?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Although structural equation model (SEM) is a powerful and widely applied tool particularly in social sciences, few studies have explored how SEM and statistical learning methods can be combined. The purpose of this paper is to explore how gradient component-wise boosting (GCB) can contribute to item selection. We ran 200 regressions with different farmer psychological variables collected to explain variation in an animal welfare indicator (AWI). The most frequently selected variables from the regressions were selected to build a SEM to explain variation in the AWI. The results show that boosting selects relevant items for a SEM.
Introduction
Structural equation models (SEMs) capture hypotheses about multivariate data by specifying relations among observed entities and hypothesized latent constructs. Generally, specifying a SEM based on substantive theory begins by defining the latent variables and then constructing items to measure them. Sometimes, however, a more exploratory approach is needed. When the substantive theory is weak and a set of variables/items exists but the exact number or meaning of the factors is unknown, specifying a SEM often starts with exploring item correlations and exploratory factor analysis (EFA). Which factors to extract and the number of factors are important steps in EFA, often followed by a confirmatory factor analysis (CFA). In datasets where there is weak correlation between an observed dependent variable and the available explanatory variables or items, it can be challenging to specify factors. Given ever richer datasets, extracting the relevant variables or items to specify factors becomes even more laborious. Commonly applied tools like EFA and CFA work well to extract factors. However, these methods offer little help when one also wants to use the factors to explain variation in a dependent variable because they do not relate the items directly to the variable they are supposed to predict. Thus, we agree with Jacobucci et al. (Citation2019) that there is an increasing need for efficient item selection when constructing a SEM.
Maybe the newcomer ESEM (Exploratory structural equation modeling; Asparouhov and Muthén (Citation2009)), where an EFA measurement model with rotation can be used in a SEM has the potential to help select items.
A commonly used phrase to describe these activities within SEM is conducting a specification search (Long, Citation1983). The basic idea behind conducting a specification search is to find a better fitting model after comparing distinct SEM models using a chi-square difference test or other selected fit indices (Marcoulides & Falk, Citation2018). Rudimentary forms of specification searches in SEM may consist of manually fitting alternative models and comparing fit, with model changes based on substantive theory: modification indices to determine which parameters to free; Wald-based tests to determine which parameters to fix; or some synthesis of these approaches (Marcoulides & Falk, Citation2018).
In parallel with the search for better specification methods in SEM, statistical learning methods (SL) are increasingly used to analyze or find patterns in very large data sets. Hence, there is a need for statistical techniques to select the most informative features or items out of a large set of predictor variables. SL refers to a vast set of tools for modeling and understanding data (James et al., Citation2017). Thus, SL is concerned with model validity, accurate estimation of model parameters and inference from the model (Fawcett & Hardin, Citation2017). In the field of SL, a key issue is the development of algorithms for model building and variable selection (Hastie et al., Citation2009). Much attention has been dedicated to the topic of how predictors can be optimally selected when little or no prior knowledge exists. Boosting algorithms represent one of the most promising methodological approaches for data analysis developed in the last two decades (Mayr et al., Citation2014). Originally developed in the machine learning community (Freund & Schapire, Citation1996; Schapire, Citation1990) primarily to handle classification problems, boosting has been successfully translated into the statistical field (Breiman, Citation1998; Friedman et al., Citation2000). In recent years, its use has been extended to many statistical problems. Because boosting fits an additive model or an ensemble in a forward-stagewise manner, some similarities with forward stepwise regression exist. In boosting, however, at each iteration a weak learner, or a learner that performs only slightly better than random, is introduced to compensate for the “shortcomings” of existing weak learners. Gradient component-wise boosting (GCB) (Breiman, Citation2000; Friedman, Citation2001), is an example of an innovation that enjoys good predictive performance, e.g., in regressions. GCB performs well in variable selection because it discards the least important ones, and also ranks variables or items by decreasing importance (Efron & Hastie, Citation2016), both important factors in specifying a SEM. The aim of this paper is to show how GCB can be applied to select relevant items for inclusion in a SEM with only one observed dependent variable.
To more efficiently specify a SEM, recent research has suggested integrating regularization techniques like the Lasso, Ridge and elastic net and SEM (Jacobucci et al., Citation2019). Various methods for identifying group differences (Frick et al., Citation2015; Kim & von Oertzen, Citation2018; Tutz & Schauberger, Citation2015) together with combinations of SEM and decision trees (Brandmaier et al., Citation2013) and forests (Brandmaier et al., Citation2016) have also been suggested. However, few proponents of SEM have explored the merits of boosting in variable selection. Our goal in this paper is to present an example of how boosting, specifically GCB, can help to answer the question: “What subset of items is most predictive for my outcome variable and at the same time gives rise to a reasonable measurement model? “. To answer this question, we use a dataset from dairy farming where an observed continuous outcome variable, an animal welfare indicator (AWI), is only weakly correlated with all items applied (Hansen & Österås, Citation2019).
To arrive at the SEM suggested in Hansen and Österås (Citation2019), (see in the Appendix for an overview), the authors first followed the traditional procedure and started with an EFA to extract factors. Although they managed to extract meaningful factors representing underlying theoretical constructs, the extracted factors showed only weak relationships with AWI, the dependent variable. The problem is that neither the EFA nor the frequently applied principal component analysis (PCA) selects subsets of the predictors or items based on their association with the outcome. Item selection is based solely on the associations among them. Therefore, selecting the relevant items to include in a SEM required significant trial and error. Hansen and Österås (Citation2019) experiences inspired the authors of this paper to use the same dataset as an example of how the specification process can be more streamlined. Our strategy was to combine GCB, CFA and SEM. A similar procedure has been used in other studies to select the most relevant variables to feed a Gaussian Process regression model (Friederich et al., Citation2020). To provide a contrast, we also included an ESEM. Because the aim of this study was not to compare Boosting and ESEM, the ESEM results are reported in the appendix and only briefly described in the results section. The remainder of the paper is organized as follows: First we introduce the example context with material and methods, and thereafter results, discussion and conclusion. An on-line appendix with R-code and an anonymized and masked data set are also included.
Example and analysis
Hansen and Österås (Citation2019):” Animal welfare is a term used to express ethical concerns about the quality of life experienced by animals, particularly animals that are used by human beings in production agriculture (Duncan & Fraser, Citation1997; Fraser & Weary, Citation2004; Tannenbaum, Citation1991). The characteristics of farmers that may influence the animal welfare standards include knowing and being skilled at the techniques they use, job motivation and satisfaction, and attitudes (Hemsworth & Coleman, Citation2009)”. Hansen and Österås (Citation2019) showed that a relationship exists between farmers’ occupational well-being and stress, and an animal welfare indicator. In as SEM, ibid. reported a significant positive relationship between the AWI and farmer occupational well-being (FOW), and a negative relationship between AWI and and farmer stress (FS). in a SEM. FOW included the items 3, 7, 8 and 12 reported in this study, and FS included items 14, 15 and 18, see . For a thorough description of the context, we refer to Hansen and Österås (Citation2019).
Table 1. The items in Hansen and Österås (Citation2019), except one item that was removed due to little relevance, with median values. All items range from 1 to 11, and items 10, 11 and 12 to 18 are reverse-coded
The dataset used in Hansen and Österås (Citation2019) was collected for a larger study about automatic milking systems (AMS), also called milking robots. The aim of the study was to explore how farmers perceive their quality of life, their working situation and mental health, the future of their farm, work division among family members, income, etc. To compare farmers with and without AMS, their questionnaire was distributed to all 1700 farmers registered with an AMS autumn 2017, and to 1700 randomly selected dairy farmers with conventional milking systems. Hansen and Österås (Citation2019) merged the 1288 answers from the web-survey with the participant’s AWI. Because not all 1288 farmers had an AWI available when the study was conducted, the final sample included 914 dairy farmers.
The items used to collect data on dairy farmer’s quality of life (items 1 to 7), working situation (items 8 to 12) and mental health (items 13 to 18) are shown in . All items were coded on an 11-point Likert scale ranging from to a smaller degree to a larger degree (Items 1–7), strongly agree/strongly disagree (Items 8–12), and very often/very seldom (Items 13–18). The items were treated as ordinal variables, and the negatively phrased items 10, 11 and 12 to 18 were reverse-coded to avoid negative factor loadings.
Boosting and modeling
In this section, we first give a brief overview of gradient component-wise boosting and the package ‘mboost’ in the statistical software R (CRAN, Citation2020). Then we describe the specification of the CFA and the SEM. Our point of departure is that several explanatory variables are needed to explain the relationship between farmer welfare and animal welfare. Therefore, we think SEM is a good tool to model this relationship. By using the CFA approach, as a measurement model, on the “right-hand side” of the equation one can benefit from minimizing bias in the regression parameter estimates, since we control for the measurement error. This is one of many strengths – and a very important one—of SEM relative to e.g., OLS regression. However, specifying a SEM item selection is crucial, and this is where boosting is particularly useful.
Gradient component-wise boosting and the R-package mboost
While several variants of boosting exist, here we focus on gradient component-wise boosting. To select relevant variables to include in a SEM, the task is to model the relationship between the AWI () and a vector consisting of the different items
=
depicted in , to obtain an “optimal” prediction of
given
. This is accomplished by minimizing a loss function over a prediction function (depending on
). Here the loss function corresponds to the least squares objective function and
is a linear function of
. If we use squared error loss, the negative gradient vector scaled by 1/2 equals the residuals. Gradient boosting tries to find the direction in space with the largest decrease in the squared error loss function
, i.e. to find the negative gradient
. Component-wise boosting means that the model calculates updates for each dimension
, and selects the best update among them. The following algorithm based on Hofner et al. (Citation2014) is used to minimize the loss function over
:
Initialize the function estimate
with offset values.
is a vector of length n. Let the vector of function estimates at iteration m be
Specify a set of base-learners. Base-learners are simple regression estimators with a fixed set of input variables and a univariate response. The sets of input variables may differ among the base-learners. Usually, the input variables of the base-learners are small sets of the predictor variables. In the simplest case like in this paper, there is exactly one base-learner for each predictor variable, and the base-learners are just simple linear models using the predictor variables as input variables. Each base-learner represents a modeling alternative for the statistical model. We denote the number of base-learners, P, and set m = 0.
Increase m by 1, where m is the number of iterations.
a) Compute the negative gradient
of the loss function and evaluate it at
(
),
(i.e. at the estimate of the previous iteration). This yields the negative gradient vector
=
:=
b) Fit each of the P-base-learners to the negative gradient vector, i.e. use each of the regression estimators specified in step 2 separately to fit the negative gradient. The resulting P regression fits yield P vectors of predicted values, where each vector is an estimate of the negative gradient vector
c) Select the base-learner that fits best according to the residual sum of squares criterion and set
equal to the fitted values of the best-fitting base-learner.
d) Update the current estimate by setting =
+
, where 0
is a real-valued step length factor.
(5) Iterate steps 3 and 4 until the stopping criterion
is reached.
From 4c and 4d we can see that the algorithm sequentially carries out variable selection and model choice, as only one base-learner is selected for updating in each iteration. Boosting is a way of fitting an additive expansion in a set of elementary “basis” functions. The final boosting iteration has similarities to the additive predictor of a generalized additive model
=
+ … … .+
, where
, … .,
correspond to the functions specified by the base-learners. Consequently,
, … .,
depend on the predictor variables that were used as input variables of the respective base-learners. A base-learner can be selected multiple times. In contrast to the choice of the stopping iteration, the choice of the step length factor
has been shown to be of minor importance for the predictive performance of a boosting algorithm. The only requirement is that the value of
is small, e.g.,
= 0.1 as applied in this paper (see Schmid & Hothorn, Citation2008b).
The R package mboost offers an easy entry into the world of boosting. It implements a model-based boosting approach that results in interpretable structured additive models in a form that will feel familiar for most researchers. The interfaces of fitting functions are quite similar to standard implementations like lm() or glm() and are hence relatively easy to use (Hofner et al., Citation2014). Because of its user-friendly formula interface, mboost can be used in a manner similar to classical functions for statistical modeling in R (Hofner et al., Citation2014). In the linear model applied in this paper the regression coefficients can be interpreted similarly to ordinary least squares coefficients. In addition to linear effects, the package also offers possibilities for modeling non-linear or interaction effects with other predictor variables. The structural assumption is given as a formula using base-learners. The mboost package also offers high flexibility when it comes to the type of risk function to be optimized. The loss function, as specified by the family argument is independent of the estimation of the base-learners. As one can see in the GCB algorithm, the loss function is used to compute the negative gradient in each boosting step only. The predictors are then related to these values by penalized ordinary least-squares estimation, irrespective of the loss function. The function glmboost() provides an interface to fit (generalized) linear models. The resulting models from glmboost() can essentially be interpreted the same way as models that are derived from glm(). The only difference is that the boosted generalized linear model additionally performs variable selection (Hofner et al., Citation2014).
To load mboost in R write library(mboost). It contains the function glmboost with the following interface:
model<−glmboost (formula, data = list(), weights = NULL, center = TRUE, control = boost_control(), …). The model is specified using a formula as in glm() of the form response ~ predictor1 + predictor2, and the data set is provided as a data.frame via the data argument. Optionally, weights can be given for weighted regression estimation. The argument center is specific for glmboost(). It controls whether the data is internally centered. Centering of predictors is of great importance as this allows much faster “convergence” of the algorithm or even ensures that the algorithm converges in the direction of the true value at all. The second boosting-specific argument, control, allows to define the hyper-parameters of the boosting algorithm. This is done using the function boost_control(). For example, one may specify:
boost_control(mstop = 200, # Initial number of boosting #iterations. default: 100
+ nu = 0.05), # step length, default: 0.1. Finally, the user is allowed to specify the distributional assumption via a family, which is “hidden” in the ‘ … ’ argument (see ?mboost_fit for details and other possible parameters). The default family is gaussian(), which is applied in this study. In the following example-script we set y equal to the AWI, and define X as a matrix containing all the items in :
model <− glmboost (y=AWI, x=X, family = gaussian(), control=boost_control, center=TRUE)
Cross-validation is crucial to determine the optimal number of iterations mstop. The following procedure invokes a ten-fold cross-validation:
cv <− cvrisk (model, folds = cv(model.weights(model), type = “kfold”, B = 10))
mstop(cv) # shows the optimal number of boosting iterations
Finally, summary yields the coefficients and the items selected in the final model:
summary(model[mstop(cv)]).
We regressed the AWI on all 18 items with the function glmboost and ordinary least squares base-learners. To rank items according to importance in the regressions we used the “selection frequency” reported in the mboost-package, which reflects point 4 c) that a base learner can be choosen several times during the fitting process.
Structural equation modeling (SEM) and Confirmatory Factor analysis (CFA)
Like SEM, CFA is a well-established research tool in social science and business. For a thorough mathematical description, we refer to Bollen (Citation1989) and Jöreskog et al. (Citation2016). A CFA was used to estimate and test a model of the latent variables. It begins by defining the latent variables one would like to measure, based on substantive theory and/or previous knowledge (Jöreskog et al., Citation2016). The CFA was built on the most frequently selected items from the GCB and tested for goodness of fit. Finally, a GCB-SEM was specified with the factors from the CFA as predictors of the dependent variable animal welfare. The CFA and the GCB-SEM were modeled using the lavaan package in R (CRAN, Citation2020).
The analysis of the dataset was done as follows:
We randomly divided the whole dataset into two equal-sized parts.
Then, one of these parts was used to run a GCB regression to select items associated with the AWI. This was done to avoid using exactly the same dataset for boosting and for SEM, and to make the task more difficult for the boosting algorithm.
Procedures 1) and 2) were repeated 200 times. For each GCB regression, we registered which items the GCB selected, and their relative importance.
Finally, the remaining part of the data set and the most frequently selected items from the GCB regressions were used to specify a CFA and a SEM to explain the variation in the AWI.
Results
The coefficients of the Spearman rank correlation are shown in .
Table 2. Spearman rank correlation correlations between the AWI and the items and between items (N = 914)
We observe that the correlations between the AWI and the different items are weak, in particular for items related to mental health (items 13 to 18). We also notice that the correlation between these items are moderate or strong (Shortell, Citation2001). The low correlations with the dependent variable suggests that it will be challenging to select relevant items and specify reliable factors. The results from the 200 GCB-regressions are shown in .
Table 3. How often each item was selected in the 200 GCB regressions on the AWI together with relative importance of the different items in each regression. The items are ranked after decreasing frequency of selection. For each regression, the GCB shows the relative importance of each variable
In we can see that the items differ significantly in how often they are selected by the GCB. A group of seven items stand out as the most important ones; items 2, 6, 5, 11, 12, 9 and 13. We expect these to be the most relevant ones to include in a CFA and a SEM. Contrary, items 17, 7, 8, 18, 1, 4, 10 and 14 appear to be of little relevance in explaining variation in the AWI. Thus, we have managed to exclude a group of eight items with a weak relationship to the AWI. The remaining items appear to be of medium importance. In each run of 200 regressions, the GCB ranks the items by decreasing importance, and the median importance for all items over the 200 GCB-runs are shown in column 3 of . The numbers in this column mirror how often the items are selected. Thus, the items most often selected; 2, 6, 5, 11, 12, 9 and 13, are all ranked the five most important ones. Conversely, items 17, 7, 8, 18, 1, 4, 10 and 14 are all ranked between six and eight. Therefore, they are of little interest. To summarize, the 200 GCB regressions have reduced the number of relevant items to less than half of the original items, indicating that boosting has contributed to both item selection and item ranking.
To specify a CFA we started at the top of the list with the most frequently selected items in , and decided to use items 2, 6, 11, 12, 13 and 10. The first five items are among the seven most frequently selected, while item 10 is ranked 12. Item 10 was chosen in spite of its low ranking in the 200 regressions in order to include at least three items per factor. It appeared that item 10 functioned well with the items 12 and 13 in a factor denoted farmer stress, see . Thus, item 10 was needed to create a coherent factor including three items.
Figure 1. The CFA with two factors, farmer occupational well-being FOW and farmer stress (FS). The dotted lines and box to the right illustrates how the CFA extends to the GCB-SEM (An arrow for the disturbance term is not included).
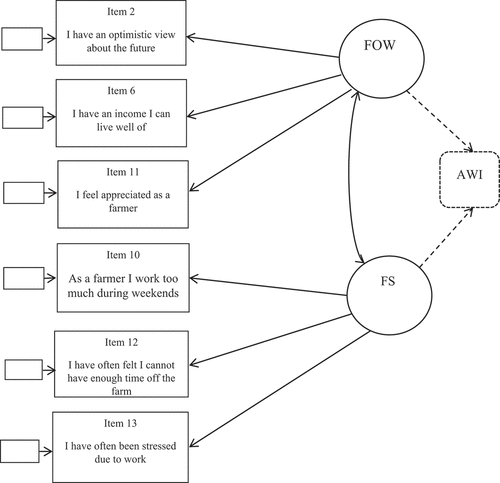
In the CFA, all standardized factor loadings are significantly different from zero and within the interval [0.483, 0.680] (see ). FOW includes items related to working situation and quality of life, while FS includes two items related to mental health and one related to quality of life. Cronbach’s alpha is calculated as 0.682 and average composite reliability is 0.611.
Table 4. Items and standardized factor loadings in CFA and SEM with standard errors
Finally, we specified the CFA and the GCB-SEM, both of which are shown in . The dotted part (dotted lines and the box AWI) extend the CFA to a SEM with a single observed dependent variable.
In the SEM, the estimated regression coefficient for FOW on AWI was 0.377*** (0.105), and the regression coefficient for FS on the AWI was −0.291***(0.107). The correlation between FOW and FS was 0.611.
FOW is positively associated with the AWI, while FS is negatively associated, and the signs are as expected. FS contributes negatively to animal welfare, while occupational wellbeing contributes positively to animal welfare. Both factors yield moderate relationships with the AWI. The theoretical models provide a good fit to the observed data (See ).
Table 5. Goodness of fit measures and degrees of freedom (df) for the CFA and the GCB-SEM
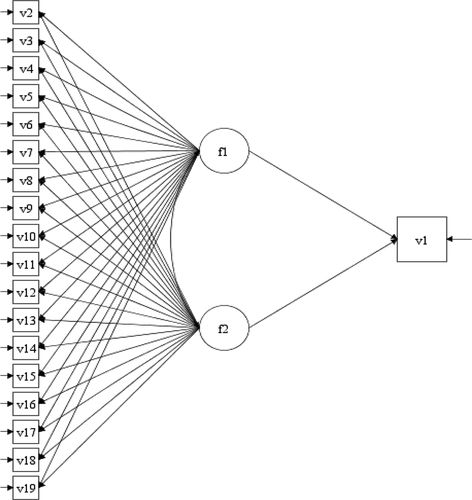
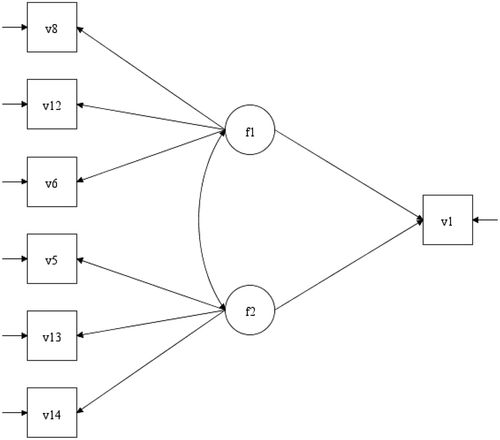
In the appendix, the section “ESEM vs Boosting” compares a SEM derived from using ESEM and the GCB-SEM derived from Boosting. As mentioned earlier, the intention of this paper is not to compare these two methods. The focus is to show how Boosting can be applied to select relevant items. Given the objective of ESEM (Asparouhov & Muthén, Citation2009) we were tempted to use ESEM on the same data set with the same goal, to build a SEM. So, we conducted a quick ESEM analysis, using Mplus 8.3: 1) In , the original ESEM is depicted, with all 18 items for two common factors. 2) In and A3 we can see the SEMs where all items with factor loadings <0.4 and <0.5, respectively, are deleted. We ended up with two distinct factors with no cross loading. This is an interesting finding. The “final” model (SEM2) depicted in was re-estimated in laavan by using the whole sample (N = 914). The same was done with the GCB-SEM (SEM3) derived from Boosting, but now including the whole sample. The results are in respectively. As we can see for SEM 3 (3 items per factor), the fit is good (Chi sq. =19.389 (12) p =.08)), the factor loadings are in the interval 0.560 to 0.705, and the regression parameters are 0.326 and −0.226, respectively. However, for SEM2 (four items per factor), the fit is not as good (Chi sq. = 54.292 (25) p = .001), the factor loadings are in the interval 0.534 to 0.866, and the regression parameters are 0.169 and −0.076, respectively. The results are interesting, however, in factor 2, three items that reflect stress due to lack of time and work overload (items 13,17 and 18) are mixed with an item reflecting stress due to health worries (item 16). Overall, the results in SEM2 are probably as one should expect, given the different “philosophy” of the two approaches. A thorough simulation study that compares the two approaches could provide more specific information.
Discussion and conclusion
This paper and the R-code provided demonstrate an example of how boosting with R can be used as a tool to facilitate a SEM specification search. That is, boosting is not used to choose among different SEM-models, but to select possible items to include in a SEM. At the outset of this study we had 18 items, and with three items per factor, there are 816 possible combinations. The GCB reduced the number of relevant items to eight, which reduces the number of possible combinations to 56. Our example, therefore, demonstrates how GCB can contribute to item selection, and thus to SEM specification. At the very least, the GCB algorithm can be effective as an initial exploratory tool that can be used in modeling complex behavioral, educational, and social phenomena. In this study, GCB selects and ranks the items and simultaneously ensures that there is a relationship between the items and the dependent variable. Compared to the SEM in Hansen and Österås (Citation2019, p. 6), the GCB-SEM in this study exhibits a stronger relationship between FOW, FS and the AWI (see ). This is as expected, since the GCB was trained in a regression with the AWI as the dependent variable, the item selection in Hansen and Österås (Citation2019) was based on a trial and error approach involving all 18 items. It is noteworthy that the GCB-SEM includes only four of the seven items used in Hansen and Österås (Citation2019) (see ). This finding suggests that not all items in Hansen and Österås (Citation2019) were the optimal ones to explain the variation in the AWI, and boosting has improved item selection. Taken together, our example shows that GCB has the potential to streamline the task of selecting items and specifying a SEM when the task is to explain the variation in an observed variable. That being said, we acknowledge that settings might exist where boosting is not necessarily the best tool to facilitate a SEM- specification search.
What can be considered a reliable factor loading remains a topic for discussion. For example, the software package SPSS uses 0.4 as the cutoff criteria. However, all factor loadings in this paper are above this value. The strength of the relationships between FOW, FS and the AWI in Figure 2, while moderate, are within the range frequently found in studies of job satisfaction and stress versus job performance (Judge et al., Citation2001). How farmers thrive at work is pivotal to productivity and to to keep up dairy farming. Our findings show that there is a positive relationship between FOW and the AWI, and a negative relationship between FS and the AWI (Figure 2). Thus, a relationship exists between farmers’ occupational well-being and stress on one side, and how well they take care of their animals on the other, which supports the findings in Hansen and Österås (Citation2019) (see ).
It is important to recognize that results from a computerized specification search like the one applied in this paper, may reflect an element of chance. Therefore, without a theoretical background related to the question being examined or preliminary research conducted in the field, there is no way one can distinguish between different ways to determine relationships among variables (Tarka, Citation2018). Furthermore, our example shows that when specifying factors there is some degree of subjectivity involved in determining the number and interpretation of the factors. Thus, we had to include item 10, ranked as number 12 by the GCB, to specify a meaningful factor. The GCB algorithm is developed for supervised statistical learning, and does not group items based on their degree of association. This task is left, instead, to the researcher. An ideal solution would be to end up with sets of highly ranked grouped items ready to include in factors. To achieve this, one possible strategy could be to integrate the GCB algorithm in the SEM framework. However, considering the complexity involved in merging the two algorithms, the authors have some doubt about whether this is the right path to follow. A more practical approach is, perhaps, to run an EFA including the items from the GCB to extract factors before one specifies a SEM. The reduced set of items would significantly facilitate both the interpretation of the EFA, and the specification of factors. Furthermore, a procedure that generates 200 GCB regressions and produces a list of items ranked by importance could easily be implemented in R, and could prove a valuable tool.
In conclusion, the example analyzed in this study demonstrates that GCB can contribute significantly to simplifying the specification search in SEM. The findings suggest that boosting, and in particular GCB, has a potential to facilitate the task of selecting items to specify a SEM when the correlation between the dependent manifest variable(s) and the items is low. More research is needed, however, to determine the full merits of employing boosting in specifying a SEM.
Acknowledgments
Ruralis-Institute for rural and regional research and Renate Butlie Hårstad are acknowledged for facilitating the data set. Professor Ingrid Kristine Glad at the University of Oslo is also acknowledged for valuable comments.
References
- Asparouhov, T., & Muthén, B. (2009). Exploratory structural equation modeling. Structural Equation Modeling, 16, 397–438. https://doi.org/https://doi.org/10.1080/10705510903008204
- Bollen, K. A. (1989). Structural equations with latent variables. John Wiley & Sons.
- Brandmaier, A. M., Prindle, J. J., McArdle, J. J., & Lindenberger, U. (2016). Theory-guided exploration with structural equation model forests. Psychological Methods, 21, 566–582. https://doi.org/https://doi.org/10.1037/met0000090
- Brandmaier, A. M., von Oertzen, T., McArdle, J. J., & Lindenberger, U. (2013). Structural equation model trees. Psychological Methods, 18, 71–86. https://doi.org/https://doi.org/10.1037/a0030001
- Breiman, L. (1998). Arcing classifier (with discussion and a rejoinder by the author). Annals of Statistics, 26, 801–849. https://doi.org/https://doi.org/10.1214/aos/1024691079
- Breiman, L. (2000). Special invited paper. additive logistic regression: A statistical view of boosting: Discussion. TheAnnals of Statistics, 28, 374–377. JSTOR. Retrieved June 11, 2021, from www.jstor.org/stable/2674029
- CRAN. (2020). The comprehensive R archive network. Retrieved October 29, 2020, from https://cran.r-project.org/.
- Duncan, I. J. H., & Fraser, D. (1997). Understanding animal welfare. In M. C. Appleby & B. O. Hughes (Eds.), Animal Welfare (pp. 19–31). CAB International.
- Efron, B., & Hastie, T. (2016). Computer age statistical inference: Algorithms, evidence, and data science. Cambridge University Press.
- Fawcett, T., & Hardin, D. (2017). Machine learning vs. statistics. The Texas death match of data science, august 10 2017. Retrieved February 2, 2019, from https://fullstackfeed.com/machine-learning-vs-statistics-the-texas-death-match-of-data-science/
- Fraser, D., & Weary, D. M. (2004). Quality of life for farm animals: Linking science, ethics, and animal welfare. In M. C. Appleby & B. O. Hughes (Eds.), The well-being of farm animals: Challenges and solutions (pp. 39–60). Blackwell.
- Freund, Y., & Schapire, R. (1996). Experiments with a new boosting algorithm. In Proceedings of the 13th international conference on machine learning: 148–156. Morgan Kaufmann Publishers Inc.
- Frick, H., Strobl, C., & Zeileis, A. (2015). Rasch mixture models for DIF detection: A comparison of old and new score specifications. Educational and Psychological Measurement, 75, 208–234. https://doi.org/https://doi.org/10.1177/0013164414536183
- Friederich, P., Gomes, G. P., DeBin, R., Aspuru-Guzik, A., & Balcells, D. (2020). Machine learning dihydrogen activation in the chemical space surrounding Vaska’s complex. Chemical Science, 11, 4584–4601. https://doi.org/https://doi.org/10.1039/D0SC00445F
- Friedman, J., Hastie, T., & Tibshirani, R. (2000). Additive logistic regression: A statistical view of boosting (With discussion and a rejoinder by the authors). Annals of Statistics, 28, 337–407. https://doi.org/https://doi.org/10.1214/aos/1016218223
- Friedman, J. H. (2001). Greedy function approximation: A gradient boosting machine. Annals of Statistics, 29, 1189–1232. https://doi.org/https://doi.org/10.1214/aos/1013203451
- Hansen, B. G., & Österås, O. (2019). Farmer welfare and animal welfare- Exploring the relationship between farmer’s occupational well-being and stress, farm expansion and animal welfare. Preventive Veterinary Medicine, 170, 104741. https://doi.org/https://doi.org/10.1016/j.prevetmed.2019.104741
- Hastie, T., Tibshirani, R., & Friedman, J. (2009). The elements of statistical learning datamining, inference and prediction (2nd ed.). Springer.
- Hemsworth, P. H., & Coleman, D. L. (2009). Animal welfare and management. In F. J. M. Smulders & B. Algers (Eds.), Welfare of production animals: Assessment and management of risks (1st ed., pp. 133–147). Wageningen Academic publishers.
- Hofner, B., Mayr, A., Robinzonov, N., & Schmid, M. (2014). Model- based Boosting in R. A hands-on tutorial using the R package mboost. The Comprehensive R Archive Network (CRAN). Retrieved September 22, 2019, from https://cran.r-project.org/web/packages/mboost/vignettes/mboost_tutorial.pdf
- Jacobucci, R., Brandmaier, A. M., & Kievit, R. A. (2019). A practical guide to variable selection in structural equation modeling by using regularized multiple-indicators, multiple-causes models. Advances in Methods and Practices in Psychological Science, 2, 55–76. https://doi.org/https://doi.org/10.1177/2515245919826527
- James, G., Witten, D., Hastie, T., & Tibshirani, R. (2017). An introduction to statistica learning with applications in R. Springer.
- Jöreskog, K. G., Olsson, U. H., & Wallentin, F. (2016). Multivariate analysis with Lisrel. Springer.
- Judge, T. A., Thoresen, C. J., Boneo, J. E., & Patton, G. K. (2001). The job-satisfaction- job performance relationship: A qualitative and quantitative review. Psychological Bulletine, 127, 376–407. https://doi.org/https://doi.org/10.1037/0033-2909.127.3.376
- Kim, B., & von Oertzen, T. (2018). Classifiers as a model-free group comparison test. Behavior Research Methods, 50, 416–426. https://doi.org/https://doi.org/10.3758/s13428-017-0880-z
- Long, J. S. (1983). Covariance structure models: An introduction to LISREL. Sage Publications.
- Marcoulides, K. M., & Falk, C. F. (2018). Model specification searches in structural equation modeling with R. Structural Equation Modeling, 25, 484–491. https://doi.org/https://doi.org/10.1080/10705511.2017.1409074
- Mayr, A., Binder, H., Gefeller, O., & Schmid, M. (2014). The evolution of boosting algorithms– from machine learning to statistical modelling. Methods of Information in Medicine, 53, 419–427. https://doi.org/https://doi.org/10.3414/ME13-01-0122
- OIE. (2016). World organization for animal health. Animal welfare and dairy cattle production systems. World Organisation for Animal Health (OIE). Retrieved June 5, 2019, from http://www.oie.int/fileadmin/Home/eng/Health_standards/tahc/current/chapitre_aw_dairy_cattle.pdf.
- Schapire, R. E. (1990). The strength of weak learnability. Machine Learning, 5, 197–227. https://doi.org/https://doi.org/10.1007/BF00116037
- Schmid, M., & Hothorn, T. (2008b). Boosting additive models using component-wise P-splines. Computational Statistics & Data Analysis, 53, 298–311. https://doi.org/https://doi.org/10.1016/j.csda.2008.09.009
- Shortell, T. (2001). An introduction to data analysis & presentation. Retrieved April 29, 2019, from http://academic.brooklyn.cuny.edu/soc/courses/712/chap18.html.
- Tannenbaum, J. (1991). Ethics and animal welfare: The inextricable connection. Journal of the American Veterinary Medical Association, 198, 1360–1376. PMID: 2061152.
- Tarka, P. (2018). An overview of structural equation modeling: Its beginnings, historical development, usefulness and controversies in the social sciences. Quality & Quantity, 52, 313–354. https://doi.org/https://doi.org/10.1007/s11135-017-0469-8
- Tutz, G., & Schauberger, G. (2015). A penalty approach to differential item functioning in Rasch models. Psychometrika, 80, 21–43. https://doi.org/https://doi.org/10.1007/s11336-013-9377-6
Appendix
Table A1. The main results from the SEM in Hansen and Österås (Citation2019) with the two factors FOW and FS, the corresponding items and the regression coefficients between the factors and the AWI
Table A2. Goodness of fit measures for the SEM in Hansen and Österås (Citation2019)
Table A3. Overview over variables used to calculate the AWI, reprinted from Hansen and Österås (Citation2019)
The R-code for boosting
library(mboost)
set.seed(123) # To get repeatable solutions
model <- glmboost(y = y.train, x = X.train, family = Gaussian(),control = boost_control(),center = TRUE)
# This is the model specification with linear base-learners. family specifies the distribution of the
#dependent variable while control is to specify the hyperparameters in the model, see boost_control in
#the package vignette for detailed info. Here one can adjust e.g., the penalizing parameter, the initial
#number of boosting iterations etc., center = TRUE is a logical indicating whether the predictor
#variables should be centered before fitting or not.
summary(model)
cv <- cvrisk(model, folds = cv(model.weights(model), type = “kfold”, B = 10))
# Cross-validation to #find the optimal number of boosting iterations.
mstop(cv)
# the optimal number of boosting iterations selected
summary(model[mstop(cv)])
# A summary of the model
round(coefficients(model[mstop(cv)]), 3)
#Rounding the coefficients to three digits
Here y.train is the AWI for half of the dataset, and X.train is a matrix containing all items and half of the observations in the dataset.
Then a simple lavaan-code with AWI as the dependent variable, and the most frequently chosen items by the GCB grouped in the two factors were run.
ESEM vs BOOSTING
Variable names: v1 = AWI; v2 = item 7; v3 = item 8; v4 = item 9; v5 = item 10, v6 = item 11; v7 = item 1;
V8 = item 2; v9 = item 3; v10 = item 4; v11 = item 5; v12 = item 6; v13 = item 12; v14 = item 13;
V15 = item14; v16 = item 15; v 17 = item 16; v18 = item 17 and v19 = item 18. Sample size = 914.
Figure A2. SEM1 based on ESEM and deleting all items where standardized factor loading < 0.4, absolute value.
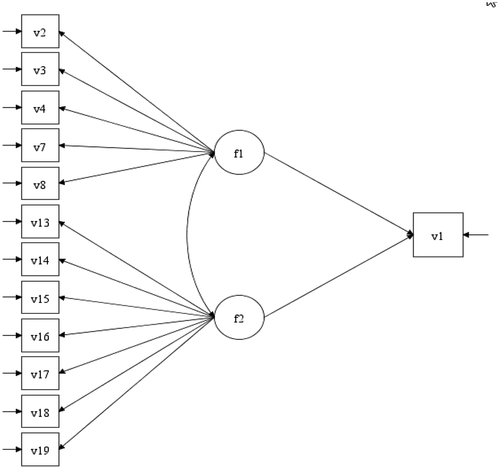
Figure A3. SEM2 based on ESEM and deleting all items where standardized factor loading < 0.5, absolute value.
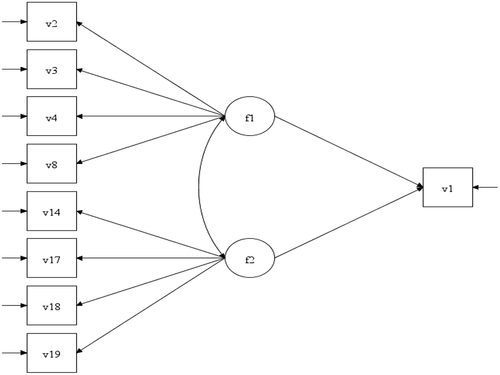
Table A4. SEM 2 depicted in . Estimator DWLS
Table A5. SEM 3 depicted in . Estimator DWLS