Abstract
The economic efficiency of sheep breeding, aiming to enhance productivity, is a focal point for improvement of sheep breeding. Recent studies highlight the involvement of the Early Region 2 Binding Factor transcription factor 8 (E2F8) gene in female reproduction. Our group’s recent genome-wide association study (GWAS) emphasizes the potential impact of the E2F8 gene on prolificacy traits in Australian White sheep (AUW). Herein, the purpose of this study was to assess the correlation of the E2F8 gene with litter size in AUW sheep breed. This work encompassed 659 AUW sheep, subject to genotyping through PCR-based genotyping technology. Furthermore, the results of PCR-based genotyping showed significant associations between the P1-del-32bp bp InDel and the fourth and fifth parities litter size in AUW sheep; the litter size of those with genotype ID were superior compared to those with DD and II genotypes. Thus, these results indicate that the P1-del-32bp InDel within the E2F8 gene can be useful in marker-assisted selection (MAS) in sheep.
Introduction
Sheep farming, deeply ingrained in global agricultural traditions, remains a vital practice with ancient roots. Originating in antiquity, the domestication of sheep has long been central to agricultural pursuits.Citation1 This enduring tradition encompasses the commercial cultivation of sheep for economic gain through the sale of meat, milk and wool, as well as personal use.Citation2 Notably, sheep farming is known for its profitability, owing to the resilient and low-maintenance nature of these animals, which primarily rely on nutrient-rich pasture fodder. This resilience not only contributes to the economic viability of sheep farming but also underscores its sustainability and efficiency. In response to rapid population growth, there is a growing need to modernize agricultural sectors, including livestock breeding, to ensure the production of high-quality products in significant quantities within optimal timeframes.
Hence, improving fertility and litter size is one of the main ways for profitable sheep farming, as underscored by breeders.Citation3 However, the conventional direct selection method has exhibited sluggish progress in enhancing litter size, primarily due to its inherent low heritability in ewes. In response, the adoption of marker-assisted selection (MAS), genomic sequencing and genomic association studies (genome-wide association study [GWAS]) is gaining prominence. These methodologies are increasingly applied to investigate genetic polymorphisms intricately linked to performance traits.Citation4,Citation5
Moreover, the predominant forms of mutations include insertions/deletions (InDels) and single-nucleotide substitutions (SNPs). Typically, InDels exert a more pronounced impact on a DNA sequence compared to single nucleotide substitutions. The latter are more inclined to influence the function within a specific region of the genome. Significantly, the existence of InDels holds significance in disease pathogenesis, leading to modifications in gene expression and protein functionality, as highlighted by Lin et al. and Akhatayeva et al..Citation6,Citation7
Recent developments within our team include the completion of a (GWAS), wherein various genes were analysed (data is not published yet). Notably, the early region 2 binding factor transcription factor 8 (E2F8) gene emerged as a key player, with predictions indicating its potential influence on sheep fertility traits. This intriguing finding has sparked a thorough investigation into the hypothesis, driving our team to delve deeper into the genetic intricacies that may contribute to sheep fertility characteristics. Moreover, to the best of our knowledge, there are no existing reports regarding the relationship between the E2F8 gene and the reproductive performance of sheep, highlighting the innovative and pioneering nature of our research. The E2F8 gene represents a vital component within the E2F family, serving as a foundational element in the transcriptional machinery. This machinery plays a critical role in orchestrating cellular processes such as the proliferation, cell cycle, differentiation, and apoptosis.Citation8 A recent investigation by Chen et al. has brought to light the potential significance of the E2F8 gene in gluconeogenesis and insulin resistance, positing it as a prospective therapeutic target for the prevention of type 2 diabetes mellitus (T2DM).Citation9 Within the mammalian context, glucose assumes a pivotal role as a primary energy source essential for both embryonic development and the metabolic needs of the gestating mother. Maintaining optimal glucose levels becomes imperative for sustaining foetal growth and meeting the maternal energy requirements during pregnancy. The perturbation of glucose metabolism, exemplified by conditions such as diabetes or insulin resistance, emerges as a factor with the potential to influence reproductive outcomes, including alterations in litter size. However, establishing a direct and unequivocal link between glucose metabolism and litter size proves intricate, given the intricate interplay of various determinants. Factors such as hormonal regulation, maternal health status, and genetic influences intricately contribute to the modulation of litter size. Therefore, a comprehensive understanding of the relationship between glucose metabolism and litter size necessitates meticulous consideration of the specific contextual factors inherent to each species and the individual health conditions of the subjects under scrutiny. In addition to this, the study conducted by Via Y Rada et al. has revealed the effects of glucose and insulin, which are like growth factors, on the development of embryos. In this study, they found that high levels of glucose stimulate the growth of blastocysts (early stage of embryos).Citation10The influence of E2F8, known for upregulating the expression of gluconeogenic genes and fostering glucose output in vitro, may impact embryonic development. This is supported by the observed phenomenon where elevated glucose concentrations fostered the growth of blastocysts, representing an early stage in embryonic development, as previously described.
Moreover, a recent study has shown that the E2F8 gene is clustered in biological processes in bovine. The terms associated with the E2F8 gene in bovine are linked to embryo development and utero embryonic development. Furthermore, this study has unveiled novel hormonal and developmental regulations of E2F8 mRNA during bovine follicular growth.Citation11Additionally, the latest study found that in female primordial germ cells of pigs, the E2F8 gene exhibited high expression levels during the later stages of development.Citation12In recent investigations, the E2F8 gene has been identified as a candidate gene for a quantitative trait locus (QTL) in pigs associated with mean platelet volume (MPV). MPV serves as a measure of the average size of platelets in the blood, commonly utilized as an indicator of platelet function and activity.Citation13 Platelets, small cell fragments, play a critical role in the immune and inflammatory responses to microbial organisms during infectious diseases.Citation14
Thus, the primary objective of our study is to investigate the potential impact of the E2F8 gene on the litter size of AUW sheep. Through a thorough investigation, this research endeavors to address an existing gap regarding the role of the E2F8 gene in sheep prolificacy. In summary, the revealed potential influence of the E2F8 gene on sheep litter size through a GWAS study, combined with its critical function and the absence of studies emphasizing its impact on sheep prolificacy traits, has motivated us to further explore its potential effects on AUW sheep litter size.
Materials and methods
Ethics statement
All experimental procedures were performed in accordance with the guidelines of the Faculty Animal Policy and Welfare Committee of Northwest A&F University (protocol no. NWAFU-314020038) for the use and care of animals in research.
Animals and phenotypic data
In exploring polymorphisms within the sheep E2F8 gene, a cohort of 659 AUW sheep spanning various parities was randomly selected from a population of AUW sheep in Tianjin province. This dataset was subsequently classified based on parity, enabling a more comprehensive analysis. From this group, litter size data were accessible for 496 out of the total 659 AUW ewes.
Genomic DNA extraction
Genomic DNA (gDNA) was obtained from ear tissues samples through the widely utilized phenol-chloroform method. The concentration and purity of the extracted DNA were meticulously evaluated using a NanoDrop 2000 spectrophotometer, a standard tool for DNA analysis. To maintain uniformity, each DNA sample was diluted to a standardized concentration of 20 ng/μL, following established protocols. For prolonged preservation, the samples were stored at −40 °C, a commonly employed temperature for ensuring DNA stability.Citation15,Citation16
Primer design and PCR - based genotyping
Utilizing the reference sequence NC_056074.1 from NCBI, primer sequences for the ovine E2F8 gene were designed through NCBI Primer Blast software, and predictions regarding insertions/deletions were extracted from Ensembl databases. Subsequently, five primer pairs (), synthesized by Sangon Biotech Company (Shanghai, China), were employed to identify novel InDel loci within the sheep E2F8 gene. A DNA pool, constructed from a random selection of 50 individuals, facilitated the detection of these InDel loci. Employing a touch-down PCR programme with a reaction volume of 13 μL, the amplification protocol comprised an initial denaturation step at 95 °C for 5 min, followed by 18 cycles of denaturation at 94 °C for 30 s, annealing at 68 °C for 30 s, and extension at a rate of 1000 bp/min at 72 °C. Subsequently, 30 cycles included denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 20 s, concluding with a final extension step at 72 °C for 10 min. Post-amplification, the reaction mixture was cooled to 4 °C. Subsequent to separation on a 3.5% agarose gel, the PCR products underwent sequencing analysis by Sangon Company (Shanghai, China).
Table 1. PCR primer sequences of the sheep E2F8 gene for amplification.
Statistical analysis
Analysis of genotypic frequencies, allelic frequencies, Hardy–Weinberg equilibrium (HWE), and genetic parameters pertaining to the identified InDel loci was undertaken utilizing the SHEsis platform, accessible at http://analysis.bio-x.cn and http://analysis.bio-x.cn/myAnalysis.php. The evaluation of the association between the polymorphic site and litter size employed a general linear model: Yijk = μ + Pi + Gj + eijk. In this model, Yijk represents the phenotypic value of litter size, μ signifies the overall population mean, Pi signifies the fixed effect of parity, Gj signifies the fixed effect of genotype, and eijk signifies the random error. The association analysis was executed through either the one-way ANOVA analysis utilizing SPSS software (version 25.0, IBM Corporation, Armonk, NY).Citation17
Results
InDel genotyping
A total of five primer pairs were designed from the NCBI database for the identification of mutation sites within the E2F8 gene (). Subsequent to analysis based on the DNA pool, only one (P1-del-32bp) out of the five InDels displayed polymorphism. The observed mutations manifested in three distinct genotypes: insertion (II), insertion/deletion (ID) and deletion/deletion (DD). Furthermore, At the P1-del-32bp site, the size of the amplified fragment for the II genotype was 391 bp, for the ID genotype it was 359/391 bp, and for the DD genotype it was 359 bp ().
Figure 1. The research design: the identification of the sheep E2F8 gene was achieved through a comprehensive genome-wide association study (GWAS). Subsequently, we conducted an exhaustive search across the ensemble and NCBI databases to explore potential genetic loci and designed five primer pares. This comprehensive approach led to the discovery of the P1-del-32bp InDel variant. Furthermore, this was followed by PCR analysis to confirm our findings. Agarose gel electrophoresis (3.5%) of allele specific polymerase chain reaction (PCR) product and sequencing map of the P1-del-32bp InDel variant within the E2F8 gene in AUW sheep. II: homozygous insertion genotype; DD: homozygous deletion genotype; ID: heterozygous insertion/deletion genotype. The sequence with the black border is the difference sequence. The top band in the electrophoresis pattern of ID represented heteroduplex.
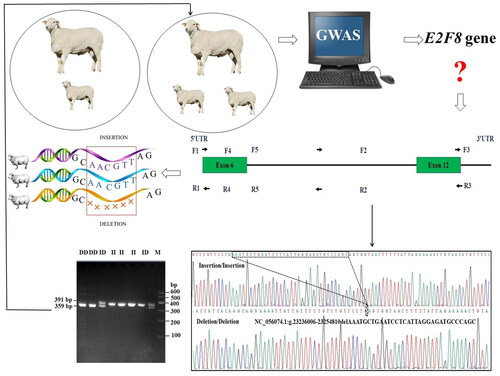
Genotypic and allelic frequencies
Genotype and allele frequencies, along with heterozygosity (He), homozygosity (Ho), effective allele numbers (Ne) and polymorphism information content (PIC), linked to the ovine E2F8 locus, were computed to ascertain the genotype distribution within AUW ewes. The results indicated a predominance of the II genotype (0.480) over the ID and DD genotypes in AUW sheep (). Notably, the ‘I’ allele exhibited a higher frequency than the ‘D’ allele in the P1-del-32bp variant of the E2F8 gene. The observed genotype distribution for the P1-del-32bp variant conformed to the HWE (p > 0.05). Furthermore, the assessment of the PIC value revealed a substantial level of variability in the AUW sheep population attributable to the InDels (PIC > 0.25).
Table 2. Genetic diversity parameters for E2F8-P1-del-32bp locus in AUW sheep population.
Association of the E2F8 gene with litter size in AUW sheep
We examined associations between P1-del-32bp, InDel locus and litter size in different parity groups of AUW ewes. Our findings showed that the P1-del-32bp variant within the E2F8 gene was significantly correlated to the litter size in the fourth (p = 0.011) and fifth parities (p = 1.2914E-9) in tested population. Furthermore, the individuals with genotype ID displayed higher litter sizes compared to those with insertion/insertion (II) and DD genotypes ().
Table 3. Associations of E2F8-P1-del-32bp with litter size in different parity groups in AUW ewes.
Discussion
Sheep stand as a species with multifaceted positive contributions to human life. The objectives of sheep husbandry have evolved significantly, encompassing diverse dimensions. In addition to this, sheep yield various advantageous products. In the realm of sheep breeding, the acquisition of wool, skins, meat and milk represents some of the healthiest and most sought-after options.Citation18 Undoubtedly, these commodities represent lucrative sources of income when traded over time. However, there are some enormous challenges to address, such as enhancing the economic efficiency of sheep breeding. Consequently, attention has shifted towards MAS breeding, offering advantages over traditional methods, including time efficiency and the ability to select animals based on their genotype.Citation3 In addition, researchers globally are actively engaged in exploring diverse genetic variants within candidate genes that influence litter size. A deep understanding of the variations in genes linked to sheep fecundity is essential for unraveling the complex molecular mechanisms that underlie their exceptional reproductive performance. The objective of these investigations is to identify molecular markers that can improve the effectiveness of MAS breeding practices in sheep.Citation19,Citation20,Citation21 Furthermore, as previously discussed, our team conducted a GWAS study, leading us to select the E2F8 candidate gene for further examination of its effect on sheep litter size. This gene stands out as a promising candidate for additional investigation and potential application in MAS within sheep breeding. Thus, our study is the first to investigate the association of the E2F8 gene with litter size in sheep, making it an interesting subject for future researches. To substantiate our hypothesis, we conducted an experiment employing PCR genotyping, unveiling a specific InDel variant, namely P1-del-32bp, within the E2F8 gene. Our findings demonstrated that polymorphism within the E2F8 gene had three genotypes: II, ID, and DD. Furthermore, association analyses demonstrated a significant correlation between the P1-del-32bp InDel and the fertility traits in the fourth and fifth parities (p < 0.05).
Besides, the significant effects of these polymorphisms can be explained by functions of the E2F8 gene. Interestingly, recent study by Morrell et al. has revealed, for the first time, that E2F8 in follicles undergoes developmental and hormonal regulation. This finding suggests that E2F8 gene plays a role in follicular development.Citation22
Theca cells (TC) are crucial endocrine components within the ovary. Among their functions are the stimulation of folliculogenesis and active participation in the ovulation process for individual follicles. Additionally, TC play a pivotal role in androgen synthesis.Citation23,Citation24 In a study conducted by Schütz et al., it was found that E2F8 gene is expressed in bovine TC. Furthermore, Morrell and colleagues propose that E2F8 stands out as a key candidate gene implicated in the proliferation of TC, playing a significant role in folliculogenesis.Citation25Notably, a study led by Putowski and colleagues indicates that within the ovary, certain E2F members have been recognized for their potential roles. Specifically, in human and rat granulosa cells, E2F1 overexpression has been linked to a decrease in FSH receptor transcription, while E2F5 overexpression has the opposite effect, increasing FSH receptor transcription. This suggests that E2F proteins, aside from E2F8, may play roles in follicular growth across different species.Citation26
Moreover, it has been indicated that E2F8 plays an important role in placental development.Citation27 Of interest, the placenta serves as a pivotal organ, integrating the physiological systems of both mother and foetus, therefore it is essential for reproduction.Citation28 Moreover, the placenta’s indispensable role in facilitating the transfer of oxygen and nutrients underscores its significance. Disruptions in foetal-placental circulation wield a profound impact on maternal-foetal exchange, thereby influencing foetal growth. Variations in the structure and functionality of the placenta can influence the reproductive output of a species.Citation29,Citation30 Therefore, the E2F8 gene, which has been recognized for its involvement in placenta development, is considered crucial. Investigating its associations provides valuable insights into the diverse adaptations of reproductive strategies across species.
Furthermore, the identified variant (P1-del-32 bp) is situated outside the coding region of the gene, thereby not inducing any amino acid alterations in the gene’s protein structure. Nonetheless, it is imperative to note that polymorphisms situated in non-coding regions such as introns, promoters, and regulatory regions are not inherently less functionally significant.Citation31,Citation32 These non-coding variations can contribute to the establishment or complete disruption of a site essential for the binding of a transcription factor, crucial for the regulation of a specific gene. They may also markedly impact the affinity of regulatory protein binding to their respective sites.Citation32
In summary, this study represents the first exploration into the influence of the InDel polymorphism within the E2F8 gene on sheep litter size. The revealed connections between this polymorphism and litter size, coupled with the functions of the E2F8 genes, offer valuable insights, laying the foundation for future sheep breeding strategies. Notably, this underscores the potential of MAS as a promising avenue for advancing breeding practices in the realm of sheep husbandry.
Conclusion
To conclude, according to the study findings, the P1-del-32bp variant within the sheep E2F8 gene displayed a significant association with litter size in AUW sheep. Remarkably, these outcomes align with the experimental findings from the GWAS study, which initially identified the E2F8 gene as a candidate gene for sheep fertility traits, thereby corroborating and expanding upon those earlier results.
Author contributions
Leijing Zhu and Nazar Akhmet: Contributed to formal analysis, software implementation, investigation, and original draft writing. Engaged in reviewing and editing the manuscript; Didi Bo: Participated in formal analysis, software development, and investigative efforts; Chuanying Pan: Oversaw project administration and took part in its conceptualization; Participated in formal analysis, software development, and investigative efforts; Jiyao Wu and Xianyong Lan: Exercised supervision over the project, engaging in conceptualization, validation, funding acquisition, project administration, and participated in reviewing and editing the manuscript.
Acknowledgements
We extend our heartfelt appreciation to Ph.D. Qingfeng Zhang and the dedicated staff of the Tianjin Aoqun Sheep Industry Academy Company for their invaluable contribution in providing both samples and comprehensive phenotypic traits data. Our gratitude further extends to all those involved in the meticulous care and management of the sheep, as well as the diligent collection of the essential samples. Moreover, we express our sincere thanks to the esteemed team at the Shaanxi Key Laboratory of Molecular Biology for Agriculture and The Life Science Research Core Services (LSRCS) of Northwest A&F University (Northern Campus). Their unwavering cooperation and support have played a pivotal role in the successful execution of our research endeavours. The collaborative efforts of these organizations and individuals have significantly enriched the scope and depth of our study, and for that, we are truly grateful.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Rocha J, Chen S, Beja-Pereira A. Molecular evidence for fat-tailed sheep domestication. Trop Anim Health Prod. 2011;43(7):1–8.
- Zhu J, Moawad AR, Wang CY, Li HF, Ren JY, Dai YF. Advances in in vitro production of sheep embryos. Int J Vet Sci Med. 2018;6:S15–S26.
- Yang Y, Hu H, Mao C, et al. Detection of the 23-bp nucleotide sequence mutation in retinoid acid receptor related orphan receptor alpha (RORA) gene and its effect on sheep litter size. Anim Biotechnol. 2022;33(1):70–78.
- Våge DI, Husdal M, Kent MP, Klemetsdal G, Boman IA. A missense mutation in growth differentiation factor 9 (GDF9) is strongly associated with litter size in sheep. BMC Genet. 2013;14(1):1.
- Demars J, Fabre S, Sarry J, et al. Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep. PLoS Genet. 2013;9(4):e1003482.
- Lin M, Whitmire S, Chen J, Farrel A, Shi X, Guo JT. Effects of short indels on protein structure and function in human genomes. Sci Rep. 2017;7(1):9313.
- Akhatayeva Z, Mao C, Jiang F, et al. Indel variants within the PRL and GHR genes associated with sheep litter size. Reprod Domest Anim. 2020;55(11):1470–1478.
- Lee DY, Chun JN, Cho M, So I, Jeon JH. Emerging role of E2F8 in human cancer. Biochim Biophys Acta Mol Basis Dis. 2023;1869(6):166745.
- Chen Y, Yu D, Wang L, Du S. Identification of E2F8 as a transcriptional regulator of gluconeogenesis in primary mouse hepatocytes. Biochemistry (Mosc). 2019;84(12):1529–1536.
- Via Y Rada R, Daniel N, Archilla C, et al. Identification of the inner cell mass and the trophectoderm responses after an in vitro exposure to glucose and insulin during the preimplantation period in the rabbit embryo. Cells. 2022;11(23):3766.
- Nazar M, Lu X, Abdalla IM, et al. Genome-wide association study candidate genes on mammary system-related teat-shape conformation traits in Chinese holstein cattle. Genes (Basel). 2021;12(12):2020.
- Chen M, Long X, Chen M, et al. Integration of single-cell transcriptome and chromatin accessibility of early gonads development among goats, pigs, macaques, and humans. Cell Rep. 2022;41(5):111587.
- Yang S, Ren J, Yan X, et al. Quantitative trait loci for porcine white blood cells and platelet-related traits in a White Duroc x Erhualian F resource population. Anim Genet. 2009;40(3):273–278.
- Allaoui A, Khawaja AA, Badad O, et al. Platelet function in viral immunity and SARS-CoV-2 infection. Semin Thromb Hemost. 2021;47(4):419–426.
- Akhmet N, Zhu L, Song J, et al. Exploring the sheep MAST4 gene variants and their associations with litter size. Animals (Basel). 2024;14(4):591.
- Davis GH, Balakrishnan L, Ross IK, et al. Investigation of the Booroola (FecB) and Inverdale (FecX(I)) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim Reprod Sci. 2006;92(1–2):87–96.
- Akhatayeva Z, Li H, Mao C, et al. Detecting novel Indel variants within the GHR gene and their associations with growth traits in Luxi Blackhead sheep. Anim Biotechnol. 2022;33(2):214–222.
- Tao L, Wang X, Zhong Y, et al. Combined approaches identify known and novel genes associated with sheep litter size and non-seasonal breeding. Anim Genet. 2021;52(6):857–867.
- Su P, Luo Y, Huang Y, et al. Short variation of the sheep PDGFD gene is correlated with litter size. Gene. 2022;844:146797.
- Abd El-Hack ME, Abdelnour SA, Swelum AA, Arif M. The application of gene marker-assisted selection and proteomics for the best meat quality criteria and body measurements in Qinchuan cattle breed. Mol Biol Rep. 2018;45(5):1445–1456.
- Cao C, Zhou Q, Kang Y, et al. A repertoire of single nucleotide polymorphisms (SNPs) of major fecundity BMPR1B gene among 75 sheep breeds worldwide. Theriogenology 2024;219:59–64.
- Morrell BC, Zhang L, Schütz LF, Perego MC, Maylem ERS, Spicer LJ. Regulation of the transcription factor E2F8 gene expression in bovine ovarian cells. Mol Cell Endocrinol. 2019;498:110572.
- Liu T, Qin QY, Qu JX, Wang HY, Yan J. Where are the theca cells from: the mechanism of theca cells derivation and differentiation. Chin Med J (Engl). 2020;133(14):1711–1718.
- Magoffin DA. Ovarian theca cell. Int J Biochem Cell Biol. 2005;37(7):1344–1349.
- Schütz LF, Hurst RE, Schreiber NB, Spicer LJ. Transcriptome profiling of bovine ovarian theca cells treated with fibroblast growth factor 9. Domest Anim Endocrinol. 2018;63:48–58.
- Putowski L, Gasior W, Gogacz M, Gagała J, Jakowicki JA. Zmiany w aktywności promotora genu receptora FSH człowieka i szczura pod wpływem czynników transkrypcyjnych E2F1, E2F4 i E2F5 [Differences in human and rat FSH receptors promote activity as a result of the transcriptional factors: E2F1, E2F4 and E2F5 overexpression]. Ginekol Pol. 2001;72(12A):1560–1566. in Polish)
- Mizuno M, Miki R, Moriyama Y, et al. The role of E2F8 in the human placenta. Mol Med Rep. 2019;19(1):293–301.
- Sheridan MA, Fernando RC, Gardner L, et al. Establishment and differentiation of long-term trophoblast organoid cultures from the human placenta. Nat Protoc. 2020;15(10):3441–3463.
- Natale BV, Gustin KN, Lee K, et al. Δ9-tetrahydrocannabinol exposure during rat pregnancy leads to symmetrical fetal growth restriction and labyrinth-specific vascular defects in the placenta. Sci Rep. 2020;10(1):544.
- Maltepe E, Fisher SJ. Placenta: the forgotten organ. Annu Rev Cell Dev Biol. 2015;31(1):523–552.
- Jo BS, Choi SS. Introns: the functional benefits of introns in genomes. Genomics Inform. 2015;13(4):112–118.
- Vitsios D, Dhindsa RS, Middleton L, Gussow AB, Petrovski S. Prioritizing non-coding regions based on human genomic constraint and sequence context with deep learning. Nat Commun. 2021;12(1):1504.
