 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Arylalkylamine-N-acetyl-transferase (AA-NAT) is one of several genes that influence sheep reproduction. Thus, the objective of this study was to investigate whether genetic variability within the AA-NAT gene influenced the reproductive performance of Awassi and Hamdani ewes. A total of 99 twin and 101 single-progeny ewes were analyzed for genomic DNA. Polymerase chain reaction (PCR) was used to produce amplicons of 300, 313, and 287 bp from exons 1, 2, and 3 of the AA-NAT gene. A 300-bp amplicon was genotyped, resulting in two genotypes: GG and GA. Through sequence analysis, a mutation 203 G > A was identified in the GA genotype. The statistical analysis revealed a strong correlation between the single nucleotide polymorphism (SNP) 203 G > A and reproductive performance. Ewes carrying this mutation showed significantly increased litter sizes, twinning rates, lambing rates, and fewer days to lambing compared to those carrying GG. These findings demonstrate that the presence of the 203 G > A SNP variant has a significant positive impact on litter sizes and enhances the fertility of Awassi and Hamdani sheep.
Introduction
Seasonal sheep reproduction is a key tool for developing breeding programs. Through genomic analysis, a variety of genes that influence seasonal reproductive traits in sheep have been successfully identified.Citation1 The Arylalkylamine-N-acetyl-transferase (AA-NAT) gene plays a role in seasonal sheep reproduction and reproductive traits.Citation1,Citation2 It is situated on chromosome 11 in sheep and comprises 4 exons (NCBI Reference Sequence NC_056064.1) and on chromosome 19 in bovines (Bos taurus).Citation3 This gene encodes an enzyme essential for melatonin biosynthesis, which regulates seasonal breeding in animals.Citation4 AA-NAT activity is directly related to the production of melatonin (MT). The enzyme AA-NAT synthesizes the melatonin hormone in the pineal gland. This hormone specifically binds to the MT1 and MT2 receptors in the hypothalamus and pituitary gland. The AA-NAT enzyme plays a critical role in animal reproductive systems because it is the rate-limiting enzyme in melatonin biosynthesis.Citation5 Melatonin, the primary messenger of photoperiodic information, influences the release of gonadotropin-releasing hormone (GnRH) through the reproductive axis.Citation6 MT can also lead to an improvement in the ovulation rate and litter size, enhance luteal function, and increase embryo viability.Citation7 Therefore, it has been suggested that genetic mutations in the AA-NAT gene could potentially influence the seasonal estrus response in the sheep population.
Genetic mutations are extensively used in genetics and livestock breeding due to their traceability, versatility, and utility.Citation3,Citation8 Numerous studies have demonstrated a significant correlation between livestock economic traits and SNPs in candidate genes.Citation3,Citation9 The EX3 486 A > G mutation in AA-NAT is associated with seasonality in Chinese sheep.Citation2 Mingxing et al.Citation10 documented the connections between the C265T polymorphism of the AA-NAT gene and litter size in high-prolific Jining Grey goats. Furthermore, novel SNPs C825T and C1249T in the AA-NAT gene are associated with reproductive traits in Indian goats.Citation11 Three SNPs in the AA-NAT gene, namely N-SNP1/g.55290169 T > C, N-SNP2/g.55289357 T > C, and N-SNP3/g.55289409 C > T, are significantly associated with milk traits in Holstein cows.Citation3 There has recently been an association between a c.486A > G mutation within the AA-NAT gene and both seasonal and non-seasonal reproduction in indigenous and exotic Iranian sheep populations.Citation4 According to these literature references, no variation has been found in the AA-NAT gene in the Awassi and Hamdani sheep breeds. In this context, the study investigated whether polymorphism in the AA-NAT gene impacts reproductive success in these sheep populations.
Material and methods
Animal population
The research was conducted between July 2022 and September 2023 and approved by the Al-Qasim Green University Animal Ethical Committee following international animal care and use guidelines (Agri, no. 01, 7, 22). This study examined 130 Awassi and 70 Hamdani ewes that were sexually mature and healthy. The ewes were aged between 3 and 4 years and weighed between 40 and 60 kg. The two breeds were genetically distinct and geographically isolated. Twin and singleton ewes were randomly selected from Babylon and Karbala stations after parturition and classified as 99 and 101, respectively. The animals were given concentrated feed equivalent to 2.5% of their body weight, consisting of 59% barley, 40% bran, and 1% salt. The animals were also given one kilogram of straw and three kilograms of alfalfa. The animals always had access to fresh water. The breeding stations monitored the rates of twinning, lambing, survival, and the time to lambing.
Genetic analysis
Genetic testing was conducted on sheep using blood samples taken from their jugular veins. Rapid salting-out of whole blood was used to isolate genomic DNA.Citation12 A total of 200 genetic sequences for each primer within the AA-NAT gene were amplified using NCBI Primer-BLAST.Citation13 The Eppendorf thermal gradient device from Germany was used to determine the optimal conditions for PCR amplification, as indicated in . The PCR amplification process was programmed as follows: pre-denaturation at 94 °C for 5 min, followed by a series of 30 cycles of denaturation at 94 °C for 30 s, annealing for 30 s, extension at 72 °C for 30 s, final extension at 72 °C for 5 min, and storage at 4 °C for 4 min.Citation14 An agarose gel electrophoresis was performed on PCR products (2%) and analyzed using a Chemidoc Gel Imager.Citation15
Table 1. The Oligonucleotide primer sets are designed for the amplification of the ovine AA-NAT gene.
The genotyping of each PCR product was performed as described by Mohammed et al.Citation16 The single-strand conformation polymorphism (SSCP) genotyping method involved combining 10 μL of each amplicon with SSCP loading dye (Bio-Rad, Hercules, CA, USA). The samples were denatured at 95 °C for 7 min and then rapidly cooled on wet ice for 10 min.Citation17 Afterward, they were promptly loaded onto acrylamide:bisacrylamide gels (37.5:1; 12%). The PCR products were electrophoresed at 200 mA and 100 V in 0.5x TBE buffer for 4 hours at 4 °C.Citation18 Then, the silver nitrate staining method was used to visualize DNA fragments.Citation19 The SSCP detection of polyacrylamide gel bands was followed by Sanger sequencing of all PCR products. Potential SNPs were identified using SnapGene Viewer and BioEdit. The novelty of the samples was assessed using the Ensemble Genome Browser 96. The impact of various genotypes on the secondary structure of the AA-NAT gene mRNA was examined using the RNA Folding Web Server, a powerful web-based program (http://rna.tbi.univie.ac.at/cgi-bin/rnawebsuite/ rnafold.cgi).
Data analysis
The genotypes and allele frequencies of the participants were determined using the PopGen32 program, version 1.31.Citation20 Following the Hardy–Weinberg equilibrium (HWE), the polymorphism information content (PIC) was calculated according to Botstein et al.Citation21 The association analysis of AA-NAT genotypes was conducted using IBM SPSS 23.0 (NY, USA) as follows:
where: Yijkl = traits phenotypically determined traits, μ = mean, Gi = fixed effect of ith genotype (i = GG, GA), Bj = fixed effect of the jth breed (j = 1, 2), Pk = fixed effect of kth parity (k = 1, 2, 3), and eijkl = random residual error. The Tukey-Kramer test revealed a significant difference at the 0.05 and 0.01 levels. An analysis of three reproductive traits was conducted using the Chi-square test: lambing rate, survival rate, and litter size. A logistic regression analysis was conducted to investigate the association between the AA-NAT polymorphism and litter size. The potential effects of factors such as lambing season, age, and factor interaction were evaluated. If they were found to be insignificant, they were excluded from the analysis.
Results
Genetic analysis
The coding regions of the AA-NAT gene were amplified, along with their flanking regions, resulting in three DNA fragments of 300 bp, 313 bp, and 287 bp (). The PCR products spanning exons 2 and 3 displayed a single distinct PCR-SSCP banding pattern. In contrast, exon 1 was analyzed using PCR-SSCP, revealing two distinct genotypic patterns (). Confirmation of the presence of the SNP 203 G > A was obtained through sequencing analysis of the SSCP variants, particularly the 300 bp amplicons. Genetic analysis revealed two variants of SSCP: GG and GA, as indicated by the homozygous G/G and heterozygous G/A patterns (). The RNA fold Web Server was used to predict the secondary structure of AA-NAT mRNA using highly reliable online prediction software. SNP 203 G > A mutations revealed modified secondary structures of the AA-NAT gene ().
Figure 1. An overview of the PCR-SSCP-sequencing technique applied to the analysis of the AA-NAT gene in Awassi and Hamdani ewes. (A) Primers were designed to amplify 300 bp, 313 bp, and 287 bp fragments in exon 1, exon 2, and exon 3, respectively. (B) PCR-SSCP genotyping revealed homozygous and heterozygous variations in exon 1. (C) Electropherograms of DNA sequencing were obtained for the GA genotype, which showed the SNP 203 G > a in exon 1. (D) Secondary structure prediction of different genotypes of mRNA in the AA-NAT gene.
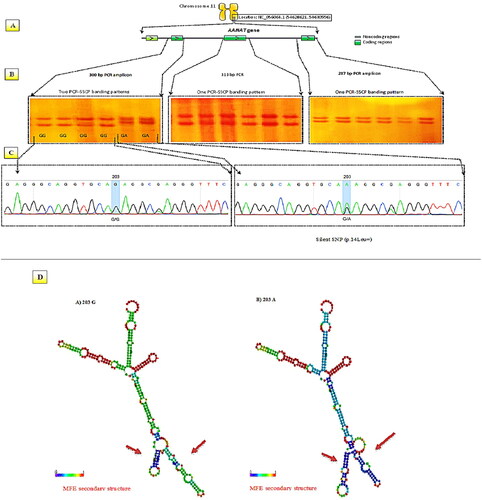
Regarding genetic diversity, the Hardy-Weinberg results for Awassi and Hamdani sheep in terms of AA-NAT genotypes and allele frequencies are shown in . Significant deviation from Hardy–Weinberg equilibrium was observed in both the Awassi and Hamdani populations, as demonstrated by chi-square values with significance levels of P ≤ 0.05. A moderate level of polymorphic information content was also observed within the ovine AA-NAT gene in both sheep breeds examined in the current analysis. A low level was classified by PIC values under 0.25, a medium level by 0.25 to 0.5, and a high level by values above 0.5.
Table 2. Genetic diversity of the AA-NAT gene in Awassi and Hamdani ewes detected by PCR-SSCP.
Association analysis
A statistical association analysis of the 203 G > A SNP in both breeds revealed no significant difference (P ≥ 0.01) in survival rates between individuals with GG and GA genotypes. At the same genetic locus, GA genotypes were significantly (P ≤ 0.01) associated with higher litter size, twinning rate, lambing rate, and fewer lambing days than GG genotypes. The GA genotypes resulted in 1.70 lambs per ewe in Awassi and 1.59 lambs per ewe in Hamdani, compared to the GG genotypes. As a result, SNP 203 G > A was positively associated with these traits ( and ).
Table 3. The association between AA-NAT genetic polymorphism and reproductive performance in Awassi and Hamdani ewes.
Table 4. Logistic regression analysis of AA-NAT genotype and litter size in Awassi and Hamdani ewes.
Discussion
Identifying gene polymorphism that are associated with economically beneficial traits has significantly contributed to genetic progress. This transformative practice has opened doors to substantial advancements in the field.Citation1,Citation22 Numerous studies have shown that the AA-NAT gene exhibits genetic variability in livestock. The EX3 486 A > G mutation in Chinese sheep has been associated with seasonal patterns,Citation2 while the C825T and C1249T mutations in Indian goats have been linked to reproductive characteristics.Citation11 The influence of the g.55290169 T > C, g.55289357 T > C, and g.55289409 C > T mutations on milk traits in Holstein cows is truly remarkable.Citation3 Furthermore, the c.486A > G mutation has been observed to affect both seasonal and non-seasonal reproduction in indigenous and exotic Iranian sheep populations.Citation4 These findings underscore the crucial role of the AA-NAT gene in shaping various aspects of livestock production. However, the literature on the variation of AA-NAT in Awassi and Hamdani sheep is extremely limited. In most Middle Eastern countries, Awassi and Hamdani sheep are prevalent. These breeds have been extensively involved in selection programs for milk, meat, or carpet wool production for several years.Citation23,Citation24 Each breed has distinct morphological characteristics and variations in nucleic acid sequences in response to external environmental challenges.Citation25,Citation26 While Awassi sheep are recognized for their adaptability,Citation27 the Hamdani breed is a special animal that thrives in its natural habitat, providing a plentiful source of meat, milk, and wool. Their reproductive rates, however, are lower than those of Karakuls and Assafs.Citation23,Citation26 As a result, this study fills a knowledge gap by reporting new associations and genotypic information. SNP bioinformatics analysis revealed that the silent mutation at position 14 (Leu) in exon 1 of the AA-NAT gene had a significant impact on litter size. Although silent mutations can result in proteins with the same amino acid sequence, they can still exhibit different functions and structures.Citation28 Synonymous mutations may not affect the exact meaning, but RNA splicing can affect protein production. Furthermore, modifying regulatory sequences can significantly impact gene expression, as highlighted by Li et al. in their study.Citation29 This scenario can directly impact the mRNA translation process into AA-NAT protein, thereby affecting the transmission of the hedgehog signal. Ahmed et al.Citation30 conducted a study that revealed how a silent mutation can significantly impact the regulation of post-translational gene expression, especially in its interaction with RNA-binding proteins. This interaction has the potential to impact several critical aspects of genome functionality. The influence of these factors is remarkably significant in various processes, including protein folding, RNA interaction with functional sites, alternative splicing, and the capacity of proteins to recognize RNA binding sites.Citation31
Based on statistical analysis, the GG genotype showed decreased litter sizes, lower twinning rates, reduced lambing rates, and longer lambing intervals compared to the GA genotype. Mutation in the AA-NAT gene at p.14Leu = is beneficial for reproduction. This can be attributed to the fact that the AA-NAT enzyme is responsible for the synthesis of melatonin, which causes the pituitary gland to release reproductive hormones.Citation4 Melatonin plays a crucial role in regulating the reproductive system. It acts directly influences the gonads to control the secretion of reproductive hormones and indirectly affects the anterior pituitary, thereby regulating the secretion of follicle-stimulating hormone and luteinizing hormone.Citation32 Melatonin receptors MT1 and MT2 have been identified in granulosa cells, cumulus cells, and oocytes, indicating that melatonin affects steroidogenesis, follicular development, oocyte maturation, ovulation, and luteinization through its receptor pathway.Citation33 The presence of melatonin receptors in cumulus-oocyte complexes (COCs) indicates that melatonin contributes to oocyte maturation in bovines.Citation34 Furthermore, melatonin regulates the reproductive system of seasonal estrous animals by promoting the estrous cycle, increasing ovulation, litter size, enhancing luteal function, and improving embryo viability in ewes.Citation10,Citation35 Melatonin improves oocyte quality by acting as an antioxidant, enhances luteal function, stimulates progesterone secretion, leads to improved pregnancy rates, and ultimately increases litter size.Citation36 Fathy et al.Citation37 conducted a study that presents compelling evidence of significant correlations between AA-NAT/SNP and litter size in Egyptian Ossimi sheep. These findings suggest that the AA-NAT gene could influence the reproductive performance of sheep breeds. Different genotypes are associated with litter size in Awassi and Hamdani sheep, which supports this conclusion. The Awassi and Hamdani breeds could also have low prolificacy due to the low prevalence of a silent SNP. Therefore, AA-NAT could affect sheep reproduction and potentially lead to a significant improvement in sheep fertility rates in the future.
Conclusions
A novel SNP was identified in the heterozygous GA genotype of the AA-NAT gene, 203 G > A. The GA genotype was associated with larger litter sizes, increased twinning rates, improved lambing rates, and fewer days to lambing compared to the GG genotype in ewes. Individuals with GA genotypes exhibited better reproductive traits compared to those with GG genotypes. This study indicates that ewes with the 203 G > A SNP have a higher litter size and are also more prolific.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bozhilova-Sakova M, Dimitrova I, Petrov N. Investigation of polymorphisms on ABCG2 and AA-NAT genes in different sheep breeds in Bulgaria. Bulg J Agric Sci. 2019;25(1):1–8.
- Ding-Ping B, Cheng-Jiang Y, Yu-Lin C. Association between AA-NAT gene polymorphism and reproductive performance in sheep. Electro Journal of Biotech. 2012;15(2):7–7.
- Yao S, Liu Y, Liu X, Liu G. Effects of SNPs in AANAT and ASMT Genes on Milk and Peripheral Blood Melatonin Concentrations in Holstein Cows (Bos taurus.) Genes. 2022;13(7):1196.
- Emanizadeh S, Alnajm H, Javanmard A, Shoja J, Rafat A. Allele frequency of c. 486A > G polymorphism of the AA-NAT gene in Iranian indigenous and exotic sheep populations. Iran J Appl Anim Sci. 2023;13(2):287–295.
- Abecia JA, Casao A, Canto F, et al. Polymorphisms of the melatonin receptor 1A (MTNR1A) gene do not affect sexual activity, plasma testosterone concentrations or testicular characteristics of 8-month-old ram-lambs born in autumn. JABB. 2022;10(4):1–6.
- Tsutsui K, Ubuka T. GnIH control of feeding and reproductive behaviors. Front Endocrinol (Lausanne). 2016;7:170.
- Song Y, Wu H, Wang X, et al. Melatonin improves the efficiency of super-ovulation and timed artificial insemination in sheep. Peer J. 2019;7:e6750.
- Ali MA, Kadhim AH, Al-Thuwaini TM. Genetic variants of the bone morphogenetic protein gene and its association with estrogen and progesterone levels with litter size in Awassi ewes. IJVS. 2022;36(4):1017–1022.
- Mamutse J, Purwantini D, Susanto A, Sodiq A. Determining the Polymorphism of V397I SNP of Growth Differentiation Factor 9 (GDF9) gene in Indonesian Saanen Goats. Adv Biol Sci Res. 2022;20:155–158.
- Mingxing C, Yan Y, Pingqing WU, et al. Polymorphism of AA-NAT gene and its relationship with litter size of Jining Grey goat of China. Anim Sci Paper Rep. 2013;31(1):15–26.
- Sharma R, Ahlawat S, Tantia MS. Novel polymorphism of AA-NAT gene in Indian goat breeds differing in reproductive traits. Iran J Vet Res. 2015;16(4):377–380.
- Al-Shuhaib MBSA. A universal, rapid, and inexpensive method for genomic DNA isolation from the whole blood of mammals and birds. J Genet. 2017;96(1):171–176.
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13(1):1–11.
- Al-Thuwaini TM, Kareem ZA. Novel missense variant L46Q of fatty acid synthase gene and fatty acids content in Awassi sheep. Acta Sci Anim Sci. 2022;44:e56273–e56273.
- Al-Thuwaini T. Association between polymorphism in BMP15 and GDF9 genes and impairing female fecundity in diabetes type 2. Middle East Fertil Soc J. 2020;25(1):1–10.
- Mohammed MM, Al-Thuwaini TM, Al-Shuhaib MBS. A novel p. K116Q SNP in the OLR1 gene and its relation to fecundity in Awassi ewes. Theriogenology. 2022;184:185–190.
- Alkhammas AH, Al-Thuwaini TM. Association of birth type and LHX4 gene polymorphism with reproductive hormones, growth hormone, and prolactin in Awassi ewes. Mol Biol Rep. 2023;50(4):3951–3956.
- Al-Shuhaib MBS, Al-Fihan RA, Al-Qutbi AA, Al-Thuwaini TM. Potential consequences of DGAT2 and BTN genes polymorphism in Iraqi Holstein cattle. Sci Agric Bohem. 2017;48(3):127–141.
- Byun SO, Fang Q, Zhou H, Hickford JGH. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal Biochem. 2009;385(1):174–175.
- Yeh FC, Yang RC, Boyle T. Microsoft Window-Based Freeware for Population Genetic Analysis (POPGENE). ver. 1.31. University of Alberta: Canada, 1999.
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32(3):314–331.
- Ajafar MH, Al-Thuwaini TM, Dakhel HH. Association of OLR1 gene polymorphism with live body weight and body morphometric traits in Awassi ewes. Mol Biol Rep. 2022;49(5):4149–4153.
- Al-Barzinji YM. Molecular analysis of FecGH gene in Hamdani sheep breed in Iraqi Kurdistan region. Iraqi J Agric Sci. 2022;53(1):1–1.
- Al-Thuwaini TM, Al-Hadi ABA. Association of lamb sex with body measurements in single and twin on the Awassi ewes. AAVS. 2022;10(8):1849–1853.
- Bingöl E, Bingöl M. Some growth, reproduction and lactation characteristics of hamdani sheep. Yuzuncu Yil Univ J Agric Sci. 2018;28(2):161–167.
- Ajafar MH, Kadhim AH, Al-Thuwaini TM. The reproductive traits of sheep and their influencing factors. Rev Agric Sci. 2022;10:82–89.
- Al-Thuwaini TM. The relationship of hematological parameters with adaptation and reproduction in sheep; A review study. IJVS. 2021;35(3):575–580.
- Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67(20):9609–9612.
- Li D, McIntosh CS, Mastaglia FL, Wilton SD, Aung-Htut MT. Neurodegenerative diseases: a hotbed for splicing defects and the potential therapies. Transl Neurodegener. 2021;10(1):16.
- Ahmed RO, Bello SF, Shu’aibu I, Hegarty MJ. An investigation of polymorphism in SMO and LMF1 genes and their association with body size in white Fulani and Muturu cattle breeds. ABB. 2020;11(07):319–344.
- Li MJ, Yan B, Sham PC, Wang J. Exploring the function of genetic variants in the non-coding genomic regions: approaches for identifying human regulatory variants affecting gene expression. Brief Bioinform. 2015;16(3):393–412.
- Hu W, Tang J, Zhang Z, et al. Polymorphisms in the ASMT and ADAMTS1 gene may increase litter size in goats. Vet Med Sci. 2020;6(4):775–787.
- Xiao L, Hu J, Song L, et al. Profile of melatonin and its receptors and synthesizing enzymes in cumulus–oocyte complexes of the developing sheep antral follicle—a potential estradiol-mediated mechanism. Reprod Biol Endocrinol. 2019;17(1):1–9.
- El-Raey M, Geshi M, Somfai T, et al. Evidence of melatonin synthesis in the cumulus oocyte complexes and its role in enhancing oocyte maturation in vitro in cattle. Mol Reprod Dev. 2011;78(4):250–262.
- Younis LS, Hatif SA. AA-NAT melatonin gene polymorphism in ewes lambing out of season. Onl J Vet Res. 2017;21(3):118–125.
- Wu H, Ma W, Yan L, et al. Investigation of SNP markers for the melatonin production trait in the Hu sheep with bulked segregant analysis. BMC Genom. 2023;24(1):502.
- Fathy HA, Gouda EM, Gafer JA, Galal MK, Nowier AM. Genetic polymorphism in melatonin receptor 1A and arylalkylamine N-acetyltransferase and its impact on seasonal reproduction in Egyptian sheep breeds. Arch Anim Breed. 2018;61(4):505–516.
