?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Increasing the number of teats in sheep helps to improve the survival rate of sheep lambs after birth. In order to analyze the candidate genes related to the formation of multiple teats in Hu sheep, the present study was conducted to investigate the genetic pattern of multiple teats in Hu sheep. In this study, based on genome-wide data from 157 Hu sheep, Fst, xp-EHH, Pi and iHS signaling were performed, and the top 5% signal regions of each analyzed result were annotated based on the Oar_v4.0 for sheep. The results show that a total of 142 SNP loci were selected. We found that PTPRG, TMEM117 and LRP1B genes were closely associated with polypodium formation in Hu sheep, in addition, among the candidate genes related to polypodium we found genes such as TMEM117, SLC25A21 and NCKAP5 related to milk traits. The present study screened out candidate genes for the formation of multiple teats at the genomic level in Hu sheep.
Introduction
Sheep is an important breed of livestock After thousands of years of domestication, artificial selection has led to the diversity of sheep populations, the Hu sheep is China’s famous Taihu breed, with high fertility, year-round estrus, its lactation function is excellent, and its nutritional value is worth studying, Chen et al. found that the Hu sheep’s milk contains a unique lactic acid bacterium, which has vertical transmission advantages.Citation1 Traditionally, the greater the number of teats in a mammal, the greater the number of children it produces. In sheep breeding, sheep udder traits are correlated with growth rates of weaned lambs.Citation2 Optimal udder and teat traits in ewes can significantly improve lamb survival and growth.Citation3 The optimal udder and teat traits in ewes significantly improve lamb survival and growth.Citation3 In sheep, polydactylous rams and ewes generally have 2–6 teats with a small anterior and large posterior distribution. Lactation performance studies have shown that some polydactyls can lactate normally.Citation4 Some studies have found that some polydactyls can lactate normally.Citation5 A study on removing multiple teats in sheep found that lactation yield was significantly reduced, but there was no difference in lactation yield between 2 and 4 teat.Citation6 The results showed a significant reduction in milk production, but no difference in milk production between 2 and 4 teats.Citation6 The localization of functional genes for multiple teats in sheep will facilitate the pace of scientific research on new lines of multiple teats in sheep and goats, as well as breeding selection. In sheep, teats are a highly heritable trait, and the development of extra teats is a complex multigenic trait.Citation7 In pig production, teat number influences growth productivity.Citation7 And teat number of pigs can be improved by selective breeding.Citation8 This trait can be utilized to increase the number of teats in sheep to enhance the milk production performance of sheep.
Mammalian teats can be classified into functional and nonfunctional teats. Zhao et al. found that LHFP, DPYSL2 and TDP-43 genes were associated with the formation of teat number in Hu sheep through a GWAS study of HD chip data from populations with different teat counts.Citation8 Peng et al. conducted GWAS analysis of HD SNP chip data from sheep and screened out BBX, CD47 and CASK genes associated with the formation of paramecium in sheep, and found that Wnt, Oxt ytcoin, MAPK and axon guidance pathway were important signaling pathways affecting paramecium formation in sheep.Citation4 Laura et al. showed that sheep redundant teats have similar immune cells and similar structures such as lymphoid follicles as normal teats, but there is a significant increase in the length and throughput diameter of the redundant teats compared to normal teats.Citation7 In a study of commercial sows, Audrey et al. found that elevated numbers of functional teats increased the number of weaned piglets.Citation9 The genes associated with teat number are different in different animals. BOVO et al. found that NUDT and NAR6A1 genes were associated with teat number in African rabbits through GWAS study and Fst analysis of 50K chip data in African rabbits, which differed from the candidate genes associated with teat number in pigs.Citation10 GWAS analysis of 60K SNP chip data from three pig species by Tang et al. found that the VRTN and KDM6B genes were closely related to teat number formation in pigs.Citation11 Yang et al. GWAS analysis of 50K chip data from pigs revealed that VRTN and ABCD4 genes influenced teat number formation in pigs.Citation12
The extreme imbalance between high fertility and low lactation in Hu sheep results in lambs with light birth quality, low survival rate and growth retardation, which becomes a bottleneck restricting the breed’s ability to maximize farming profitability, and death in the form of lambs is the greatest loss to sheep farming enterprises. Currently, most studies on mammalian teat number use GWAS and other analytical methods. In this study, we analyzed the selection signals of HD chip data of 157 Hu sheep from a multiple teats population and a two-teat population in order to screen out candidate genes related to the formation of multiple teat number in Hu sheep through different analytical methods in anticipation of providing data references for the selection and breeding of Hu sheep.
Materials and methods
Experimental materials
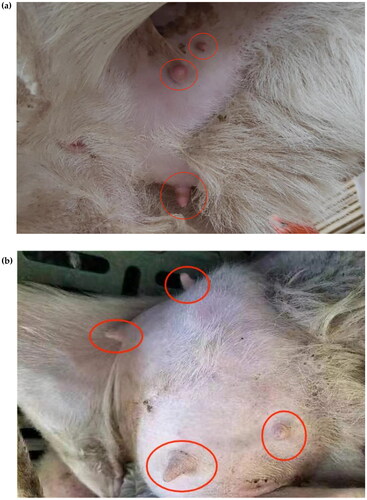
In this study, genome-wide data of 157 Hu sheep from a normal group of 76 two-teaters () and a group of 81 with extra teats were used (), which were obtained from https://figshare.com/s/197c20e3229490ccb5d6. Phenotypic data were obtained from https://figshare.com/s/0b326ab8632bbd1c82d6.Citation8 The data set was filtered by the following criteria: (i) absence of SNPs for chromosomal and physical location; (ii) SNPs >0.1 for genotypic deletions; (iii) SNPs <0.01 for minor allele frequencies; (iv) individuals with genotyping rates <90%; (v) p-values for Fisher’s exact test Hardy–Weinberg equilibrium <0.00001.
Test methods
Different biological analysis tools have different results, but no single method can completely detect all biological results in genome-wide selective scanning analysis.Citation13 In order to capture genomic selection signals more accurately, multiple methods are often required.Citation13 For this purpose, we chose four complementary statistical analysis tests: Fst (fixation index), xp-EHH (cross population extended haplotype homozygosity), iHS (integrated haplotype homozygosity score) and Pi (Population nucleic acid diversity). In this study, Fst, xp-EHH were analyzed between the two-teat and multi-teat populations, and Pi, iHS were analyzed separately for the multi-teat population.
Fst analysis
Calculation of Fst (Fixation Index) for two-teat and multi-teat groups using VCFTOOLS.Citation14 It is a measure of genetic differences between populations and is used to determine the degree of genetic differentiation between populations; higher Fst values mean greater genetic differences between populations and vice versa. Fst analysis allows for the study of genetic diversity between populations or subspecies. We used VCFTOOLS for Fst analysis, set the sliding window to 50,000 bp and the step size to 12,500 bp. The formula for Fst analysis is:
MSG is the mean square of error for detected within-population loci, MSP is the mean squared deviation of loci between tested populations, and NC denotes the corrected mean between overall sample sizes.
PI analysis
The multiple teat population of Hu sheep was analyzed for population nucleic acid diversity (Pi) using VCFTOOLS. We set the sliding window of the Pi analysis to 500 kb set the step size to 50 kb, and the SNP loci with 5% of the final Pi value were considered as significant candidatesCitation15 that were used to analyze polymorphism in the multiple teat groups in Hu sheep.
xpEHH analysis
We used rehh Hu sheep genomic data to do heterozygous extended haplotype purity tests (xpEHH),Citation16 Rehh (R package for detecting recent positive selection using extended haplotype homozygosity) is a statistical tool for detecting selection signals based on extended haplotype homozygosity (EHH). It can be used to identify positive selection events occurring in genetic variants in the human genome.
The xpEHH statistic was calculated by comparing the two-teat group to the multiple-teat group with the formula:
where unxpEHH is set:
iESpop1 integrates EHH genetic distance statistics, and iESpop2 integrates genetic distances for EHH statistics for multi-teat populations.
iHS analysis
Separate polypodium clusters for integrated haplotype purity scoring (iHS),Citation17 a single marker locus was used to replace the core haplotypes in the EHH statistic, defining them as core loci. And their ratios were calculated to select the signal detection statistic used to complement the Fst analysis, calculated as:
where uniHS is set:
IHH is the integration of genetic distances of EHH (integrated EHH), a represents the ancestral (progenitor) allele, and D represents the new mutant (derived) allele.
SNP annotation and analysis
For each scan the results were visualized using-R. We took the first 5% of SNP sites for Fst, xpEHH, and iHS values, and only the last 5% of SNP sites for Pi analysis were taken as positively selected sites to identify the selected regions in the genome, and all the obtained SNPs were annotated based on Sheep Oar_v4.0 (https://www.sheephapmap.org/). Genes were functionally analyzed with reference to the NCBI database (http://www.ncbi.nlm.nih.gov/gene), and visual intersection analysis was performed on individual analytical annotations using Draw Venn Diagram (Draw Venn Diagram (ugent.be)), using the DAVID tool (http://david.Abcc.ncifcrf.gov/) for gene ontology database (GO, http://geneontology.org)Citation18 and Kyoto Encyclopedia of Genes and Genomes (KEGG, Kyoto Encyclopedia of Genes and Genomes)Citation19 analysis. Related candidate genes for protein interactions analyzed on the STRING website (https://cn.string-db.org/).
Results
Fst analysis results
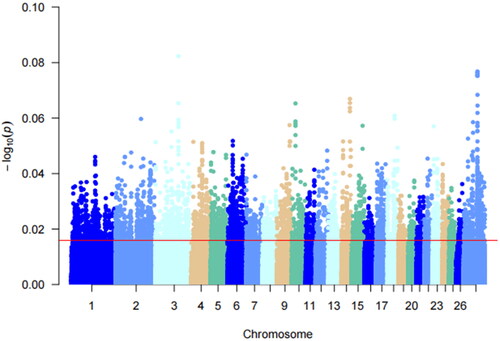
Multiple teats are a special phenotypic feature of Hu sheep compared to two teats. Multiple teats population favors the survival of lambs with multiple births. Using PLINK 19.0 software, Fst analysis was performed on the two-teat and multi-teat populations of Hu sheep, and the top 5% of Fst values. And the regions with Fst values >0.012 were considered as candidate regions for the multi-teat population and its significant loci were mainly distributed on chromosomes 3, 14, 2 and 8. A total of 10,278 loci were selected, and gene annotation was performed on the selected regions (), resulting in 2088 genes.
Results of Pi analyses
Nucleic acid polymorphisms were analyzed in the multi-teat population of Hu sheep, and the top 5% of the Pi values, with a Pi value >0.000106168, were considered as the selected regions, and 5140 SNP loci were selected (), and annotation analyses were performed on the selected regions, and 3,557 genes were obtained.
Results of xpEHH analyses
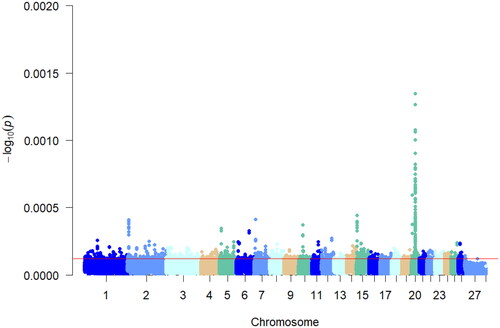
The xpEHH analyses were performed using VCFTOOLS on the multi-teat and two-teat populations of Hu sheep by comparing the extended haplotype purity differences between the two populations, with the two-teat population serving as the reference population and the multi-teat population serving as the test population for the selection signals analyses. As shown in , the top 5% of the absolute value of the xpEHH analysis results were selected as the loci in the multiple teat population, and 1779 SNP loci were found in the multi-teat population, and 1779 genes were obtained after annotation analysis of the relevant loci.
Results of iHS analyses
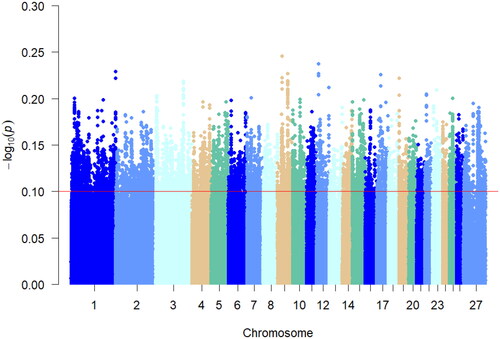
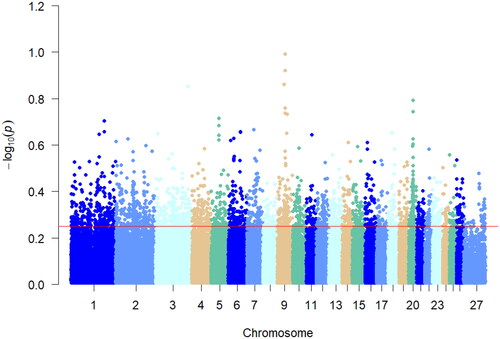
iHS detects strong selection signals in favor of alleles that have not yet reached fixation.Citation17 In this study, iHS analysis was performed individually on the Hu sheep multiple teat population, as shown in . The top 5% of the absolute value of the iHS results were considered as the region of selection, and 23,558 were the selected loci. Chromosomes 1, 7, 10, 20, and 21 showed peaks indicating loci on these chromosomes that were partially not yet fixed. Annotation analysis of the significant candidate loci yielded 3968 genes.
Veen analysis and gene enrichment
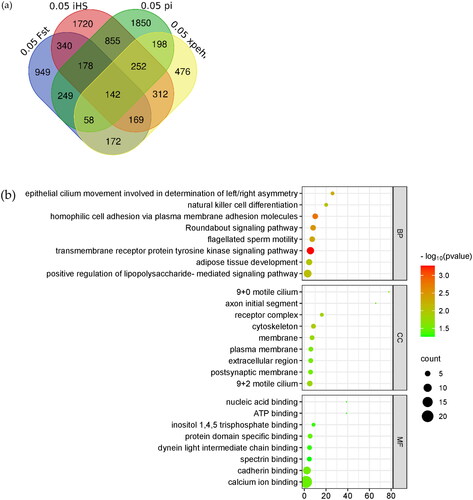
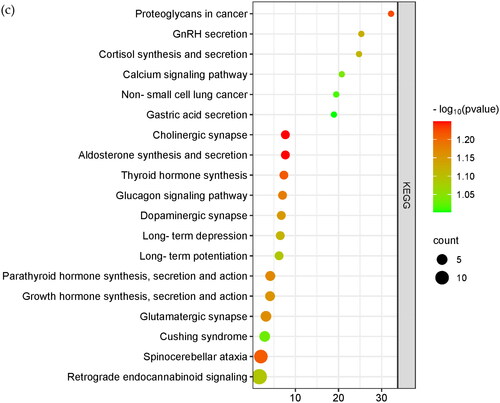
The sheep 4.0 genome was used as a reference to annotate the candidate-selected regions analyzed by Fst, Pi, xpEHH and iHS, as shown in , and 142 genes were overlapped (). For further functional analysis of the overlapped genes, we used DAVID online software to analyze the GO and KEGG pathways of the 142 genes, as shown in .
Figure 7. Veen diagram and enrichment analysis. Shows the common genes annotated from the candidate SNP loci for the four analyses. The 0.05FST in blue in the figure represents the genes annotated for the top 5% of SNPs loci analyzed by Fst. The light red part 0.05 iHS analyzed for the first 5% of SNPs loci, 0.05 PI in green in the figure represents the genes annotated from the first 5% of SNPs loci analyzed by Pi, and 0.05 xpEHH in yellow in the figure represents the genes annotated from the first 5% of SNPs loci analyzed by xpEHH. (b) Shows the bubble plot of GO enrichment analysis of candidate genes. (c) Shows the bubble plot of KEGG enrichment analysis of candidate genes.
Table 1. Candidate gene correlation analysis values. The genes presented in the table are candidate genes common to the four selection signal analyses in the text.
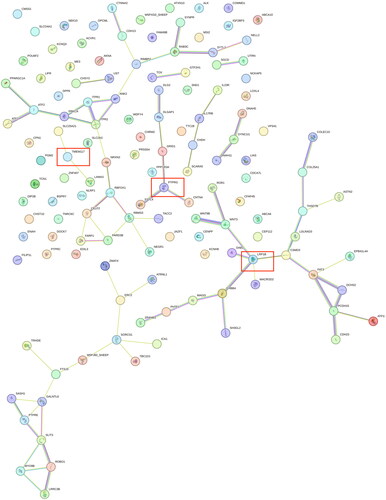
Peng et al. showed that paraplasia in sheep may be mediated by regulating cell proliferation, and that the selective signaling pathway of its occurrence is involved in breast cancer development.Citation6 In this study, calcium binding pathway (GO:0005509) was shown and NELL2, CDH23, PCDH15, CDH13, SLIT3, FAT3, EDIL3, LRP1B, FSTL4 genes were shown. In order to screen out the candidate genes among the further candidate genes related to polydactyly in Hu sheep, we carried out gene interactions analyses on the related genes. As shown in , the PTPRG, TMEM117 and LRP1B genes were shown to be associated with multiple genes.
Discussion
Detection of genomic regions under positive selection specific to a population is essential to reveal the genetic basis of variation in localized adaptive traits.Citation20 In this study, we genotyped genomic data between a multiple teat population and a two teats population of Hu sheep for Fst analysis. Fixation indices analysis is a measure of whether the actual frequency of genotypes in a population deviates from the theoretical proportion of genetic equilibrium (Harwin’s equilibrium). The genetic structure did not show a large genetic differentiation between the multi-teat population and the two-teat population of Hu sheep. In order to reduce the interfering factors of Fst analysis and increase the accuracy of selection signal detection. Screened the selected genes for the two populations of Hu sheep based on the theory of linkage disequilibrium as well as the extended haplotype purity test for the positive characteristics of the haplotypes. Analyzed the allele frequency analysis between the multi-teat and two-teat populations of Hu sheep using xp-EHH analysis as well as the pure haplotypes of multi-teat populations of Hu sheep for iHS analyses were complemented. The correlation analysis of its haplotypes revealed that the xpEHH analysis of its haplotypes revealed that the candidate xpEHH values ranged from 0.08 to 0.25 and the highest xpEHH value was found on chromosome 9. The same iHS analysis showed that the locus associated with multi-teat was fixed on chromosome 9. Its nucleotide diversity showed that loci with higher diversity were mainly located on chromosome 9. In addition, Pi analysis was used to screen the genomic selection signals of the multi-teat population of the Hu sheep. The highest Pi value was 0.014, which revealed that the subpopulation diversity of the polyposis population of the Hu sheep was not high. And based on the above analyses, we considered that the heritability of the multi-teat population of the Hu sheep was high.
In order to further identify the candidate genes related to multi-teat formation in Hu sheep, we annotated the overlapping regions of the four analysis methods, and among the 143 candidate genes screened for association with multi-teat. Based on the results of the gene interactions map we identified the mammary gland-related PTPRG,Citation21 TMEM117Citation22 and LRP1B genes.
Protein tyrosine phosphatase receptor type G (PTPRG) gene, located on sheep chromosome 19 at position NC_056072.1 (39155807.39930537, complement). This gene has been reported to inhibit mammary epithelial cell motilityCitation23 and has been associated with growth traits in sheep.Citation24 In addition PTPRG has been identified as a tumor suppressor gene in breast cancer. The expression of PTPRG is reduced during breast carcinogenesis and is particularly low in high malignancy grade breast cancer.Citation25 PTPRG is involved in the control of FGFR1 activity and influences the sensitivity of sarcoma cells to FGFR kinase inhibitors, and is important for cytostasis in breast cancer.Citation26
The transmembrane protein 117 (TMEM117) gene, located on sheep chromosome 3 NC_056056.1 (142302614.142937834, complement), was reported to be associated with posterior udder attachment width in hesitant cows.Citation22 Zhu et al.Citation27 also conducted a GWAS study and found that the TMEM117 gene was associated with saturated fatty acid composition in Simmental cows. Xia et al. find gene linked to beef traitsCitation28 the gene has also been reported to be associated with fatty acid formation in Ethiopian Indigenous Goat. This gene has also been reported to be associated with fatty acid formation in Ethiopian Indigenous Goat.Citation29 The gene was also reported to be associated with fatty acid formation in Ethiopian Indigenous Goat.Citation29
LDL receptor related protein 1B (LRP1B) gene, located on chromosome 2 of sheep at position NC_056055.1 (168072439.170268670). was reported to reduce malignant tumor cells in breast cancer cell lines,Citation30 in a study by Peng et al. on sheep paramammary glands, LRP1B was shown in GWAS analysis and was the significant locus.Citation6 This gene has also been reported to be associated with litter size in Greater Montenegrin sheep,Citation31 LRP1B may contribute to birth weight variability in pigsCitation32 and is involved in smooth muscle cell migration in chickens.Citation33
Extra teats in Hu sheep have similar immune cell and lymphatic structures as normal teat formation, and it has been shown that part of the extra teats in Hu sheep have the function of milk production, and the screening of individuals with extra teats in Hu sheep has certain benefits for lamb survival and production traits in Hu sheep populations.Citation7 In this study, we analyzed the selection signals of excess teats in Hu sheep, hoping to provide a molecular basis for the production as well as screening of excess teats in Hu sheep through genetic aspects. In this study, we found that the genes PTPRG, TMEM117 and LRP1B were closely associated with the production of surplus teats in Hu sheep.
In addition, this study identified genes associated with production growth among the relevant candidate genes. Genes such as DNAH11,Citation34 NELL2,Citation35 ANK2,Citation36 SLIT3,Citation37 DIP2B,Citation38 NEK10,Citation39 GRID1,Citation40 and PHTF1Citation41 associated with reproductive traits. And SH3GL2,Citation42 DOCK7,Citation43 ENAH,Citation44 SHISA9,Citation45 AGBL4,Citation46 SGCD,Citation47 COL25A1,Citation48,Citation49 DLG2,Citation5 DOCK7,Citation50 and other genes associated with milk production traits. And TMEM117,Citation22 SLC25A21,Citation51 NCKAP5,Citation52 PRKCA,Citation53 ITPR2,Citation54 NEGR1,Citation55 and other genes, and ASTN2,Citation56 GRID1,Citation57 FILIP1L,Citation58 ATRNL1,Citation59 and ERC2,Citation60 which are associated with stress tolerance.
Conclusions
In this study, candidate genes related to multi-teat in Hu sheep, such as TMEM117, PTPRG and LRP1B genes, were screened by selective signal analysis of HD chip data. These genes provide data for breed selection of Hu sheep, and also provide reference for the study of other multi-teat.
Author contributions
Conceptualization, W.Z., X.L. (Xiaopeng Li) and Z.H. (Zhipeng Han); methodology, X.L. (Xiaopeng Li), C.Z. (Cheng-long Zhang); software, X.L. (Xiaopeng Li), and R.Y. (Ruizhi Yang); validation, Z.H. (Zhipeng Han); formal analysis, W.Z.; investigation, Z.H. (Zhipeng Han) and L.Z. (Lulu Zhang) ; resources, W.Z.; data curation, W.Z.; writing—original draft preparation, W.Z.; writing—review and editing, W.Z., C.L. (Chunjie Liu); visualization, W.Z.; supervision, R.W. (Ruotong Wang) and J.W. (Jieru Wang); project administration, S.L. (Shudong Liu); All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We are thankful to all lab members for their support during the study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Li Y, Chen Z, Fang Y, et al. Runs of homozygosity revealed reproductive traits of Hu sheep. Genes. 2022;13(10):1848.
- Griffiths K, Ridler A, Compton C, Corner-Thomas R, Kenyon P. Associations between lamb growth to weaning and dam udder and teat scores. N Z Vet J. 2019;67(4):172–179.
- Smith EG, Hine BC, Acton GA, Bell AM, Doyle EK, Smith JL. Ewe udder and teat traits as potential selection criteria for improvement of Merino lamb survival and growth. Small Ruminant Research. 2023;225:107019.
- Peng W-F, Xu S-S, Ren X, et al. A genome‐wide association study reveals candidate genes for the supernumerary nipple phenotype in sheep (Ovis aries). Anim Genet. 2017;48(5):570–579.
- Banerjee P, Carmelo VAO, Kadarmideen HN. Genome-wide epistatic interaction networks affecting feed efficiency in Duroc and Landrace pigs. Front Genet. 2020;11:121.
- Lu W, Fang P, Lei Z. Mammary tissue structure and expression of insulin-like growth factor I receptor gene in multi-papillary lake sheep. J Agric Biotechnol. 2017;25:84–93.
- Hardwick LJA, Phythian CJ, Fowden AL, Hughes K. Size of supernumerary teats in sheep correlates with complexity of the anatomy and microenvironment. J Anat. 2020;236(5):954–962.
- Zhao Y, Pu Y, Liang B, et al. A study using single‐locus and multi‐locus genome‐wide association study to identify genes associated with teat number in Hu sheep. Anim Genet. 2022;53(2):203–211.
- Earnhardt-San AL, Gray KA, Knauer MT. Genetic parameter estimates for teat and mammary traits in commercial sows. Animals. 2023;13(15):2400.
- Bovo S, Schiavo G, Utzeri VJ, et al. A genome‐wide association study for the number of teats in European rabbits (Oryctolagus cuniculus) identifies several candidate genes affecting this trait. Anim Genet. 2021;52(2):237–243.
- Tang J, Zhang Z, Yang B, et al. Identification of loci affecting teat number by genome-wide association studies on three pig populations. Asian-Australas J Anim Sci. 2016;30(1):1–7.
- Yang L, Li X, Zhuang Z, et al. Genome-wide association study identifies the crucial candidate genes for teat number in crossbred commercial pigs. Animals. 2023;13(11):1880.
- Hohenlohe PA, Phillips PC, Cresko WA. Using population genomics to detect selection in natural populations: key concepts and methodological considerations. Int J Plant Sci. 2010;171(9):1059–1071.
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370.
- Li X. 2022. Research on Trait Variation of Different Coat Types and Their Molecular Genetic Mechanisms in Inner Mongolia Velvet Goats. Inner Mongolia Agricultural University.
- Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918.
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLOS Biol. 2006;4(3):e72.
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29.
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
- da Silva Ribeiro T, Galván JA, Pool JE. Maximum SNP FST outperforms full-window statistics for detecting soft sweeps in local adaptation. Genome Biol Evol. 2022;14(10):evac143.
- Lin G, Aranda V, Muthuswamy SK, Tonks NK. Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen of the “PTP-ome”. Genes Dev. 2011;25(13):1412–1425.
- Nazar M, Abdalla IM, Chen Z, et al. Genome-wide association study for udder conformation traits in Chinese Holstein cattle. Animals. 2022a;12(19):2542.
- Rahmatpanah F, Jia Z, Chen X, Jones FE, McClelland M, Mercola D. Expression of HER2 in breast cancer promotes a massive reorganization of gene activity and suggests a role for epigenetic regulation. J Data Mining Genomics Proteomics. 2012;3:e102.
- Kizilaslan M, Arzik Y, White SN, Piel LMW, Cinar MU. Genetic parameters and genomic regions underlying growth and linear type traits in Akkaraman sheep. Genes. 2022;13(8):1414.
- Sloth RA, Axelsen TV, Espejo MS, et al. Loss of RPTPγ primes breast tissue for acid extrusion, promotes malignant transformation and results in early tumour recurrence and shortened survival. Br J Cancer. 2022;127(7):1226–1238.
- Pandey G, Borcherding N, Kolb R, et al. ROR1 potentiates FGFR signaling in basal-like breast cancer. Cancers. 2019;11(5):718.
- Zhu B, Niu H, Zhang W, et al. Genome wide association study and genomic prediction for fatty acid composition in Chinese Simmental beef cattle using high density SNP array. BMC Genomics. 2017;18(1):464.
- Xia J, Qi X, Wu Y, et al. Genome-wide association study identifies loci and candidate genes for meat quality traits in Simmental beef cattle. Mamm Genome. 2016;27(5–6):246–255.
- Berihulay H, Li Y, Gebrekidan B, et al. Whole genome resequencing reveals selection signatures associated with important traits in ethiopian indigenous goat populations. Front Genet. 2019;10:1190.
- Kadota M, Yang HH, Gomez B, et al. Delineating genetic alterations for tumor progression in the MCF10A series of breast cancer cell lines. PLOS One. 2010;5(2):e9201.
- E. G-X, Duan X-H, Zhang J-H, et al. Genome-wide selection signatures analysis of litter size in Dazu black goats using single-nucleotide polymorphism. 3 Biotech. 2019;9(9):336.
- Wang L, Sun X, Guo F, Zhao Y, Zhang J, Zhao Z. Transcriptome analysis of the uniparous and multiparous goats ovaries. Reprod Domest Anim. 2016;51(6):877–885.
- Tanaga K, Bujo H, Zhu Y, et al. LRP1B attenuates the migration of smooth muscle cells by reducing membrane localization of urokinase and PDGF receptors. Arterioscler Thromb Vasc Biol. 2004;24(8):1422–1428.
- Trujano-Chavez MZ, Ruíz-Flores A, López-Ordaz R, Pérez-Rodríguez P. Genetic diversity in reproductive traits of Braunvieh cattle determined with SNP markers. Vet Med Sci. 2022;8(4):1709–1720.
- Ryu BJ, Kim HR, Jeong JK, Lee BJ. Regulation of the female rat estrous cycle by a neural cell-specific epidermal growth factor-like repeat domain containing protein, NELL2. Mol Cells. 2011;32(2):203–207.
- Tetkova A, Jansova D, Susor A. Spatio-temporal expression of ANK2 promotes cytokinesis in oocytes. Sci Rep. 2019;9(1):13121.
- Dickinson RE, Hryhorskyj L, Tremewan H, et al. Involvement of the SLIT/ROBO pathway in follicle development in the fetal ovary. Reproduction. 2010;139(2):395–407.
- Adlat S, Sah RK, Hayel F, et al. Global transcriptome study of Dip2B-deficient mouse embryonic lung fibroblast reveals its important roles in cell proliferation and development. Comput Struct Biotechnol J. 2020;18:2381–2390.
- He LC, Li PH, Ma X, et al. Identification of new single nucleotide polymorphisms affecting total number born and candidate genes related to ovulation rate in Chinese Erhualian pigs. Anim Genet. 2017;48(1):48–54.
- Sánchez-Ramos R, Trujano-Chavez MZ, Gallegos-Sánchez J, Becerril-Pérez CM, Cadena-Villegas S, Cortez-Romero C. Detection of candidate genes associated with fecundity through genome-wide selection signatures of Katahdin Ewes. Animals. 2023;13(2):272.
- Pryce JE, Haile-Mariam M, Goddard ME, Hayes BJ. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet Sel Evol. 2014;46(1):71.
- El Nagar AG, Heddi I, Sosa-Madrid BS, Blasco A, Hernández P, Ibáñez-Escriche N. Genome-wide association study of maternal genetic effects on intramuscular fat and fatty acid composition in rabbits. Animals. 2023;13(19):3071.
- Menin L, Weber J, Villa S, et al. A planar polarized MYO6-DOCK7-RAC1 axis promotes tissue fluidification in mammary epithelia. Cell Rep. 2023b;42(8):113001.
- Ma X, Cheng H, Liu Y, et al. Assessing genomic diversity and selective pressures in Bohai black cattle using whole-genome sequencing data. Animals. 2022;12(5):665.
- Duan X, An B, Du L, et al. Genome-wide association analysis of growth curve parameters in Chinese Simmental beef cattle. Animals. 2021;11(1):192.
- Wang K, Wu P, Wang S, et al. Genome-wide DNA methylation analysis in Chinese Chenghua and Yorkshire pigs. BMC Genom Data. 2021;22(1):21.
- Li D, Huang M, Zhuang Z, et al. Genomic analyses revealed the genetic difference and potential selection genes of growth traits in two duroc lines. Front Vet Sci. 2021;8:725367.
- Mancini G, Gargani M, Chillemi G, et al. Signatures of selection in five Italian cattle breeds detected by a 54K SNP panel. Mol Biol Rep. 2014;41(2):957–965.
- Purfield DC, Evans RD, Berry DP. Reaffirmation of known major genes and the identification of novel candidate genes associated with carcass-related metrics based on whole genome sequence within a large multi-breed cattle population. BMC Genomics. 2019;20(1):720.
- Wang K, Liu D, Hernandez-Sanchez J, et al. Genome wide association analysis reveals new production trait genes in a male Duroc population. PLOS One. 2015;10(9):e0139207.
- Zhou C, Li C, Cai W, et al. Genome-wide association study for milk protein composition traits in a Chinese Holstein population using a single-step approach. Front Genet. 2019;10:72.
- Jiang J, Ma L, Prakapenka D, VanRaden PM, Cole JB, Da Y. A large-scale genome-wide association study in U.S. Holstein cattle. Front Genet. 2019;10:412.
- Wang H, Zhong J, Zhang C, et al. The whole-transcriptome landscape of muscle and adipose tissues reveals the ceRNA regulation network related to intramuscular fat deposition in yak. BMC Genomics. 2020;21(1):347.
- Chen Z, Yao Y, Ma P, Wang Q, Pan Y. Haplotype-based genome-wide association study identifies loci and candidate genes for milk yield in Holsteins. PLOS One. 2018;13(2):e0192695.
- Mohammadi H, Farahani AHK, Moradi MH, et al. Weighted single-step genome-wide association study uncovers known and novel candidate genomic regions for milk production traits and somatic cell score in Valle del Belice dairy sheep. Animals. 2022;12(9):1155.
- Yudin NS, Larkin DM. Candidate genes for domestication and resistance to cold climate according to whole genome sequencing data of Russian cattle and sheep breeds. Vavilovskii Zhurnal Genet Selektsii. 2023;27(5):463–470.
- Ai H, Yang B, Li J, Xie X, Chen H, Ren J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics. 2014;15(1):834.
- Sun L, Qu K, Ma X, et al. Whole-genome analyses reveal genomic characteristics and selection signatures of Lincang humped cattle at the China-Myanmar border. Front Genet. 2022;13:833503.
- Kim K-S, Seibert JT, Edea Z, et al. Characterization of the acute heat stress response in gilts: III. Genome-wide association studies of thermotolerance traits in pigs. J Anim Sci. 2018;96(6):2074–2085.
- Tian D, Han B, Li X, et al. Genetic diversity and selection of Tibetan sheep breeds revealed by whole-genome resequencing. Anim Biosci. 2023;36(7):991–1002.