Abstract
Nucleic acid aptamers have been used in the past for the development of diagnostic methods against a number of targets such as bacteria, pesticides, cancer cells etc. In the present study, six rounds of Cell-SELEX were performed on a ssDNA aptamer library against X-enriched sperm cells from Sahiwal breed cattle. Sequencing was used to examine the aptamer sequences that shown affinity for sperm carrying the X chromosome in order to find any possible X-sperm-specific sequences. Out of 35 identified sequences, 14 were selected based on bioinformatics analysis like G-Score and Mfold structures. Further validation of their specificity was done via fluorescence microscopy. The interaction of biotinylated-aptamer with sperm was also determined by visualizing the binding of streptavidin coated magnetic beads on the head region of the sperm under bright field microscopy. Finally, a real-time experiment was designed for the validation of X-sperm enrichment by synthesized aptamer sequences. Among the studied sequences, aptamer 29a exhibited a higher affinity for X sperm compared to Y sperm in a mixed population of sperm cells. By using aptamer sequence 29a, we obtained an enrichment of 70% for X chromosome bearing sperm cells.
Introduction
In animal husbandry, gender selection is the deliberate attempt to manipulate the sex of offspring. Gender selection facilitates the efficient utilization of resources by livestock farmers and produces offspring with desirable traits for meeting ever-increasing market demand. Pre-sexed spermCitation1 or embryo-mediated livestock productionCitation2 offer a promising strategy to meet the rising demand for food production. Sperm sexing involves the separation of X and Y chromosome-bearing sperm cells, and for a long time, it has been a major area of interest among researchers and livestock industries.Citation3 It represents a promising breeding technique that allows for greater control over the gender distribution of offspring. Because it allows for targeted growth in the population of either female or male animals, depending on the need for meat and milk, this development is especially advantageous.Citation4 In India’s context, the dairy industry, even after achieving new heights in milk production, is still far from fulfilling the growing demand. In order to meet the demands of the dairy market, it is necessary to skew the female cattle population with the help of sexed semen straws.Citation5 Additionally, they play a vital role in the production of calves, ensuring the sustainability and growth of the industry. Traditional methods for obtaining sex-sorted semen have varying advantages and disadvantages.Citation6 Previous studies are generally based on the detection of sperm DNA contentCitation7,Citation8 or the assessment of certain sperm parameters like their motility,Citation9,Citation10 integrity,Citation11 volumetric differences,Citation12 centrifugal counter-current distribution,Citation13 and relevant immunological sperm characteristics.Citation14–16
However, none of the previously studied strategies have been able to achieve a statistically significant separation of viable sperm populations, nor have they been shown to be reproducible.Citation17 Flow cytometric sperm sorters, which separate spermatozoa with X and Y chromosomes based on DNA content, remain the sole well-established and commercially available approach.Citation7,Citation18–20 Nevertheless, challenges such as low sorting efficiency, low conception rates, high equipment and maintenance costs, and high operational skill persist with sperm sexing. Additionally, the cost of semen straws is also one of the discouraging factors for livestock farmers to opt for sexed semen straws. So, the dream of becoming self-sufficient in the field of sex-sorted semen remains unfulfilled. However, the recent advancements in scientific understanding and the availability of cutting-edge technology are encouraging enough to allow researchers to pursue alternate methods for separating sperm into the X and Y types. The identification of differentially and certain uniquely expressed proteins between X and Y sperm cells opened new dimensions for research in sperm sorting.Citation21,Citation22 Using this information, Umehara et al.Citation23 developed a ligand-based approach targeting the TLR7/8 receptor, which was uniquely expressed on X sperm cells. Several immunological approaches utilizing ScFvCitation24 or monoclonal antibodies against the uniquely expressed protein as targets are also being developed.Citation25,Citation26 However, antibodies are difficult as well as expensive to synthesize. In contrast to antibodies, aptamers present themselves as a more promising alternative.Citation27 Aptamers, composed of single-stranded oligonucleotides, have emerged as potentially useful tools for recognizing specific target molecules, including the whole cell.Citation28 Aptamers are easier as well as cheaper to synthesize and have a higher shelf life than antibodies.Citation29 The concept behind the development of aptamers is known as the Systematic Evolution of Ligands by Exponential Enrichment, also known as SELEX.Citation30,Citation31
Variations in cell surface characteristics between two cell types can facilitate their identification. Aptamers are composed of a vast variety of fixed-length nucleotides (n = 40–70) that can interact in several different ways with the various molecules found on the cell surface.Citation32 Our approach builds upon a similar concept, aiming to leverage differences between X and Y chromosome-bearing sperm using a sizable aptamer library. Nucleic acid aptamers are created by selecting them from a large random sequence pool by a process called SELEX.Citation33 SELEX is a combinatorial chemistry procedure that allows rapid selection, starting from a large initial library of oligonucleotides that have an appropriate binding affinity to a given molecular target.Citation34 These are typically composed of single-stranded DNA (ssDNA) or RNA.Citation35 Aptamers exhibit specificity and avidity comparable to or exceeding that of antibodies and can be generated against most targets. The application of aptamers in various fields, such as delivery, therapy, and diagnosis, has been demonstrated in previous studies.Citation36,Citation37 In this context, attempts have been made to generate aptamers with selectivity for X or Y spermatozoa in boar sperm,Citation38 but there is no report of their application. In 2020, Vinod et al.Citation39 generated and validated the aptamer against mixed sperm, but no sorting experiments were performed. At present, there is no information available on the feasibility of capitalizing on the capacity of aptamers for identification of bovine sperms that contain the X or Y chromosomes, respectively.
Material and methods
Cell-SELEX
Production and selection of aptamers
The aptamers used in this experiment were purchased from TriLink Biotechnology Ltd USA. In brief, a random library of DNA sequences, biotinylated, containing 86 nucleotides, 40 nucleotides of these random was used for selection. The remaining 46 nucleotide sequence known to have identifying primers involving 23 and 23 nucleotides at each end of the sequence. In the SELEX process, commercially sexed female sperm was used to obtain aptamers with particular affinity for X-chromosome (Female) spermatozoa.
Preparation of commercial bull semen
Briefly, five Sahiwal-sorted semen straws (∼2 Million sperm cells/straw), were thawed at 37 °C for 20 s and allowed to stabilize at room temperature (35–37 °C) for 5 min (all the following procedures were performed at this temperature). The semen from the straws was transferred to a 15-ml tube containing 4.5 ml of PBS. The semen was centrifuged at 500×g for 5 min after gentle stirring for homogenization, and the supernatant was discarded. The semen pellet was gently resuspended with 4.5 ml of PBS (1X) and centrifuged again for 5 min at 500×g. The supernatant was discarded, and the pellet was gently re-suspended in 1.2 ml of PBS (1X). A volume of 200 µl was removed for control group, and it was instantly frozen at −20 °C for performing DNA extraction and PCR later for enrichment in real-time by quantitation.
Incubation of spermatozoa with DNA library pool
A 330 μl aliquot of the binding buffer with 10 × 106 spermatozoa was used for binding with the oligonucleotide library (in a 370 μl binding buffer). After that, the mixture was incubated on a rotary shaker for one hour at room temperature. The binding buffer, composed of 1 liter of DPBS containing 4.5 g of glucose, 100 mg of tRNA, 1 g BSA and 5 ml of 1 M MgCl2 was stored at 4 °C for up to 1 month. Note: All the above reagents were obtained from M/S Sigma Aldrich, INC. (India)
Extraction of spermatozoa bound sequences
Following incubation, ssDNA aptamer bound sperm cells were centrifuged at 13,000 rpm for 5 min at 37 °C. The supernatant containing unbound sequences was removed and a second wash using 1 ml of washing buffer was given to the pellet. Washing buffer is composed of 1 liter of DPBS, containing 4.5 g of glucose and 5 ml of 1 M MgCl2 and was stored at 4 °C for up to 3 months. For the extraction of ssDNA aptamer, the pellet was suspended in nuclease free water for 10 min at 95 °C followed by centrifugation for 5 min at 13,000 rpm. The extracted ssDNA aptamers were then used for PCR amplification in consecutive SELEX rounds and optimization.
Aptamers amplification and optimization by polymerase chain reaction (PCR)
PCR amplification of the eluted aptamers pool was done using primers specific to the 5′ and 3′ ends of aptamers (). Mix the reaction mixer thoroughly and pipette 12.5 μl into 10 individual tubes (using 48-well thermal cycler) for 25 μl per reaction. The amplification conditions were optimized for the optimum cycle number, resulting in enough aptamers for the subsequent round of SELEX. The PCR amplification cycle consisted of 95 °C for 5 min, 18–22 cycles each of 95 °C for 30 s, temperature for annealing 50 °C for 30 s and 72 °C for 30 s with a final extension for 5 min at 72 °C. Following PCR, the amplicons were electrophoresed and the findings were reported in a 4% agarose gel in TBE with Safe View Classic dye (Himedia incorporation) at 80 V/cm.
Table 1. Primers designed for selected aptamer sequence.
We utilized the streptavidin/biotin method to convert double-stranded DNA (dsDNA) into single-stranded DNA (ssDNA). Initially, an empty DNA synthesis column (obtained from Glen research cat no. 20-0021-01) was prepared by placing a filter at one end of the column. Subsequently, the plunger was removed from a 10 ml syringe and inserted into the opposite end of the DNA synthesis column. This entire setup was then securely positioned on a clamp.
Next, a suspension of streptavidin-Sepharose beads (from GE healthcare) was poured into the syringe. The column was prepared by allowing the controlled passage of the storage buffer by gently inserting and pressing the plunger into the syringe. Subsequently, the PCR product obtained from previous SELEX round was guided through the column and eluate containing ssDNA was collected.
Cloning and sequencing of the selected aptamer
After the 6th round of SELEX, we cloned the aptamer in E. coli (TOP 10) cells by using Clone JET PCR Cloning Kit. The thirty-five (35) clones were sent for forwarding and backward sequencing to the First Base sequencing company.
Fluorescence microscopy analysis for aptamer-sperm interactions
From vast number of sequences, six aptamers were selected and synthesized (Integrated DNA Technologies, Inc. 1710 Commercial Park Coralville, Iowa 52241 USA). The synthesized aptamers were labeled with fluorescent markers (FAM) attached to their 5′ ends. Afterward, the aptamers were incubated for 1 hours with sperm to confirm their binding ability.
Bioinformatics analysis
The secondary structures were identified in the Mfold web server (http://unafold.rna.albany.edu/?q=mfold/DNA-Folding-Form). To determine their binding potential to bovine spermatozoa, the representative sequences from the last rounds of SELEX were synthesized with 5′ modification (FAM tag). The Mfold study was carried out at 28 ͦ C, NaCl 140 mM conc. Mg2+ conc. was in the range of 5 mM. The most stable structure was taken into further consideration.
Separation of aptamer bound sperm cells using magnetic beads
The semen was washed and diluted with 1.0 ml PBS in 2.0 ml tubes as stated above. In each replicate, 1.0 µM of aptamer was added. The biotinylated aptamers and mixed sperm cell population were incubated with constant gentle agitation for 30 min at room temperature. After this duration, sperm were incubated with 10 µl of magnetic beads conjugated to streptavidin for 30 min under gentle stirring at room temperature. After the incubation, the tubes containing the mixture of sperm cells, aptamer and streptavidin coated magnetic beads were placed on the magnetic rack (Sigma-Aldrich, Inc.) for 5 min. After placing in a magnetic rack, these sperm cells were pulled toward the wall of the tube. After that, 1.0 ml of the supernatant (also referred to as the unbound fraction) was carefully removed using a micropipette from the opposite side of the magnet. The volume remains in the tube’s bottom (∼ 200 µl), was retrieved and discarded. The aptamer bound sperm cells were retained due to interaction with streptavidin coated magnetic beads. This second fraction of sperm that was recovered was referred to as the retained fraction (also referred to as bound fraction) (refer to ). After being recovered, the sperm that had been retained at the tube wall were washed with 200 ml of PBS and placed to a new 1.5 ml tube. The sperm were obtained by centrifuging samples of the free fraction at 1500×g for 5 min and discarding 800 µl of the supernatant. They were re-suspended using the pipette, and the resulting volume was approximately 200 µl. All samples were immediately frozen at −20 °C following the separation procedures. After the sperm separation process was completed, the following sample groups were obtained: one fraction containing the free spermatozoa (also referred to as unbound fraction), one fraction of retained or bound sperm cells and the control sample.
Enrichment analysis for the sperm cells separated via aptamers-magnetic bead-based approach
Extraction and quantification of DNA
Semen straws were thawed gently at room temperature and DNA was extraction and purification was performed with the Genomic DNA Mini kit (Thermo Fisher Scientific Inc. Pure Link™ Genomic DNA Mini Kit Catalog number: K182002). The purified DNA was eluted in 50 µl of elution buffer (provided with the kit) and was quantitated by a spectrophotometer (Nano Drop 2000, Thermo Fisher Scientific Inc.). A 200 µl aliquots of each DNA sample was prepared with a concentration of 50 ng DNA/µl of ultrapure water. These aliquots were used for the quantification and for the real time PCR.
Primer designing
Primer pairs specific for the bovine X chromosome and chromosome Y were designed using the Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The X chromosome-specific primer pair was designed within the sequence deposited for MAOA Gene. The pair of primers specific for the Y chromosome was designed from a conserved region of the sex-determining gene Y (SRY, Sex-determining region Y).
The nucleotide sequences of the primers used are shown in .
Table 2. Gene name, sequence and product size for X and Y specific primers.
Quantitative real-time PCR of dsDNA
Quantitative RT-PCR was performed using an optimized protocol. A 96-well real-time PCR plate was taken and in each well, 10 μl reaction mixture was prepared by adding the sequentially the following components- 0.25 μl Forward primer, 0.25 μl reverse primer, 5 μl SYBR Green (cat no. K0221). Separate reaction mixtures were made for both types of primers- PLP and SRY. No template control (NTC) which contained no dsDNA template in the mixture was used as a negative control. An initial pre-incubation at 95 °C was carried out for 2 min, followed by 40 repeated cycles of denaturation for 15 s at 95 °C, annealing for 60 s at 60 °C and extension for 72 °C for 30 s.
Statistical analysis
A paired student’s t-test was used to compare the enrichment or either X or Y chromosome bearing sperm cell in either bound or unbound fractions after aptamer treatment. The enrichment was analyzed between the control group and aptamer treated group as well as between bound and unbound fractions. The difference was considered significant at p < 0.05.
Results
Selection of aptamers by cell SELEX
For the selection and amplification of the suitable aptamer sequences, the single-stranded aptamer library was incubated with a mixed population of both X and Y chromosome bearing sperm cells. In this experiment, 86 bp aptamers were used, showing that aptamers were amplified primarily in the positive incubation and not in the negative incubation (). Lanes 2 and 4 show the amplified library near to 100 bp instead of 86 bp due to the labeling of the primers. There was no nonspecific amplification as observed in lanes 3 and 5.
Figure 1. (a) Extraction and PCR based amplification of DNA aptamers by SELEX: Lane 1–5: 50 bp ladder (thermo scientific; cat#SM0371), amplified aptamers library, no template control (NTC), amplified aptamers library and NTC respectively; (b) PCR amplification product of eluted aptamers after each round of cell-SELEX.
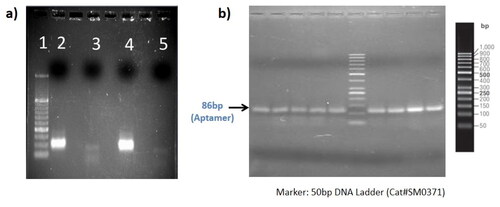
After the amplification, the aptamer library (10 nM) was incubated with X -sperm enriched semen sample. We performed Cell SELEX for 6 rounds and the results obtained are described in . After each round of SELEX, the band was obtained at 86 bp position, suggesting that only the desired sequence of aptamer has been selected, amplified and converted back to a single strand of the aptamer for the next round of SELEX.
Based on our experiments, it was observed that 18 cycles were most suitable for proper amplification without nonspecific amplicons. We used this cycle number for subsequent amplifications, to produce an ample amount of PCR products which can be used in the later SELEX rounds. The amplified aptamers were sequenced and the data was analyzed. Only the top 14 aptamers were selected for the further selection process.
In silico analysis for the selection of suitable aptamer sequences
ssDNA and RNA aptamers have G-rich sequences or motifs and have a tendency of forming G quadruplex structures which in turn are essential for their functioning. The G-score for the selected ssDNA aptamer was calculated using QGRS mapper, a web-based server. The G-score was calculated using random region of the aptamer sequence as input and the bases that were analyzed for each sequence are mentioned in . A G-score defines the quadruplex forming ability of the aptamer sequences. It is believed that a higher G-score has a good affinity for the target. Subsequently, based on the G-score, aptamer 29a was selected as the candidate aptamer sequence for further analysis and enrichment experiments.
Table 3. G Scores for the shortlisted aptamers.
Moreover, to characterize the interactions between the aptamer and the target protein, it is imperative to ascertain the secondary structures formed by the chosen aptamer sequences. The secondary structure analysis helps in identifying the nucleotides that are involved in loop formation, Watson-Crick pairing and stem formation. The 2D and 3D structures for 29a DNA aptamer were generated using Mfold. represents the selected 2D structure having the minimum Gibbs energy value (ΔG = −2.57).
Interaction analysis of FAM-labeled aptamers with sperm using fluorescence microscopy
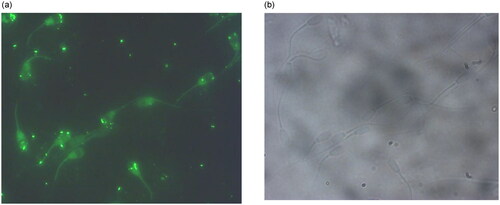
In order to determine the binding location of the aptamer 29a on spermatozoa, FAM labeled aptamers were incubated with the mixed sperm population and were visualized using fluorescence microscopy. The selected aptamer predominantly showed binding at the acrosomal membrane and post acrosomal membrane of sperm cells (), providing unambiguous proof that aptamers bind to the sperm cells in a specific manner.
Enrichment of X sperm population using aptamer 29a
The magnetic bead-based separation of X chromosome bearing sperm cells was carried out by treating a mixed sperm population with the X-chromosome specific aptamers (). The amount of enrichment achieved by this aptamer based approach was evaluated by using qRT-PCR of SRY and MAOA gene’s Ct values in bound and unbound fractions. After Comparing the Cycle threshold value of the unbound fraction to the bound fraction, we found that the concentration of X sperm in the unbound fraction decrease by more than 30%, indicating a more than 70% enrichment of X sperm in the bound fraction ().
Figure 4. A Schematic representation of (a) binding of selected aptamer to sperm cells through interaction with sperm surface proteins and (b) magnetic separation of aptamer treated sperm cells bound and unbound fractions after aptamer treatment.
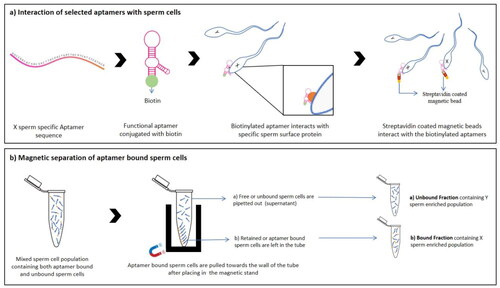
Figure 5. Real-time PCR analysis for determining the level of enrichment after treatment with X sperm specific aptamer 29a. # the difference between the two groups were significant at p < 0.05 and *** difference w.r.t control group were significant at p < 0.05.
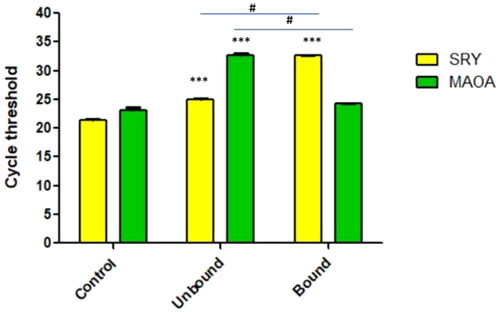
The level of enrichment was significantly different as compared to each other as well as control. The qRT-PCR data was analyzed using Graphpad prism software, followed student’s t-test statistical analysis for comparing the means of different fraction.
Discussion
India attained a milk production of 221.06 million tons in 2022 with the cattle population reaching 306.9 million heads as compared to 305.5 million in 2021. In recent years, the demand for milk and milk related products has increased exponentially, and this trend is presumed to continue in the future. One way to achieve the production goal is by increasing the number of female animals which can be accomplished through pre-selection of sex, enabling the selective birth of only female offspring. Male bulls produce semen carrying approximately 50% of X and 50% of Y chromosome bearing sperm cells. Over the years, researchers have identified several physical differences between X and Y chromosome bearing sperm cells, including differences in their size,Citation40 weight, density, velocity of swimming,Citation41 surface electric charges as well as differences in their surface proteins.Citation42,Citation43 (Dan et al., 2021) However, no evidence has yet been found suggesting that these differences can be used to efficiently separate the two types of sperm cells. FACS assisted separation of sperm cells based on the sex chromosome has the disadvantage of being expensive and cytotoxic to the sperm cells. In light of current challenges, there is a growing need for the development of innovative sorting methods that exhibit both simplicity and cost-effectiveness.Citation44
In a recent study conducted by Umehara et al.Citation23 a promising breakthrough involving the drug resiquimod (R848) has been achieved, which targets the X chromosome bearing sperm cells through their TLR7 and 8 receptors. This drug has demonstrated the ability to significantly impede the motility of X sperm, thereby facilitating their separation from Y sperm. Separation of X chromosome bearing sperm cells using the drug R848 has been achieved in many other organisms including mouse,Citation23 cattleCitation45 and goat.Citation46 In another study, magnetic beads with a negative charge were used in an attempt for developing an innovative approach for separating X and Y sperm cells.Citation47 In this approach, beads were added to a mixture of X and Y donkey sperm in order to separate them by using the difference in their zeta potentials. The negative zeta potential of Y sperm enabled effective interaction with the beads and isolation using a micro centrifuge. This approach offers you an opportunity to keep X sperm in suspension for use in artificial insemination (AI) procedures in the future.
Aptamers are short, single-stranded oligonucleotides that have the ability to bind to a group of target molecules.Citation48 Several studies have highlighted the importance of aptamers in diagnostics due to their higher specificity and affinity toward their cognate target.Citation49,Citation50 Aptamers are capable of binding to a wide range of target molecules, like proteins,Citation51–53 bacteria,Citation54,Citation55 whole cells,Citation56,Citation57 etc. The focus of this study was to identify aptamers selectively interacting with only the sperm cells carrying the X-chromosome. After multiple rounds of SELEX and sequencing, a subset of aptamers showing the highest affinity to X sperm cells were selected as candidate aptamer sequences for further validation. To validate the proper binding of aptamers to sperm cells, fluorescence microscopy was performed using FAM-labeled aptamers. The X-specific aptamer, named 29a, was used for the enrichment of X-chromosome-bearing sperm from the mixed population of sperm cells. The secondary and tertiary structures of the selected aptamer were determined using Mfold, and the structure having the lowest Gibbs energy was selected as the most stable structure. The selected aptamer sequence also has a G score of 58, suggesting a tendency to form G quadraplexes. In addition to this, docking as a computational method to predict the binding interactions between aptamers and the X and Y sperm-specific proteins can also be used, however, the accuracy of computational approach as a predictive tool is still dependent on experimental validation. Furthermore, the enrichment of X and Y sperms in different fractions was validated using qRT-PCR. The separation of X-chromosome-carrying sperms by using X-sperm-specific aptamers yielded good results, with the remaining free or unbound fraction carrying a higher proportion of Y-chromosome-bearing sperms with respect to unsexed or control sperm. Our experiments revealed an enrichment of X sperms by 70% after sorting using aptamer 29a. By using molecular biology and fluorescence tools, we found that selected aptamers bind to sperm strongly and specifically. Sperm sorting by flow cytometry is an effective method that is commercially available for cattle and other mammalian species. Despite its limitations and imperfections, this approach has about 90% accuracy in the separation of sperm for sex.Citation58 Through the above-described experiments, the separation of a single type of sperm using the selected aptamers is more practical, as depicted by the obtained results. The confirmation of an enrichment of one sex in a fraction after treatment with aptamers suggests that good results have been achieved and this technique can be used in the near future for semen sexing in cattle or buffalo. Through this study, a proof of concept (PoC) has been successfully established, demonstrating the feasibility of selecting aptamers that not only exhibit binding affinity toward X- sperm but also possess the potential for application in sperm sorting procedures. Moreover, an important advantage of Cell-SELEX is in its ability to identify a potential distinction between X and Y-sperm, even in the absence of prior knowledge regarding a specific target. While the technique is still in its developmental phase, it holds the potential to revolutionize sperm sorting for gender selection purposes.
Conclusion
Six rounds of aptamer selection through Cell SELEX were performed in order to validate the selection procedure and determine the ability of the selected aptamers to bind to X chromosome bearing sperm cells in a mixed population of sperm cells. In conclusion, the selection procedure used here is sufficient for the development of aptamers for enrichment of X-sperm cells.
Authors contributions
Sumit Kumar Singh: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft. Rohit Kumar: Data curation, Methodology, Writing – review & editing. Manya Mathur: Data curation, Software, Writing – original draft. Himanshu Kamboj: Data curation, Software, Writing – original draft. Jai Kaushik: Writing – review & editing. Ashok Mohanty: Conceptualization, Funding acquisition, Writing – review & editing. Sudarshan Kumar: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
Acknowledgements
We express our gratitude to ICAR for funding the project and National Dairy Research Institute, Karnal. We are grateful to members of Cell Biology and proteomics Lab for their support in carrying out the experiments for completing this work. Sumit Kumar Singh would like to thank Department of Biotechnology, Government of India for providing financial support from DBT-JRF program in Biotechnology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Brito L, Vishwanath R, Heuer C, Evans K. Bovine sexed semen production and utilization. Clin Theriogenol. 2019;11:297–315.
- Chowdhury MMR, Lianguang X, Kong R, et al. In vitro production of sex preselected cattle embryos using a monoclonal antibody raised against bull sperm epitopes. Anim Reprod Sci. 2019;205:156–164.
- Bhalakiya N, Haque N, Patel D, et al. Sperm sexing an its application in livestock sector. Int J Curr Microbiol Appl Sci. 2018;7:259–272.
- Boro P, Naha BC, Madkar A, Prakash C. Sexing of semen in bulls: a mini review. Int J Appl Res. 2016;2(4):460–462.
- Holden SA, Butler ST. Review: Applications and benefits of sexed semen in dairy and beef herds. Animal. 2018;12(s1):s97–s103.
- Rahman MS, Pang MG. New biological insights on X and Y chromosome-bearing spermatozoa. Front Cell Dev Biol. 2019;7:388.
- Johnson LA. Sex preselection by flow cytometric separation of X and Y chromosome-bearing sperm based on DNA difference: a review. Reprod Fertil Dev. 1995;7(4):893–903.
- Wang HX, Flaherty SP, Swann NJ, Matthews CD. Genetics: Discontinuous Percoll gradients enrich X- bearing human spermatozoa: a study using double-label fluorescence int-situ hybridization. Hum Reprod. 1994;9(7):1265–1270.
- Li J, Zhu S, He X, et al. Application of a microfluidic sperm sorter to in vitro production of dairy cattle sex-sorted embryos. Theriogenology. 2016;85(7):1211–1218.
- Sarkar S, Jolly DJ, Friedman T, Jones OW. Swimming behaviour of X and Y human sperm. Differentiation. 1984;27(2):120–125.
- Kaneko S, Iizuka R, Oshio S, Nakajima H, Oshio S, Mohri H. Separation of human X- and Y-bearing sperm using free-flow electrophoresis. Proc Jpn Acad, Ser B. 1983;59(8):276–279.
- Van Munster EB, Stap J, Hoebe RA, Te Meerman GJ, Aten JA. Difference in volume of X‐and Y‐chromosome‐bearing bovine sperm heads matches difference in DNA content. J Int Soc. 1999;35(2):125–128.
- Ollero M, Pérez‐pé R, Gargallo I, et al. Separation of ram spermatozoa bearing X and Y chromosome by centrifugal countercurrent distribution in an aqueous two‐phase system. J Androl. 2000;21(6):921–928.
- Hoppe PC, Koo GC. Reacting mouse sperm with monoclonal HY antibodies does not influence sex ratio of eggs fertilized in vitro. J Reprod Immunol. 1984;6(1):1–9.
- Howes EA, Miller NG, Dolby C, Hutchings A, Butcher GW, Jones R. A search for sex-specific antigens on bovine spermatozoa using immunological and biochemical techniques to compare the protein profiles of X and Y chromosome-bearing sperm populations separated by fluorescence-activated cell sorting. J Reprod Fertil. 1997;110(2):195–204.
- Yadav SK, Gangwar DK, Singh J, et al. An immunological approach of sperm sexing and different methods for identification of X- and Y-chromosome bearing sperm. Vet World. 2017;10(5):498–504. May
- Joshi H, Mathur M, Mohanty A, et al. Semen sexing in bovine: current status and the need to develop alternative techniques. ARU. 2021; 1(1):17–31. 3
- Garner DL. Flow cytometric sexing of mammalian sperm. Theriogenology. 2006;65(5):943–957.
- Garner DL, Evans KM, Seidel GE. Sex-sorting sperm using flow cytometry/cell sorting. In: Spermatogenesis. Totowa, NJ: Humana Press; 2013:279–295.
- Garner DL, Gledhill BL, Pinkel D, et al. Quantification of the X-and Y-chromosome-bearing spermatozoa of domestic animals by flow cytometry. Biol Reprod. 1983;28(2):312–321.
- Chen X, Zhu H, Wu C, et al. Identification of differentially expressed proteins between bull X and Y spermatozoa. J Proteomics. 2012;77:59–67.
- Shen D, Zhou C, Cao M, et al. Differential membrane protein profile in bovine X- and Y-sperm. J Proteome Res. 2021;20(6):3031–3042.
- Umehara T, Tsujita N, Shimada M. Activation of Toll-like receptor 7/8 encoded by the X chromosome alters sperm motility and provides a novel simple technology for sexing sperm. PLoS Biol. 2019;17(8):e3000398.
- Sringarm K, Thongkham M, Mekchay S, et al. High-efficiency bovine sperm sexing used magnetic-activated cell sorting by coupling scFv antibodies specific to Y-chromosome-bearing sperm on magnetic microbeads. Biology. 2022;11(5):715.
- Quelhas J, Santiago J, Matos B, Rocha A, Lopes G, Fardilha M. Bovine semen sexing: Sperm membrane proteomics as candidates for immunological selection of X- and Y-chromosome-bearing sperm. Vet Med Sci. 2021;7(5):1633–1641.
- Uhm SJ, Heo YT, Yu DM, Kim DK, Gupta MK. Pre-implantation development of cattle embryos produced from fresh bull semen enriched for X-chromosome-bearing spermatozoa using a monoclonal antibody. Vet Res Commun. 2023;47(4):2101–2109. Published online July 11,
- Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45(9):1628–1650.
- Mairal T, Ozalp VC, Lozano Sánchez P, Mir M, Katakis I, O'Sullivan CK. Aptamers: molecular tools for analytical applications. Anal Bioanal Chem. 2008;390(4):989–1007.
- Schlecht U, Malavé A, Gronewold T, Tewes M, Löhndorf M. Comparison of antibody and aptamer receptors for the specific detection of thrombin with a nanometer gap-sized impedance biosensor. Anal Chim Acta. 2006;573–574:65–68.
- Guo KT, Ziemer G, Paul A, Wendel HP. CELL-SELEX: novel perspectives of aptamer-based therapeutics. Int J Mol Sci. 2008;9(4):668–678.
- Ohuchi S. Cell-SELEX technology. Biores Open Access. 2012;1(6):265–272.
- Gooch J, Tungsirisurp S, Costanzo H, Napier R, Frascione N. Generating aptamers towards human sperm cells using massively parallel sequencing. Anal Bioanal Chem. 2021;413(23):5821–5834.
- Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5(6):1169–1185.
- Zhang Y, Hu Y, Deng S, et al. Engineering multivalence aptamer probes for amplified and label-free detection of antibiotics in aquatic products. J Agric Food Chem. 2020;68(8):2554–2561.
- Ulrich H, Magdesian MH, Alves MJ, Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J Biol Chem. 2002;277(23):20756–20762.
- Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19(1):60–71.
- Thiviyanathan V, Gorenstein DG. Aptamers and the next generation of diagnostic reagents. Proteomics Clin Appl. 2012;6(11–12):563–573.
- Colley AJ, Buhr M, Golovan SP. Development of sperm-specific aptamers to boar sperm. Ontario Swine Res Rev. 2007.
- Vinod SP, Vignesh R, Priyanka M, Tirumurugaan KG, Sivaselvam SN, Raj GD. Generation of single stranded DNA with selective affinity to bovine spermatozoa. Anim Biosci. 2021;34(10):1579–1589.
- Carvalho O, Silva R, Sartori R, Dode MAN. Nanoscale differences in the size and shape of X and Y chromosome-bearing bovine sperm heads assessed by atomic force microscopy. PLoS One. 2013;8(3):e59387.
- Check JH, Shanis BS, Cooper SO, Bollendorf A. Male sex preselection: swim-up technique and insemination of women after ovulation induction. Arch Androl. 1989;23(2):165–166.
- Chen X, Yue Y, He Y, et al. Identification and characterization of genes differentially expressed in X and Y sperm using suppression subtractive hybridization and cDNA microarray. Mol Reprod Dev. 2014;81(10):908–917.
- De Canio M, Soggiu A, Piras C, et al. Differential protein profile in sexed bovine semen: shotgun proteomics investigation. Mol Biosyst. 2014;10(6):1264–1271.
- Gaur P, Saini G, Saharan P, Bisla A, Yadav V. Sex sorted semen-methods, constraints and future perspective. Vet Res Int. 2020;8:368–375.
- Umehara T, Tsujita N, Zhu Z, Ikedo M, Shimada M. A simple sperm-sexing method that activates TLR7/8 on X sperm for the efficient production of sexed mouse or cattle embryos. Nat Protoc. 2020;15(8):2645–2667.
- Huang M, Cao XY, He QF, et al. Alkaline semen diluent combined with R848 for separation and enrichment of dairy goat X-sperm. J Dairy Sci. 2022;105(12):10020–10032.
- Domínguez E, Moreno-Irusta A, Castex HR, et al. Sperm sexing mediated by magnetic nanoparticles in donkeys, a preliminary in vitro study. J Equine Vet Sci. 2018;65:123–127.
- Tombelli S, Mascini M. Aptamers as molecular tools for bioanalytical methods. Curr Opin Mol Ther. 2009;11(2):179–188.
- Devi S, Sharma N, Ahmed T, et al. Aptamer-based diagnostic and therapeutic approaches in animals: current potential and challenges. Saudi J Biol Sci. 2021;28(9):5081–5093.
- Zhang Y, Chen Y, Han D, Ocsoy I, Tan W. Aptamers selected by cell-SELEX for application in cancer studies. Bioanalysis. 2010;2(5):907–918.
- Bayat P, Nosrati R, Alibolandi M, et al. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie. 2018;154:132–155.
- Chen J, Zhao Y, Feng W. Selection and characterization of DNA aptamers targeting hLCN6 protein for sperm capture. Appl Biochem Biotechnol. 2022;194(6):2565–2580.
- Shraim AS, Abdel Majeed BA, Al-Binni MA, Hunaiti A. Therapeutic potential of aptamer-protein interactions. ACS Pharmacol Transl Sci. 2022;5(12):1211–1227. Published 2022 Nov 4.
- Afrasiabi S, Pourhajibagher M, Raoofian R, Tabarzad M, Bahador A. Therapeutic applications of nucleic acid aptamers in microbial infections. J Biomed Sci. 2020;27(1):6.
- Kolm C, Cervenka I, Aschl UJ, et al. DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing. Sci Rep. 2020;10(1):20917.
- Cerchia L, Giangrande PH, McNamara JO, de Franciscis V. Cell-specific aptamers for targeted therapies. Methods Mol Biol. 2009;535:59–78.
- Shangguan D, Li Y, Tang Z, et al. From the cover: aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103(32):11838–11843.
- Garner DL, Jr, Seidel GE. Sexing bull sperm. In: Chenoweth PJ, ed. Topics in Bull Fertility. Ithaca: International Veterinary Information Service; 2000:10–12.