Abstract
The evaluation of an alkylating nitrogen mustard agent that utilizes D-alanine as a drug carrier for three chloroethyl substituents (ClCH2CH2-) is shown. Various important pharmacological properties were determined including polar surface area, partition coefficient, molar volume, polarizability, numbers of -OH and -NH2 groups, and aqueous solubility. The synthetic approach utilizes 1,2-dichloroethane reaction with the primary amine of D-alanine resulting in chloroethyl (ClCH2CH2-) substituents. A nitrogen mus-tard group results, and this agent showed alkylation activity in aqueous solution directed toward a nucleophilic primary amine group. Kinetics of alkylation activity is determined by placing p-chloroaniline and the nitrogen mustard agent in sodium bicarbonate buffered aqueous solution at physiological pH 7.4 and 37°C. Samples taken from the test solution are injected with fluorescamine that reacts specifically with primary amine functional groups. Absorbance measurements obtained at 400 nm by UV-Vis spectrometer indicates the relative amounts of nonalkylated p-chloroaniline in the test solution. The D-alanine nitrogen mustard agent effectively alkylated the nucleophilic primary amine of p-chloroaniline with zero-order kinetics. The rate constant was determined to be 7.445E-04 mol/l/min. Formula weight, polar surface area, Log P, molar volume, violations of Rule of 5 for D-alanine mustard agent are 276.59, 29.543 angstroms2, 1.605, 222.3 cm3, and zero violations, respectively. Cluster analysis of molecular properties showed D-alanine mustard agent to be quite similar to cyclophosphamide. This agent showed good druglikeness and zero violations of the Rule of 5, indicating good bioavailability. Molecular properties calculated for the mustard agent are numerically comparable to some clinical anticancer drugs.
Keywords :
Alkylating agents are the most widely used anticancer drugs and consist of highly reactive small molecules that bind covalently to electron rich nucleophilic large or small molecules. More nucleophilic atoms include sulfur, nitrogen, oxygen, and phosporus. The preferred sites of alkylation on DNA are the N-7, O-6, N-1 of guanine,N-7, N-3, N-1 of adenine, N-3 of cytosine, and O-4 of thymidine (Silverman Citation1992). Nitrogen mustards are highly reactive vesicants, strong electrophiles, radiomimetic (their action on DNA resembles radiation), proliferation dependent, and not cell cycle specific (Nogrady Citation1988; Gringauz Citation1997). Nitrogen mustard agents are used for the clinical treatment of tumors from leukemias to solid tumors, in which cyclophospha- mide is the most widely used.
Previous studies have shown that nitrogen mustards alkylate DNA primarily at the N-7 position of guanine (Mattes, Hartley, and Kohn Citation1986) can inhibit enzymes such as prostatic tumor acid phospatase (Workman Citation1978) and cause DNA lesions that induce premature in vitro transcription (Peiper and Erickson Citation1990). Nitrogen mustards (N-mustards) have various levels of effectiveness on leukemia lines (Burchenal, Burchenal, and Johnston Citation1951), favorable results on malignancies of lymphoid and reticular systems (Zulik Citation1958), and Hodgkin's disease (Sampey Citation1963). Various molecular structure designs and carrier drugs have been utilized with N-mustard agents, such as cycloalkyl types (Popp, Roth, and Kirby Citation1963), coumarins (Elderfield and Roy Citation1967), amides (Keitel and Poege 1974), and polyaromatic structures (Mathur, Gupta, and Gupta Citation1977). Previous studies have shown that the simultaneous application of radiation along with a N-mustard agent will enhance the beneficial medicinal effects of certain N-mustards (Conners et al. Citation1965).
N-mustards react via a one-step bimolecular substitution (SN2) or a two-step unimolecular nucleophilic substitution reaction (SN1) (Gringauz Citation1997). For the SN1 reaction the total rate of reaction does not depend on the concentration or nature of the nucleophile. For the SN2 mechanism the rate of reaction depends on both the concentration of the N-mustard agent and nucleophile (bimolecular). Aromatic N-mustards generally will alkylate through a two step unimolecular nucleophilic substitution reaction mechanism, whereas aliphatic N-mustards initially will form an ethylene immonium ion intermediate (Gringauz Citation1997).
This work describes the synthesis, molecular properties, and alkylation activity of a N-mustard agent that uses a D-amino acid as a scaffold. This type of structure has favorable pharmacological properties. Utilizing D-amino acids as drug carriers can produce drugs that have distinct pharmacological advantages, including resistance to normal metabolic degradation (Bladon Citation2002).
MATERIALS AND METHODS
Chemicals and Instrumentation
All reagents were obtained from Aldrich (Milwaukee, WI, USA). Infrared spectra was obtained in dimethylsulfoxide dried over molecular sieves with a Galaxy FTIR spectrometer. UV-Vis spectra were determined using a Spectronic 21D single-beam spectrometer with 1 cm glass cuvettes.
Software and Computations
Molecular modeling and determination of molecular properties were accomplished by ChemSketch (ACD, Toronto, Ontario, Canada). Other molecular properties were determined by Molinspiration (Bratislava, Slovak Republic), Episuite (©2000 U.S. EPA, Washington, DC, USA), Actelion (Allschwil, Switerland), Daylight Chemical Information (Mission Viejo, CA, USA), and MolSoft (La Jolla, CA, USA). Cluster analysis was accomplished by Amada version 2.0.3 (Xia and Xie Citation2001) and KyPlot version 2.0 beta 15 (©Koichi Yoshioka 1997–2001). Multiple regression analysis was accomplished by Statistica (StatSoft, Tulsa, OK, USA). Backpropagation neural network analysis was accomplished by Tiberius v. 1.4.7 (©2001–2003 Philip Brierley; Andrew Lewis). Druglikeness was determined by the methods of Actelion and MolSoft.
Synthesis of D-Alanine Nitrogen Mustard
Place 0.300 g of D-alanine (Aldrich Chemical, St Louis, MO, USA) into acetonitrile previously dried over molecular sieves. Add 1 to 3 ml triethylamine to serve as a proton sink and act as catalyst for reaction. Then administer a flow of ammonia (NH3) gas through the mixture for 30 to 60 min. Degas with nitrogen or air flow thoroughly for ≥30 min to remove residual ammonia (add additional acetonitrile as needed to compensate for evaporation). Add an amount of 1,2-dichloroethane to equal three times the molar amount of D-alanine plus 15% excess to drive reaction. Reflux mildly for 1 to 2 hr. Precipitate product at -10°C, filter out, wash with -10°C diethyl ether, and store at this temperature.
Determination of Alkylation Kinetics
Place 25 mg of D-alanine nitrogen mustard into a total volume of 800 μL of aqueous 0.100 molar NaHCO3 at pH 7.4 having 1.0 mg of p-chloroaniline. Incubate at 37°C removing 60 μL aliquots at known time intervals. Immediately add 60 μL fluorescamine (5 mg/mL in methanol), mix, and read absorbance at 400 nm. The absorbance peak of the fluorescamine-amine derivative occurs at 400 nm (Bartzatt and Kasher Citation2002). This allows monitoring of unreacted p-chloroaniline and thereby the determination of reaction order.
RESULTS AND DISCUSSION
Differences in activity observed among drugs having similar structure is considered to be due to structure activity relationships. These studies help determine characteristics of the pharmacophore or site of biological activity. The differences in effective activity among alkylating agents is due to differences in lipid solubility, penetration of central nervous system, membrane transport, detoxification efficiency, and DNA repair activity (Silverman Citation1992). The use of amino acids as a carrier for anticancer agents has distinct advantages and is the basis of the design of melphalan, which is a phenylalanine construct. Such drug designs may access the membrane transport machinery for amino acids, enhance the therapeutic index, and improve selectivity (Silverman Citation1992). Utilizing D-amino acids as carriers of N-mustard groups may express these same benefits.
D-alanine is utilized for formation of a N-mustard group and an ester chloroethyl substituent. This product will have three potential sites for alkylation of a nucleophile. The molecular structure is shown in with SMILES designation. The three sites of potential alkylation activity are indicated by inset arrows (chlorine atoms leave in this process) with the N-mustard group indicated by inset square. A second structure is shown with atoms numbered for C-13 assignment (in ppm) in the lower portion of . The major steps for synthesis of this agent are presented in and described in detail in Materials and Methods.
Figure 1 The molecular structure of D-alanine N-mustard is shown with the N-mustard group indicated by inset rectangle and sites of alkylation to nucleophiles by inset arrows (SMILES nomenclature is shown). C-13 assignments for numbered carbon atoms are as follows (number/ppm): 2/172.0; 4/69.5; 5/44.6; 7/59.0; 9/53.1; 10/44.7; 12/14.9; 13/53.1; 14/44.7. Chlorine atoms 15, 11, and 6 are removed upon alkylation of a nucleophile.
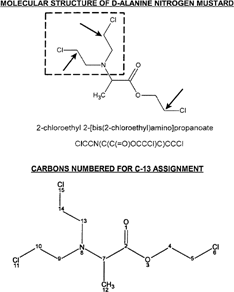
Figure 2 General synthesis represented here shows a two-step approach, where the formation of an ammonium carboxylate is the required first step. Triethylamine serves as a proton sink to protect the ester group that is formed after the second step. A mild reflux in the presence of 1,2-dichloroethane will result in the product having the chloroethyl (ClCH2CH2-) substituents.
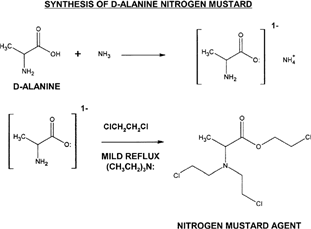
Initially, ammonia gas is circulated in the dry organic solvent containing D-alanine (this must be down in a proper chemical hood with careful attention to avoid inhalation of ammonia fumes). The ammonium carboxylate species formed is highly reactive to alkyl halides and when 1,2-dichloroethane is introduced in mild conditions there is a simultaneous double alkylation of the primary amine group and the negatively charged carboxylate anion. The product is very reactive and should be stored at a low temperature of -10°C to avoid degradation. When stored at low temperature the product was stable for more than 4 weeks. Structural features of D-alanine N-mustard were confirmed through infrared analysis, which showed clearly the chloroethyl substituents associated with N-mustard groups; chlorine atoms were indicated at wavelengths 2700 cm-1 to 2800 cm-1. The formation of an ester group was indicated with peak in wavelengths from 1000 cm-1 to 1300 cm-1.
The alkylation activity was examined by utilizing the chemical reaction of the D-alanine N-mustard with a primary amine nucleophile of p-chloroaniline. As shown in Figure (top portion), the N-mustard agent alkylates the primary amine group that then forms a secondary amine group in the product molecule (). The product formed after one alkylation reaction still retains two additional sites of potential alkylation reaction (inset arrows at product). The amount of p-chloroaniline reactant remaining can be monitored by fluorescamine that is specific for primary amine groups. A strong absorbance peak from the formed fluorescamine-amine derivative is observed at a wavelength of 400 nm (Bartzatt and Kasher Citation2002).
Figure 3 Alkylation of p-chloroaniline is shown here with zero-order plot of reaction kinetics. Note that the N-mustard agent has a total of three potential sites for reaction with nucleophiles, with two remaining (see inset arrows) after the first alkylation reaction. Absorbance versus time data falls within a 95% confidence interval (see lower portion) giving a zero-order rate constant (k=7.445E-04 mol/l/min) from the slope of the regression line.
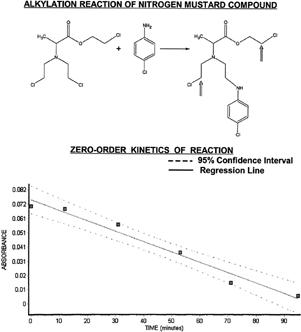
Absorbance data collected at known time periods are indicative of the remaining unreacted p-chloroaniline and extent of alkylation at those conditions (pH 7.4 and 37°C here). A plot of the absorbance versus time is a zero-order plot for determination of rate constant of the alkylation. The lower half of shows the zero-order plot of absorbance versus time, with data points falling within the 95% confidence interval. The regression line has a Pearson correlation coefficient of r=-0.9852 (very high correlation), r2=0.9706 (accounting for 97.06% of variance), and equation of line defined as: y=-0.00074452x + 0.0746774. The slope value is the rate constant -k, making the zero-order rate constant equal to 7.44E-04 mol/l/min. The reaction is zero-order when the rate of reaction is independent of the concentration of the reactants. Therefore, the actual rate of reaction is constant. When a limiting reactant is completely consumed, the reaction immediately stops. The rate law is given as (-d[A]/dt) = k, with the integrated form as [A] = -kt + [A]0.
Important properties were determined for D-alanine Nmustard and are presented in : formula weight (for consideration of the violations of the Rule of 5), the polarizability parameter parachor, polar surface area, partition coefficients, solubility, dermal permeability coefficient Kp, and druglikeness.
Table 1 Molecular properties of D-alanine nitrogen mustard
Some properties were determined by Episuite to be as follows: partition between water and organic layer Log Kow(Log P) = 2.23 (this determination assumes all species present are neutral molecules); rate of acid/base hydrolysis of the ester group having rate constant of Kb = 0.1474 L/mol · sec; melting point = 73.22°C; the rate of skin penetration indicated as Kp = 0.00151 cm/hr; and aqueous solubility at 2.119 g per liter. Having zero violations of the Rule of 5 (determined by Molinspiration) indicates that the D-alanine N-mustard agent will have good bioavailability (Lipinski et al. Citation1997).
Values of Clog P are determined from the sum of molecular fragments of the molecule. Various methods exist for the assessment of druglikeness which utilize topological descriptors. The overall assessment determines the similarity to drugs already in use. The method by Actelion assesses a numerical value of druglikeness between approximately 8 to -13. Similarly for the MolSoft method the druglikeness values range from approximately 2.3 to -2.4. The result for Actelion analysis is 4.59 and for MolSoft analysis is -0.11, which places this alkylating agent well within the ranges for drugs currently in clinical application. The value of the polar surface area at 29.54 cm2 indicates that the expected intestinal absorbance will be more than 90% of the total amount drug present (Palm et al. Citation1997). In addition, the value of polar surface area (at less than 60 cm2) indicates this N-mustard agent can penetrate the blood-brain barrier (Kelder et. al. Citation1999; Clark Citation1999). The Log BB (BB = Cbrain/Cblood) can be determined from the relationship: Log BB = -0.0148PSA + 0.152CLog P + 0.139 where PSA is polar surface area. For the D-alanine N-mustard agent the Log BB value is 0.731 (or BB = 5.38). Previous work has shown that Log BB values greater than 0.3 indicate drugs that readily cross the blood-brain barrier (Clark Citation1999). Therefore it this N-mustard agent could be utilized to attack brain tumors. The value of Clog P at 1.98 falls well within the required range of 2 + 0.5 determined to be necessary to penetrate the blood-brain barrier (Hansch and Leo, Citation1995).
Additional important molecular properties are calculated for D-alanine N-mustard and six anticancer drugs utilized clinically are presented in . These include molar refractivity, molar volume, index of refraction, number of oxygens and nitrogens, and number of hydroxyl and amine groups. Values of miLog P are partition coefficients determined by Molinspiration. The property of molar refractivity correlates the dispersion forces, having the relationship that the larger the formula weight the larger the steric effect and density.
Table 2 Molecular properties of D-alanine nitrogen (N) mustard and clinical agents
Grubb's test for outliers was applied to each set of seven formula weights, miLog P, and polar surface areas, and no outliers were identified which shows that values for D-alanine N-mustard are consistent with those of clinical drugs. Tiberius neural network analysis is a nonlinear back propagation multilayer perceptron algorithm. Utilizing the group values of molar refractivity, molar volume, parachor, index of refraction, and polarizability shown in , this network predicted an miLog P value of 1.37 for D-alanine N-mustard (calculated = 1.605) which incurs only a 14.6% error. This result suggests significant similarity between D-alanine N-mustard and the six clinical drugs included in .
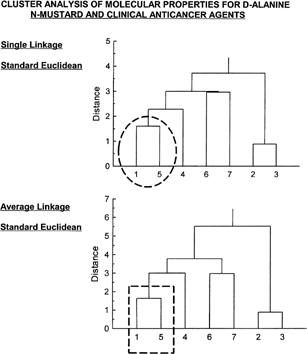
Cluster analysis is a pattern recognition technique that uses multivariate data to determine similarity of cases within the data matrix. Cluster analysis was performed utilizing the molecular properties given in for all seven compounds (indicated by numbers 1 through 7 on the far left-hand side). Cluster analysis evaluations are shown in . Both cluster analysis (single linkage and average linkage) show clearly that D-alanine N-mustard (1) is quite similar to cyclophosphamide (5) (see inset circle [single linkage] and inset rectangle [average linkage] of ), both falling within the identical cluster (cases within a cluster having greater similarity than for cases located outside the cluster). The drugs 6 and 7 (chlorambucil and melphalan) are shown to be most similar and in the same cluster. Likewise, the results show compounds 2 and 3 (mechlorethamine and mustargen) to be most similar.
Figure 4 Cluster analysis of properties shown in for the seven compounds. Both single linkage and average linkage analysis (standard Euclidean for both) show D-alanine N-mustard (1) to be highly similar to cyclophosphamide (5). Drug 6 (chlorambucil) is most similar to drug 7 (melphalan). Drug 2 (mechlorethamine) is likewise most similar to drug 3 (mustargen).
Multiple regression analysis utilizing formula weight as the dependent variable was accomplished with properties polar surface area (TPSA), miLog P, index of refraction (ind of ref), number of oxygens and nitrogens (nOnN), and hydroxyl and amine (nOHnNH) groups (independent variables). The equation accurately predicts formula weights and may predict other analogous anticancer drugs. The equation follows:
Actual formula weight of D-alanine N-mustard is 276.587 compared with the predicted value from the equation to be 276.56.
Multiple regression analysis to predict formula weight was again accomplished utilizing the following molecular properties of the seven compounds in : molar refractivity, molar volume, parachor, and polar surface area (TPSA). The equation obtained is
From this equation the predicted formula weight of D-alanine N-mustard is 276.02 compared with actual of 276.587. This demonstrates that these properties may be used to predict the formula weight of analogous drug designs.
Other properties of D-amino acids support their application as carrier drugs. Previous studies have shown that D-amino acids are highly resistant to proteolysis and they have been evaluated as substituents in antimicrobial agents (Hamamoto et al. Citation2002). D-amino acids incorporated into vaccines resist enzyme degradation and improve the specificity and effectiveness of the compound (Sela and Zisman Citation1997).
Conclusion
An alkylating N-mustard agent was synthesized using D-alanine as a scaffold. This N-mustard effectively alkylated a nucleophilic primary amine group in aqueous solution at physiological pH 7.4 and 37°C. The kinetics of alkylation reaction was zero-order with a rate constant of 7.445E-04 mol/l/min. Zero violations of the Rule of 5 indicate this N-mustard agent will have good bioavailability. The polar surface area and partition coefficient values for this agent indicate it will readily pass through the blood-brain barrier. Molecular properties of D-alanine N-mustard were determined and found to be comparable to those of clinically applied antitumor drugs. Cluster analysis of molecular properties showed D-alanine N-mustard to be quite similar to cyclophosphamide. This alkylating agent also showed good druglikeness as determined by two methods. These results support the potential of utilizing D-amino acids as carrier drugs and their application for clinical application as antineoplastic agents.
This work was funded by the College of Arts and Sciences, University of Nebraska, Omaha, Nebraska, USA.
REFERENCES
- Bartzatt R., Kasher L. Synthesis of aromatic nitrogen mustard agents and analysis of their alkylation activity at physiological pH and temperature. Physiol. Chem. Phys. Med. NMR 2002; 34: 103–117, [PUBMED], [INFOTRIEVE]
- Bladon C. Pharmaceutical Chemistry. John Wiley and Sons, West Sussex 2002; 58
- Burchenal J., Burchenal J. R., Johnston S. Chemotherapy of leukemia. III. Further studies on the effect of nitrogen mustards and related compounds on transmitted mouse leukemia. Cancer 1951; 4: 353–356, [PUBMED], [INFOTRIEVE]
- Clark D. Rapid calculation of polar molecular surface area and its application to the prediction of transport phenomena. 2. Prediction of blood-brain barrier penetration. J. Pharm Sci. 1999; 88(8)815–821, [PUBMED], [INFOTRIEVE], [CROSSREF]
- Conners T., Jeney A., Warwick G., Whisson M. Factors influencing the sensitivity of tumors to nitrogen mustards. Isotop. Exp. Pharmacol. 1965; 1965: 433–438
- Elderfield R., Roy J. Synthesis of potential anticancer agents. XVIII. Nitrogen mustards from 6-substituted coumarins. J. Med. Chem. 1967; 10(5)918–921, [PUBMED], [INFOTRIEVE], [CROSSREF]
- Gringauz A. Medicinal Chemistry. Wiley-VCH, New York 1997; 102–138
- Hamamoto K., Kida Y., Zhang Y., Shimizu T., Kuwano K. Antimicrobial activity and stability to proteolysis of small linear cationic peptides with D-amino acid substitutions. Microbiol. Immunol. 2002; 46(11)741–749, [PUBMED], [INFOTRIEVE]
- Hansch C., Leo A. Explorsing QSAR. American Chemical Society, Washington, DC 1995; 394–395
- Herrero J., Valencia A., Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 2001; 17: 126–136, [PUBMED], [INFOTRIEVE], [CROSSREF]
- Keitel R., Poege A. Testing of some nitrogen mustard compounds for cytosta-tic activity by a screening test. Archiv Fuer Geschwulstforschung. 1975; 43(2)168–171
- Kelder J., Grootenhuis P., Bayada D., Delbressine L., Ploemer J.-P. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm. Res. 1999; 16(10)1514–1519, [PUBMED], [INFOTRIEVE], [CROSSREF]
- Lipinski C., Lombardo F., Dominy B., Feeney P. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Drug Del. Rev. 1997; 23: 3–25, [CROSSREF]
- Mathur I., Gupta H., Gupta S. K. Antitumor activity of certain N-arylmethyl nitrogen mustards in Walker carcinosarcoma 256 and lymphocytic leukemia P 388. Ind. Vet. Med. J. 1977; 1(1)11–19
- Mattes W., Hartley J., Kohn K. DNA sequence selectivity of guanine N-7 alkylation by nitrogen mustards. Nucleic Acids Res. 1986; 14(7)2971–2987, [PUBMED], [INFOTRIEVE]
- Nogrady T. Medicinal Chemistry. Oxford University Press, New York 1988; pp. 412–417
- Palm K., Stenberg P., Luthman K., Artursson P. Polar molecular surface properties predict the intestinal absorption of drugs in humans. Pharm Res. 1997; 14(5)568–571, [PUBMED], [INFOTRIEVE], [CROSSREF]
- Peiper R., Erickson L. DNA adenine adducts induced by nitrogen mustards and their role in transcription termination in vitro. Carcinogenesis. 1990; 11: 1739–1746
- Popp F., Roth S., Kirby J. Synthesis of potential anticancer agents. IX. Some cycloalkyl mustards and related compounds. J. Med. Chem. 1963; 6: 83–86, [CROSSREF]
- Sampey J. Nitrogen mustards in the management of Hodgkin's disease and the leukemias. J. S. Carolina Med. Assoc. 1963; 59: 272–274
- Sela M., Zisman E. Different roles of D-amino acids in immune phenomena. FASEB J. 1997; 11: 449–456, [PUBMED], [INFOTRIEVE]
- Silverman R. The Organic Chemistry of Drug Design and Drug Action. Academic Press, San Diego, CA 1992; 244–251
- Workman P. Inhibition of human prostatic tumor acid phosphatase by N,N-p-di-2-chloroethylaminophenol, N,N-p-di-2-chloroethylaminophenyl phosphate and other difunctional nitrogen mustards. Chem. Biolog. Interactions 1978; 20(1)103–112, [CROSSREF]
- Xia X., Xie Z. AMADA: analysis of microarray data. Bioinformatics 2001; 17: 569–570, [PUBMED], [INFOTRIEVE], [CROSSREF]
- Zulik R. Treatment of malignancies of the lymphoid and reticular systems by a new nitrogen mustard derivative. Arzneimittel-Forschung 1958; 8: 360–361, [PUBMED], [INFOTRIEVE]