 2010;114(3):896–899.
- Wang F, Zhu D, Hu F, et al. CHINET surveillance of bacterial resistance in China. Chin J Infect Chemother. 2013;13(5):321–330.
- Patel K, Goldman JL. Safety concerns surrounding quinolone use in children. J Clin Pharmacol. 2016;56(9):1060–1075.
- WAGoISoAO, Resistance. Critically important antimicrobials for human medicine: Ranking of antimicrobial agents for risk management of antimicrobial resistance due to non-human use. Geneva, Switzerland: World Health Organization, 2017.
- Xu Y, Yu W, Ma Q, Zhou H. Occurrence of (fluoro) quinolones and (fluoro) quinolone resistance in soil receiving swine manure for 11 years. Sci Total Environ. 2015;530–531:191–197.
- Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(2):S120–S126.
- Vinué L, Lantero M, Sáenz Y, et al. Characterization of extended-spectrum β-lactamases and integrons in Escherichia coli isolates in a Spanish hospital. J Med Microbiol. 2008;57(Pt 7):916–920.
- Liu BT, Liao XP, Yang SS, et al. Detection of mutations in the gyrA and parC genes in Escherichia coli isolates carrying plasmid-mediated quinolone resistance genes from diseased food-producing animals. J Med Microbiol. 2012;61(Pt 11):1591–1599.
- Yang J, Luo Y, Li J, et al. Characterization of clinical Escherichia coli isolates from China containing transferable quinolone resistance determinants. J Antimicrob Chemother. 2010;65(3):453–459.
- Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281.
- Patel JB. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Berwyn, PA: Clinical and Laboratory Standards Institute; 2015.
- Colom K, Pérez J, Alonso R, Fernández-Aranguiz A, Lariño E, Cisterna R. Simple and reliable multiplex PCR assay for detection of bla TEM, bla SHV and bla OXA–1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223(2):147–151.
- Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of AAC (6C aIb-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006;50(11):3953–3955.
- Richter SN, Frasson I, Bergo C, Manganelli R, Cavallaro A, Palù G. Characterisation of qnr plasmid-mediated quinolone resistance in Enterobacteriaceae from Italy: association of the qnrB19 allele with the integron element ISCR1 in Escherichia coli. Int J Antimicrob Agents. 2010;35(6):578–583.
- Yang F, Liu L, Li X, et al. N-Acetylcysteine-mediated modulation of antibiotic susceptibility of bovine mastitis pathogens. J Dairy Sci. 2016;99(6):4300–4302.
- Dias R, Eller M, Duarte V, et al. Use of phages against antibiotic-resistant Staphylococcus aureus isolated from bovine mastitis. J Anim Sci. 2013;91(8):3930–3939.
- Begum YA, Talukder K, Azmi IJ, et al. Resistance pattern and molecular characterization of enterotoxigenic Escherichia coli (ETEC) strains isolated in Bangladesh. PLoS One 2016;11(7):e0157415.
- Xiao YH, Giske CG, Wei ZQ, Shen P, Heddini A, Li LJ. Epidemiology and characteristics of antimicrobial resistance in China. Drug Resist Updat. 2011;14(4–5):236–250.
- Saini V, McClure J, Léger D, et al. Antimicrobial resistance profiles of common mastitis pathogens on Canadian dairy farms. J Dairy Sci. 2012;95(8):4319–4332.
- Metzger S, Hogan J. Antimicrobial susceptibility and frequency of resistance genes in Escherichia coli isolated from bovine mastitis. J Dairy Sci. 2013;96(5):3044–3049.
- Wang X, Jiao Y, Wang G, et al. Occurrence of quinolones in cultured fish from Shandong Province, China and their health risk assessment. Mar Pollut Bull. 2022;180:113777.
- Zhao HX, Shen JZ, An XP, Fan HL, Cao JS, Li PF. Characterization of integrons in multiple antimicrobial resistant Escherichia coli isolates from bovine endometritis. Res Vet Sci. 2011;91(3):412–414.
- Wu CM, Wang Y, Cao XY, et al. Emergence of plasmid-mediated quinolone resistance genes in Enterobacteriaceae isolated from chickens in China. J Antimicrob Chemother. 2009;63(2):408–411.
- Wang GQ, Wu CM, Du XD, et al. Characterization of integrons-mediated antimicrobial resistance among Escherichia coli strains isolated from bovine mastitis. Vet Microbiol. 2008;127(1–2):73–78.
- Majumder S, Jung D, Ronholm J, George S. Prevalence and mechanisms of antibiotic resistance in Escherichia coli isolated from mastitic dairy cattle in Canada. BMC Microbiol. 2021;21(1):222.
- Divyakolu S, Chikkala R, Ratnakar KS, Sritharan V. Hemolysins of Staphylococcus aureus—An update on their biology, role in pathogenesis and as targets for anti-virulence therapy. AID. 2019;09(02):80–104.
- Paharik AE, Horswill AR. The staphylococcal biofilm: adhesins, regulation, and host response. Virulence Mechanisms of Bacterial Pathogens. Hoboken, NJ: Wiley; 2016:529–566.
- Awosile B, McClure J, Sanchez J, et al. Extended-spectrum cephalosporin-resistant Escherichia coli in colostrum from New Brunswick, Canada, dairy cows harbor blaCMY-2 and blaTEM resistance genes. J Dairy Sci. 2017;100(10):7901–7905.
- Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25(5):358–373.
- Sáenz Y, Zarazaga M, Briñas L, Ruiz-Larrea F, Torres C. Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J Antimicrob Chemother. 2003;51(4):1001–1005.
- Bébéar C, Renaudin H, Charron A, Clerc M, Pereyre S, Bébéar C. DNA gyrase and topoisomerase IV mutations in clinical isolates of Ureaplasma spp. and Mycoplasma hominis resistant to fluoroquinolones. Antimicrob Agents Chemother. 2003;47(10):3323–3325.
- Hawkey PM. Mechanisms of quinolone action and microbial response. J Antimicrob Chemother. 2003;51(90001):29–35.
- Hu YS, Shin S, Park YH, Park KT. Prevalence and mechanism of fluoroquinolone resistance in Escherichia coli isolated from swine feces in Korea. J Food Prot. 2017;80(7):1145–1151.
- Bansal S, Tandon V. Contribution of mutations in DNA gyrase and topoisomerase IV genes to ciprofloxacin resistance in Escherichia coli clinical isolates. Int J Antimicrob Agents. 2011;37(3):253–255.
- Sáenz Y, Ruiz J, Zarazaga M, Teixidó M, Torres C, Vila J. Effect of the efflux pump inhibitor Phe-Arg-β-naphthylamide on the MIC values of the quinolones, tetracycline and chloramphenicol, in Escherichia coli isolates of different origin. J Antimicrob Chemother. 2004;53(3):544–545.
- Ali T, Ur Rahman S, Zhang L, et al. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to IS CR1. Front Microbiol. 2016;7:1931.
- Tark D-S, Moon DC, Kang HY, et al. Antimicrobial susceptibility and characterization of extended-spectrum β-lactamases in Escherichia coli isolated from bovine mastitic milk in South Korea from 2012 to 2015. J Dairy Sci. 2017;100(5):3463–3469.
- Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48(1):1–14.