Abstract
Magnetic poly (vinyl alcohol) microspheres (MPVAMS) immobilized with iminobiotin as the ligand were prepared for the purification of streptavidin. Parameters of magnetic separation, including spacers, NHS-iminobiotin added, elution behavior, and sample treatment, were optimized to improve the purification efficiency to achieve the maximum recovery of the protein. Streptavidin was successfully purified 38-fold from culture supernatant in a single-step by iminobiotin-MPVAMS with an overall recovery of 90.15% and purity of 95.08%. Hence, this study effectively illustrated the favorable application of magnetic microcarriers for the purification of streptavidin.
INTRODUCTION
Immobilized ligand affinity chromatography is a powerful purification method, which takes advantage of the interaction between biomolecules and coupling ligand [Citation[1]]. Since the introduction of encapsulation of magnetic materials in the 1960s, there have been a broad variety of applications with magnetic microspheres as an easily separable and reproducible method in bioscience and biotechnology [Citation[2], Citation[3]]. The versatility of magnetic poly (vinyl alcohol) microspheres (MPVAMS) has been described for various types of applications including quantitative detection and magnetic affinity separation [Citation[4], Citation[5]], of which MPVAMS were mainly designed for isolation and purification of nucleic acid, proteinase and active proteins [Citation[5], Citation[6]]. To further investigate the performance of MPVAMS in separation, streptavidin, which is a versatile molecule for many in vitro and in vivo applications, was purified by using immobilized iminobiotin-MPVAMS, which could specifically bind streptavidin.
Streptavidin is a nonglycosylated and neutral protein produced by Streptomyces avidinii, which can function high affinity recognition with biotin and biotinylated molecules [Citation[7]]. Isolation and purification of streptavidin by immobilized iminobiotin affinity column, which has now been commercialized, is widely used under mild dissociation conditions [Citation[8], Citation[9]], but no magnetic affinity separation was reported at this stage. In this study, a new chromatographic matrix is prepared by immobilizing iminobiotin to MPVAMS, which displayed a new use for magnetic separation of biological materials with highly favorable practicability and efficiency. We optimized different parameters of separation and provided a single-step purification of streptavidin from culture supernatant, which extended the potential applications of biotin-streptavidin system for further development of immunological and biochemical research.
MATERIALS AND METHODS
Materials
Ferrofluids were synthesized by our laboratory. PVA-1788 (Mr 75,000–79,000, 88% hydrolyzed), glutaraldehyde (EM Grade, Merck, Germany, 25% w/w) were purchased from Tianjin Chemical Agent Co., Ltd. (Tianjin, P.R. China). Gasoline 200# was supplied from Chemical Plant of Nankai University (Tianjin, P. R. China). Sulfosuccinimidyl-6-(biotinamido) hexanoate (EZ-Link Sulfo-NHS-LC-Biotin) and N-Hydroxysuccinimidoiminobiotin trifluoroacetamide (EZ-Link NHS-Iminobiotin) (PIERCE, USA), bovine serum albumin (BSA) (electrophoresis grade, Sigma, USA) and horseradish peroxidase (HRP) (Chromatographic grade, Sigma, USA) were purchased from Tianjin Haoyang Biotechnology Manufacture (Tianjin, P.R. China). O-phenylenediamine (OPD, BR1965), diaminobenzidine (DAB) (Sigma, USA) were purchased from TBD Bio. Co. Ltd. (Tianjin, P.R. China). All other reagents were of analytical grade. Streptomyces avidinii ZG049 was used for ferment production of streptavidin.
Preparation of MPVAMS
MPVAMS were prepared by applying a suspension cross-linking procedure according to the aforementioned method [Citation[10]]. Briefly, 25% glutaraldehyde (GA) solution and 3 M HCl were added to a 10% (w/v) aqueous PVA solution (Mr 75,000–79,000) containing ferrofluids. The mixture was sonicated for 1 min in an ultrasonic bath and then added to an organic phase containing 130 mL gasoline 200#, 120 mL tetrachloromethane and 6 mL Span 80. Suspension cross-linking was carried out by stirring at 70°C for 2 h, after which MPVAMS were isolated on a sieve and washed with petroleum ether and finally with water.
Activation and Immobilization Procedures of MPVAMS
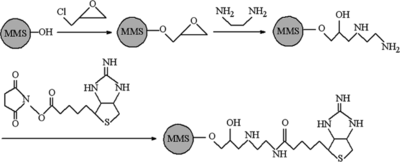
The procedure used to prepare iminobiotin immobilized MPVMS was based on the methods developed before wherein 1,2-ethylenediamine (EDA) was used to as a spacer to covalent immobilize NHS-Iminobiotin onto MPVAMS (see ) [Citation[11]]. MPVAMS were washed three times with phosphate-buffered saline (PBS) (pH 7.4), and mixed with an equal volume of epichlorohydrin (ECH) in 3 M NaOH solution at 40 for 3 h. After being washed thoroughly with acetone and distilled water, the epoxy contents of the microspheres were determined by titration of sodium thiosulfate with hydrogen chloride [Citation[12]]. Then the ECH-activated microspheres were suspended with 10% EDA solution and shaken in ambient temperature for 6 h. Excess EDA was removed by washing and the nitrogen content of the aminated microspheres were determined after pretreatment at 473 K for 2 h using a Vario EL CHNS analyzer (Elementar, Germany). To immobilize NHS-Iminobiotin, aminated microspheres were mixed with 10 mg/mL NHS-Iminobiotin dissolved in 0.1 M Na2CO3-NaHCO3 (pH 8.5). After incubation for 12 h at room temperature, iminobiotin-MPVAMS were washed and stored at 4°C for future use.
X-ray Photoelectron Spectroscopy (XPS) Analysis
XPS spectra were performed on a PHI 5300 XPS system (Perkin-Elmer, USA) using a monochromatic Al Kα radiation (1486.6 eV) at 2 × 10−7 Pa with 13 kV tension and 250 W power. The N 1s and S 2p spectra were recorded to show the evidence of activation and iminobiotin immobilization to MPVAMS. The binding energies (BE) were calibrated against the C1s peak at 284.6 eV [Citation[13]] and the curve fitting was carried out with a mixed Lorentzian–Gaussian function.
Purification of Strepavidin by Iminobiotin-MPVAMS
Streptomyces avidinii was aerobically grown in superrich medium up to 10 days at 30°C. Cells were then centrifuged at 5000 g for 30 min and the supernatant was collected for quantitative analysis by Lowry method [Citation[14]] and sandwich ELISA, respectively. The optimization of different parameters such as spacers, NHS-iminobiotin added, elution behavior, and sample treatment were conducted for magnetic separation by iminobiotin-MPVAMS. After appropriate dilution with binding buffer (50 mM ammonium carbonate buffer, pH 11, containing 500 mM NaCl), the sample was added to a glass containing iminobiotin-MPVAMS and incubated on a shaker at room temperature for 1 h. After magnetic separation, wash was done with binding buffer and elution with 0.1 M acetic acid for 30 min. The eluate was dialyzed against PBS and stored at 4°C. The iminobiotin-MPVAMS were washed with PBS containing 1% sodium hydrazoate and stored at 4°C for reuse.
Biotinylation of Protein
Biotinylation of BSA and HRP was carried out in turn according to biotinylation procedure [Citation[15]]. Briefly, 30 mg protein was first mixed with 2 mL 0.1 M Na2CO3-NaHCO3 (pH 8.5). 3 mg Sulfo-NHS-LC-Biotin was dissolved in 2 mL 0.1 M Na2CO3-NaHCO3 (pH 8.5) and added slowly. The mixture was incubated at room temperature for 2 h with end-over-end rotation. The reaction was stopped by the addition of 1 mL of 1 M glycine and incubated for 1 h. Biotinylated BSA or HRP was dialyzed against PBS and stored at 4°C.
Quantification of Streptavidin by Sandwich ELISA
Streptavidin was quantified by a sandwich ELISA: 96-well plates (Costar, USA) were coated with 100 µL of biotinylated BSA (10 µg/mL) for 2 h at 37°C in 50 mM NaHCO3-NaCO3 (pH 9.6). Then the plates were washed twice with PBST and the wells blocked overnight at 4°C with 200 µL 1% BSA in PBS. After two washes with PBST, the plates were incubated with 100 µL per well of streptavidin standard dilution samples (0.1–10 µg/mL) and serially diluted test samples for 30 min at 37°C. After six washes with PBST, the plates were finally incubated at room temperature for 30 min with 5 µg per well of biotinylated HRP. The plates were finally washed again and 100 µL of an OPD substrate solution was added to each well. After stopping the reaction by adding 50 µL 1 M H2SO4, the optical densities (OD) at 490 nm were determined using a microplate reader (ELX800, Biotech, USA).
Gel Electrophoresis and Densitometry
The qualitative measurement of purified streptavidin was conducted by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), using 8% resolving gel and 4% stacking gel under denaturing and reducing conditions, according to the method of Laemmli [Citation[16]]. The purified streptavidin were resolved on SDS-PAGE and stained with Coomassie brilliant blue R-250. Densitomertic analysis of SDS-PAGE gel was performed using Image Master VDS for data acquisition and Image Master software for integration and analysis.
Western Blot
Following the separation by SDS-PAGE, the gel was soaked in 1% Triton X-100 for 1 h at room temperature (three changes at 500 mL per wash) to remove SDS, which guaranteed the biotin binding activity of streptavidin. After renaturation, proteins were transferred onto nitrocellulose membrane for immunoblot analysis as previously described [Citation[17]]. The membranes were blocked with 5% dried skim milk in PBS overnight at 4°C, washed three times with PBST, and then incubated with 10 µg/mL Biotinylated HRP in PBST. Finally, the membranes were washed again with PBST and then developed with DAB as the substrate.
High-performance Size-exclusion Chromatography (HPSEC) Analysis
HPSEC was carried out for the chromatographic studies of streptavidin with a Shimadzu Model LC-10ATVP HPLC apparatus (Shimadzu, Japan) using a SPD-10AV UV detector, processing the samples on a TSK-GEL G2000SWXL column (30 cm × 7.8 mm i.d.) (Tosoh, Japan). The column temperature was maintained at 25°C. The detection was performed by measuring the UV absorbance at 280 nm and purity was calculated by analyzing the peak area of streptavidin. The mobile phase was 0.15 M NaCl, 0.02 M sodium phosphate, pH 7.0, with a flow rate of 0.8 mL/min.
RESULTS AND DISCUSSION
Preparation and Characterization of Iminobiotin-MPVAMS
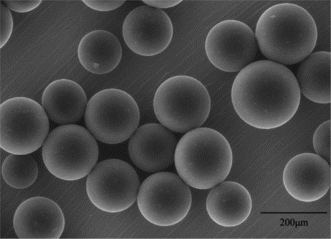
MPVAMS were successfully synthesized by suspension cross-linking technique and sieved with a size distribution from 100 to 200 µm. The morphology and intensity of microspheres were evaluated by environmental scanning electro microscopy (ESEM Quanta 200, FEI, USA) observation. As shown in , MPVAMS had a very homogenous distribution and good spherical geometry with a rather smooth surface and good intensity.
To investigate the effect of surface modification, epoxy and nitrogen contents of MPVAMS after activation by ECH and successive EDA were quantitatively determined. Both 223.83 µmol epoxy group and 477.58 µmol nitrogen content per mL MPVAMS clearly indicated the successful activation for NHS-iminobiotin immobilization.
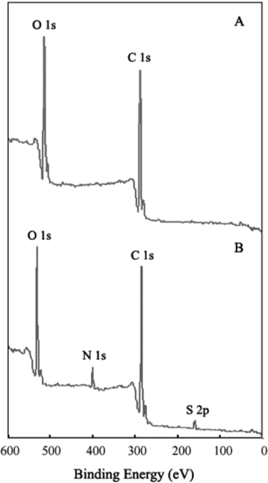
XPS spectrum is a useful tool for characterizing ligand covalent immobilizing onto solid material and also widely used to identify the existence of a particular element in materials [Citation[18]]. shows the results of XPS spectrums of MPVAMS before and after activation and NHS-iminobiotin immobilization. In comparison, it was indicated that the presence of the N 1s peak, centered at 399.98 eV, was assigned to the NH groups of immobilized iminobiotin and EDA. In addition, the appearance of the S 2p peak, at 163.72 eV, certainly identified the existence of iminobiotin as the constituent element of thiazole. Therefore, the XPS spectra provided the evidence of iminobiotin immobilization to MPVAMS, leading to the application of iminobiotin-MPVAMS and the optimization of magnetic separation.
Optimization of Different Parameters of Magnetic Separation
Iminobiotin-MPVAMS were used for the magnetic affinity separation of streptavidin and a number of parameters were optimized to improve the efficiencies of matrices. Spacers, NHS-iminobiotin added, elution behavior, and sample treatment were optimized to give the maximum yield and recovery of streptavidin. All experiments were carried out using 1 mL Iminobiotin-MPVAMS, and then the samples collected after magnetic separation were examined with Lowry method and sandwich ELISA to determine the adsorbed amount and activity of streptavidin.
Effect of Different Spacers
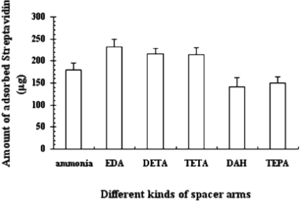
In order to investigate the adsorptive effects of length between ligand and carrier during magnetic separation, different kinds of spacers, i.e. ammonia, EDA, diethylenetriamine (DETA), triethylenetetramine (TETA), 1,6-Diaminlhexane (DAH) and tetraethylenepentamine (TEPA), were selected as amination agent to crosslink NHS-iminobiotin with ECH-MPVAMS. The amount of adsorbed streptavidin in indicates the different lengths of the cross-linking agent, which affected the purification results. From the data of adsorption amount, we could see that EDA, having the length of 8 atoms between iminobiotin and MPVAMS, is the best spacer, which corresponded to 231.43 µg of streptavidin. Further elongation of spacer structure led to decreased affinity, which could be related to deficient and ineffective support of iminobiotin with redundant arm. On the other hand, poor results were also obtained with a too short length of spacer, such as ammonia, due to insufficient surface area and apparent spatial steric effect. Therefore, the optimization of parameters and purification of streptavidin were performed with EDA as a spacer.
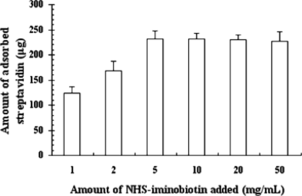
Effect of Amount of NHS-iminobiotin
It is necessary to distinguish the total amount of NHS-iminobiotin added from the binding capacity of iminobiotin-MPVAMS. To optimize the binding efficiency of NHS-iminobiotin, aminated MPVAMS immobilized with different amounts of NHS-iminobiotin were analyzed with respect to the absorbance of streptavidin during purification. The amount of NHS-iminobiotin added to the reaction mixture varied from a 1:200 molar ratio (NHS-iminobiotin to nitrogen content of aminated MPVAMS) to 50-fold higher than the beginning, which corresponded to NHS-iminobiotin added of 1 mg to 50 mg/mL MPVAMS (see ). The amount of adsorbed streptavidin rose up to 231.55 µg when 5 mg NHS-iminobiotin was added to 1 mL aminated MPVAMS. With an increasing amount of NHS-iminobiotin (≥5 mg/mL), no obvious difference in the absorbance of streptavidin was observed because of saturated binding capacity of iminobiotin-MPVAMS. Thus, the optimal amount of NHS-iminobiotin added was 5 mg/mL of MPVAMS.
Effect of Elution Behavior
The binding of streptavidin to affinity matrix was due to the specificity of molecular conformation and hydrogen bond interaction. The elution of bound protein was optimized to give a maximum amount and recovery of streptavidin. Two kinds of reported elution buffers, namely 0.1 M acetic acid and 50 mM ammonium acetate buffer (pH 4.0, containing 500 mM NaCl), were investigated. The elution was performed three times at an interval of 30 min for each. About 97.5% of the adsorbed streptavidin was eluted for the first round with 0.1 M acetic acid, which yielded 231.43 µg of streptavidin. In contrast, only 84.7% of streptavidin and 130.28 µg amount of protein were recovered in the first elution with 50 mM ammonium acetate buffer (pH 4.0, containing 500 mM NaCl). Therefore, elution condition was determined with 0.1 M acetic acid for 30 min.
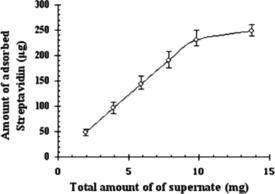
Effect of Amount of Culture Supernatant
A comparative study of adsorbed amount of streptavidin at different supernatant amount from 50 µL to 350 µL (corresponding total protein of 2 mg to 14 mg) using 1 mL iminobiotin-MPVAMS is shown in . When the total protein in the supernatant was less than 10 mg, the adsorption amount of streptavidin could be directly raised by increasing the amount of supernatant, and almost no difference in the protein absorbance was observed with increasing amount of supernatant (>10 mg), which indicated that large amount of lysate interfered with streptavidin binding onto iminobiotin-MPVAMS. The maximum adsorbed amount of streptavidin was 231.43 µg at 10 mg total protein of culture supernatant for 1 mL iminobiotin-MPVAMS in magnetic separation.
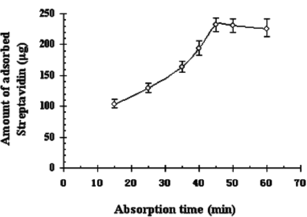
Effect of Adsorption Time
There is no doubt that adsorption time has an important influence on the dynamics of protein, including loss of capacity and rate of protein uptake. As demonstrated in , about half of the maximum amount of streptavidin was adsorbed during the first 15 min, and culmination of 231.43 µg was achieved in the following 30 min. The adsorbed streptavidin kept constant even after prolonging the time, indicating that the equilibrium reached and the time of 45 min was confirmed for adequate binding and strong interaction between streptavidin and iminobiotin-MPVAMS.
Single-step Purification of Streptavidin from Culture Supernatant
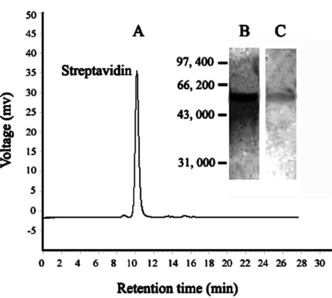
Magnetic affinity separation of streptavidin from culture supernatant after optimization of different parameters was extensively studied. The results shown in suggested that 90.15% protein activity could be recovered with 38-fold purification using iminobiotin-MPVAMS, which corresponded to about 920 mg of purified protein from 1 liter of culture supernatant in a fermenter. The identification of purified streptavidin was based on the pattern of SDS-PAGE (). A major band, about 60.0 kDa, was visualized by Coomassie blue, which agreed with the expected value of intact streptavidin. Purity analysis of streptavidin by HPSEC () showed an overall purity of 95.08%. Western blot () confirmed the identity of the purified streptavidin and the reduction of altered forms.
Table 1 Purification of Streptavidin by iminobiotin-MPVAMS from culture supernatantFootnotea
Figure 8 (A) HPSEC and (B) SDS-PAGE and (C) Western blot analysis of purified streptavidin by iminobiotin-MPVAMS.
After washing with 0.1 M acetic acid and 0.02 M PBS for three and six times, respectively, the iminobiotin-MPVAMS recovered their original capacities. Furthermore, iminobiotin-MPVAMS were reused for the purification of streptavidin for more than ten times, without showing any capacity decrease (data not shown). Good performance of iminobiotin-MPVAMS predicted long-lasting and repeated use in the purification of streptavidin from culture supernatant.
CONCLUSION
Magnetic poly (vinyl alcohol) microspheres (MPVAMS) immobilized with NHS-iminobiotin separated efficiently streptavidin from culture supernatant. Using iminobiotin-MPVAMS, a single-step method to purify streptavidin from supernatant was developed. In addition, the separating capacity was high after repetitive use of iminobiotin-MPVAMS. Preliminary studies show that it holds a promising prospect in the development of immunoassay or immunochemistry in future.
The authors acknowledge Shenqi Wang (Key Laboratory of Bioactive Materials, Ministry of Education, Nankai University) for helpful comments on the experiments.
REFERENCES
- Lee, W.C., Lee, K.H. (2004). Anal. Biochem. 324: 1–10. [INFOTRIEVE], [CSA]
- Chang, T.M.S. (1966). Trans. Am. Soc. Artif. Intern. Organs. 12: 13–19. [INFOTRIEVE], [CSA]
- Ugelstad, J., Soderberg, L., Berge, A., Bergstrom, J. (1983). Nature 303: 95–96. [CSA], [CROSSREF]
- Müller-Schulte, D., Brunner, H. (1995). J. Chromatogr. A 711: 53–60. [CSA], [CROSSREF]
- Safarik, I., Safarikova, M. (2004). Biomagn. Res. Technol. 2: 1–17. [CSA], [CROSSREF]
- Oster, J., Parker, J., Brassard, L. (2001). J. Magn. Magn. Mater. 225: 145–150. [CSA], [CROSSREF]
- Stayton, P.S., Nelson, K.E., McDevitt, T.C., Bulmus, V., Shimoboji, T., Ding, Z., Hoffman, A.S. (1999). Biomol. Eng. 16: 93–96. [INFOTRIEVE], [CSA], [CROSSREF]
- Hofmann, K., Wood, S.W., Brinton, C.C., Montibeller, J.A., Finn, F.M. (1980). Proc. Natl. Acad. Sci. USA 77: 4666–4668. [INFOTRIEVE], [CSA], [CROSSREF]
- Hylarides, M.D., Mallett, R.W., Meyer, D.L. (2001). Bioconjug. Chem. 12: 421–427. [INFOTRIEVE], [CSA], [CROSSREF]
- Zhang, K.S., Sun, J.T., He, B.L. (1999). Acta. Polym. Sin. 6: 662–667. [CSA]
- Cao, Y., Chen, J.Q., Wang, S.Q., Yang, W.B., Bai, G. (2006). Ion Exch Adsorp. 22: 112–119. [CSA]
- Jakubowski, W., Hoff, J.C., Anthony, N.C., Hill, W.F., Jr. (1974). Appl. Microbiol. 28: 501–502. [INFOTRIEVE], [CSA]
- Behra, P., Gissinger, P.B., Alnot, M., Revel, R., Ehrhardt, J.J. (2001). Langmuir 17: 3970–3979. [CSA]
- Lowry, O.H., Rosenbrough, N.H., Farr, A.L., Randall, R.J. (1951). J. Biol. Chem. 193: 265–275. [INFOTRIEVE], [CSA]
- Rosenquist, C., Fledelius, C., Christgau, S., Pedersen, B.J., Bonde, M., Qvist, P., Christiansen, C. (1998). Clin. Chem. 44: 2281–2289. [INFOTRIEVE], [CSA]
- Laemmli, U.K. (1970). Nature 227: 680–685. [INFOTRIEVE], [CSA], [CROSSREF]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K. (1995). Short Protocols in Molecular Biology, John Wiley & Sons, Inc.: New York, 3rd Ed., Chap. 10, pp. 366–373.
- Dambies, L., Guimon, C., Yiacoumi, S., Guibal, E. (2001). Colloid. Surf. A: Physicochem. Eng. Asp. 177: 203–214. [CSA], [CROSSREF]