Abstract
Active inclusion bodies of recombinant polyphosphate kinase were obtained by simple washing of Escherichia coli cells with nonionic detergent and then they were immobilized in agar/TiO2 beads. Bioenergy beads obtained are charged by polyphosphate to act as rechargeable suppply of adenosine/nucleoside triphosphates (ATP/NTP), a practical tool for synthesis of artificial receptors.
INTRODUCTION
The use of biocatalysis for industrial synthetic chemistry is growing rapidly. Most of biocatalysts in current use, however, are limited to cofactor-independent enzymes such as hydrolases, which perform relatively simple chemistry [Citation[1]]. In comparison, all living organisms are capable of using much more complex chemistry by cofactor-dependent enzymes, especially by nucleoside triphosphate (NTP) “burning” enzymes. Adenosine 5′-triphosphate (ATP) is most frequently used as an energy and phosphorylating agent. F0F1 ATP synthase represents the best biological motor for ATP synthesis during life evolution. Although partial success has been achieved in understanding its mechanism [Citation[2]] and reconstituting all the recombinant subunits into active proteoliposomes [Citation[3]], there is still a long way to go to find an efficient proton motive force to power it [Citation[4]]. Therefore, ATP-dependent biocatalytic synthesis has to be driven by other energy systems.
So far, four energy sources have been used as alternative systems, including pyruvate kinase/phosphoenolpyruvate [Citation[5]], acetate kinase/acetylphosphate [Citation[6]], creatine kinase/creatine phosphate [Citation[7]], and polyphosphate kinase (PPK)/polyphosphate (Pn) systems [Citation[8]]. Acetylphosphate, phosphoenolpyruvate, and creatine phosphate are relatively expensive substrates. It is not economically feasible to use these compounds in large-scale synthesis. Polyphosphate (Pn), on the other hand, can be obtained by cheap quenching of molten NaPO3 [Citation[9]], thereby making it the most promising substrate in NTP regeneration. E. coli PPK, the enzyme responsible for NTP regeneration from Pn, has been well characterized [Citation[10], Citation[11]] and crystallized [Citation[12]]. The protein is bound to the outer cell membrane as an active homotetramer. It is possible to overexpress the enzyme in E. coli, even in hexahistidine-tagged form, but the purification has proven to be less effective [Citation[13]].
The overexpression of PPK in E. coli cells leads to accumulation of inclusion bodies; consequently, the main part of PPK protein is in insoluble form. We found that this insoluble form is surprisingly active. This paper describes immobilization and application of PPK inclusion bodies in ATP/NTP synthesis.
EXPERIMENTAL
Materials
pET15b plasmid from Novagen (San Diego, CA, USA) harboring PPK gene from E. coli strain K-12 was prepared at previous work [Citation[13]]. Expression host strain E. coli BL21(DE3) was from Novagen (San Diego, CA). Agar (product no. 05039) from Fluka–BioChemika (Buchs, Switzerland) and titanium dioxide from Kronos, Inc. (Houston, TX, USA) were used for immobilization. Bacterial cell lysis extraction reagent (CelLytic B, product no. B3553), sodium hexametaphosphate (product no. P8510) and all other compounds were from Sigma-Aldrich (Taufkirchen, Germany).
Polyphosphate Kinase Expression and Isolation of the Inclusion Bodies
Freshly transformed E. coli BL21(DE3) harboring the recombinant plasmid were grown in 50 ml of Luria-Bertani (LB) medium (10 g/L of tryptone, 5 g/L of yeast extract, 10 g/L of NaCl) with ampicillin (150 µg/ml) overnight (30°C, 225 rpm), then transferred into fresh LB (1 L) with ampicillin for another 3 h at 37°C. When A600 reached 0.8–1.0, the culture was induced with 400 µM isopropyl-1-thio-β-D-galactopyranoside (IPTG) for 4 h at 37°C. The cells were harvested by centrifugation (4,500 g, 10 min, 4°C) and lysed with nonionic detergent (50 ml, CelLytic B or 10% Triton X-100, 0.5% ethanol, 50 mM Tris-HCl, pH 7.5). After centrifugation of lysate (20,000 g, 10 min, 4°C), the debris were washed twice in 80 ml of buffer (50 mM Tris-HCl, pH 7.5) and harvested by centrifugation. The final cell debris (mainly inclusion bodies) were then suspended in 5 ml of 50 mM Tris-HCl (pH 7.5) and refrigerated.
Immobilization
15 ml of a 2%(w/v) agar solution (60°C) was mixed with 100 mg TiO2 and 5 ml of PPK inclusion bodies (suspension at 60°C). The mixture was dispersed and cooled in 50 ml of vegetable oil (25°C).
Analysis
ATP/ADP were measured by HPLC. The chromatography was performed on a Shimadzu system (LC-10AD, SPD-10AV) with a TESSEK Separon SGX NH2 column (150 × 3.3 mm i.d.; 7 µm) using isocratic elution with 50 mM H3PO4, 10 mM MgCl2 (pH 6.4, triethylamine) at a flow rate of 0.5 ml/min.
RESULTS AND DISCUSSION
First, PPK inclusion bodies were isolated using a commercial bacterial cell lysis extraction reagent (CelLytic B, Sigma product no. B3553). Later, it was found that the undefined nonionic detergent solution can be replaced with 10% Triton X-100. The inclusion bodies had similar activity despite visual observation that the Triton X-100 samples were not so white as when using Sigma CelLytic B solution.
The overexpression of PPK leads to accumulation of adenylate kinase and phosphatases, which compensate PPK activities inside the bacterial cells. Therefore, the application of whole cells in ATP/NTP regeneration results in fast ATP/NTP degradation to AMP/NMP and to adenosine/other nucleosides. On the contrary, the inclusion bodies are almost clear PPK protein without mentioned ADP degrading activities.
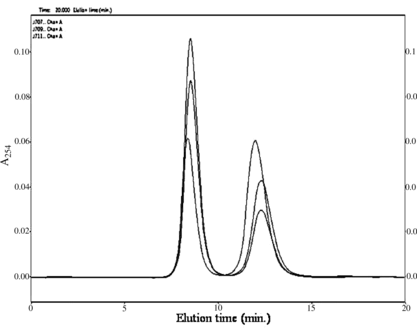
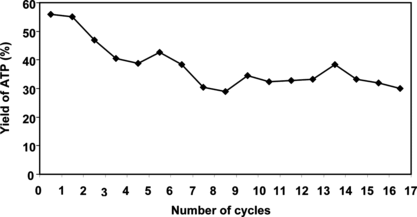
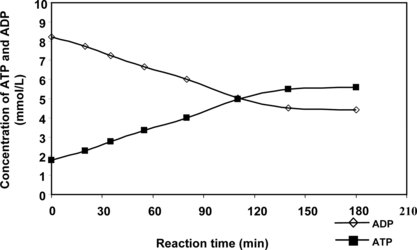
We describe here immobilization and application of PPK inclusion bodies in ATP/NTP synthesis. A simple immobilization procedure was chosen [Citation[14]]: the inclusion bodies were mixed with TiO2 powder and agar solution at 60°C, dispersed and cooled in a mixed vegetable oil. Bioenergy beads prepared like this () were then tested and their activity is depicted in . The reaction was conducted at 30°C in a mixture (10 ml) containing 10 mM ADP, 30 mM MgCl2, 1% (w/v) Pn, 50 mM Tris-HCl (pH 7.8) and 3 ml of agar beads. The progress of the reaction was monitored by HPLC (). It is possible to see from the chromatogram that no ADP dephosphorylation to AMP and adenosine occurs (the elution times of AMP and A are less than 7 min). A time course study of the first reaction cycle revealed that the reaction reached saturation within 3 h (). After 5 h of the reaction, the beads were recovered from the mixture by sedimentation, washed with Tris-HCl (50 mM, pH 7.8), and held at 4°C in refrigerator. The next day they were used for another 5 h reaction. The repeated use of beads is demonstrated in . As expected, these beads were recyclable, but lost some PPK activity during the reaction. The yield of ATP dropped from 55 to 30% during nine reaction cycles. Further conversion cycles were then conducted continuously for 24 h at 25°C. The conversion still has been stopped at 30%; however, no degradation of ADP to AMP/A was observed.
Figure 2 Time course of ATP synthesis by immobilized PPK inclusion bodies. The reaction mixture consisted of 30 mM MgCl2, 1% (w/v) Pn, 10 mM ADP and 50 mM Tris-HCl (pH 7.8) and 3 ml of agar beads in a total volume of 10 ml.
CONCLUSION
Polyphosphate kinase has been immobilized twice before. Hoffman and coauthors [Citation[15]] obtained 390 mg of PPK from 1 kg native fresh E. coli cells after laborious chromatographic separation from ATP hydrolytic enzymes and the enzyme retained only 10.6% activity on glutaraldehyde-activated (2-aminoethyl)cellulose. Liu et al. [Citation[13]] easily isolated and immobilized recombinant PPK using histidine tag on a nickel chelating resin; however, the preparation was unstable. This paper shows that the inclusion bodies formed by over-expression can be easily isolated and freed from ATP-hydrolytic enzymes and proteases. The bioenergy beads can be generated by entrapment of PPK inclusion bodies in agar/TiO2 gel and used as a practical tool in ATP-dependent biocatalytic synthesis. Work is in progress to introduce these beads into the multiple-enzyme systems for oligosaccharide synthesis, forming artificial receptors.
This work was supported by the Slovak Grant Agency for Science VEGA by grants no. 1/4452/07 and 2/4133/04.
REFERENCES
- Zhao, H., Donk, W.A. (2003). Regeneration of cofactors for use in biocatalysis. Curr. Opin. Biotechnol. 14: 583–589. [INFOTRIEVE], [CSA]
- Capaldi, R.A., Aggeler, R. (2002). Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27: 154–160. [INFOTRIEVE], [CSA], [CROSSREF]
- Ishmukhametov, R.R., Galkin, M.A., Vik, S.B. (2005). Ultrafast purification and reconstitution of His-tagged cysteine-less Escherichia coli F1F0 ATP synthase. Biochim. Biophys. Acta 1706: 110–116. [INFOTRIEVE], [CSA], [CROSSREF]
- Nam, K.Y., Struck, D.K., Holtzapple, M.T. (1996). ATP regeneration by thermostable ATP synthase. Biotechnol. Bioeng. 51: 305–316. [CSA], [CROSSREF]
- Ichikawa, Y., Wang, R., Wong, C.H. (1994). Regeneration of sugar nucleotide for enzymatic oligosaccharide synthesis. Methods in Enzymol. 247: 107–127. [CSA]
- Lee, S.G., Lee, J.O., Yi, J.K., Kim, B.G. (2002). Production of cytidine 5′-monophosphate N-acetylneuraminic acid using recombinant Escherichia coli as biocatalyst. Biotechnol. Bioeng. 80: 516–524. [INFOTRIEVE], [CSA], [CROSSREF]
- Zhang, J., Wu, B., Zhang, Y., Kowal, P., Wang, P.G. (2003). Creatine phosphate-creatine kinase in enzymatic synthesis of glycoconjugates. Org. Lett. 5: 2583–2586. [INFOTRIEVE], [CSA], [CROSSREF]
- Noguchi, T., Shiba, T. (1998). Use of Escherichia coli polyphosphate kinase for oligosaccharide synthesis. Biosci. Biotechnol. Biochem. 62: 1594–1596. [INFOTRIEVE], [CSA], [CROSSREF]
- Rashchi, F., Finch, J.A. (2000). Polyphosphates: a review their chemistry and application with particular reference to mineral processing. Miner. Eng. 13: 1019–1035. [CSA], [CROSSREF]
- Akiyama, M., Crooke, E., Kornberg, A. (1992). The polyphosphate kinase gene of Escherichia coli. J. Biol. Chem. 267: 22556–22561. [CSA]
- Tzeng, C.M., Kornberg, A. (2000). The multiple activities of polyphosphate kinase of Escherichia coli and their subunit structure determined by radiation target analysis. J. Biol. Chem. 275: 3977–3983. [INFOTRIEVE], [CSA], [CROSSREF]
- Zhu, Y., Lee, S.K., Xu, W. (2003). Crystallization and characterization of polyphosphate kinase from Escherichia coli. Biochem. Biophys. Res. Commun. 305: 997–1001. [CSA], [CROSSREF]
- Liu, Z., Zhang, J., Chen, X., Wang, P.G. (2002). Combined biosynthetic pathway for de novo production of UDP-galactose: catalysis with multiple enzymes immobilized on agarose beads. Chem. BioChem. 3: 348–355. [CSA]
- Nilsson, K., Birnbaum, S., Flygare, S., Linse, L., Schroder, U., Jeppson, U., Larsson, P.O., Mosbach, K., Brodelius, P. (1983). A general method for the immobilization of cells for preserved vitality. Eur. J. Appl. Microbiol. Biotechnol. 17: 319–326. [CSA], [CROSSREF]
- Hoffman, R.C., Wyman, P.L., Smith, L.E., Nolt, C.L., Conley, J.L., Hevel, J.M., Warren, J.P., Reiner, G.A., Moe, O.A. (1988). Immobilized polyphosphate kinase: preparation, properties, and potential for use in adenosine 5′-triphosphate regeneration. Biotechnol. Appl. Biochem. 10: 107–117. [INFOTRIEVE], [CSA]