Abstract
Numerous proteins are sumoylated in normally growing yeast and SUMO conjugation levels rise upon exposure to several stress conditions. We observe high levels of sumoylation also during early exponential growth and when nutrient-rich medium is used. However, we find that reduced sumoylation (∼75% less than normal) is remarkably well-tolerated, with no apparent growth defects under nonstress conditions or under osmotic, oxidative, or ethanol stresses. In contrast, strains with reduced activity of Ubc9, the sole SUMO conjugase, are temperature-sensitive, implicating sumoylation in the heat stress response, specifically. Aligned with this, a mild heat shock triggers increased sumoylation which requires functional levels of Ubc9, but likely also depends on decreased desumoylation, since heat shock reduces protein levels of Ulp1, the major SUMO protease. Furthermore, we find that a ubc9 mutant strain with only ∼5% of normal sumoylation levels shows a modest growth defect, has abnormal genomic distribution of RNA polymerase II (RNAPII), and displays a greatly expanded redistribution of RNAPII after heat shock. Together, our data implies that SUMO conjugations are largely dispensable under normal conditions, but a threshold level of Ubc9 activity is needed to maintain transcriptional control and to modulate the redistribution of RNAPII and promote survival when temperatures rise.
INTRODUCTION
Nearly one tenth of all human and budding yeast proteins have been identified as targets of SUMO posttranslational modification in nonstress growth conditions, a large fraction of which are involved in gene expression and chromatin maintenance or regulation.Citation1–6 The effects of protein sumoylation are largely target-specific, and include altered localization, activity, stability, and association with other proteins and chromatin.Citation2,Citation6–9 The context-specific nature of the consequences of sumoylation is exemplified by its apparently contrasting roles in regulating transcription.Citation7,Citation10 Whereas tethering SUMO or Ubc9, the SUMO conjugating enzyme, to the promoters of reporter genes dramatically reduced their expression, genome-wide chromatin immunoprecipitation (ChIP) studies showed that sumoylated proteins are found predominantly at promoters of transcriptionally active genes.Citation8,Citation11–16 Furthermore, hundreds of sequence-specific transcription factors (SSTFs) are known SUMO conjugates, and while their sumoylation is frequently associated with reduced expression of target genes, efficient gene activation by some SSTFs has been shown to depend on their sumoylation.Citation9 Sumoylation, therefore, does not appear to have general transcription-promoting or transcription-repressing roles, but instead, it likely controls expression of numerous genes in different ways.Citation7,Citation9,Citation17,Citation18
SUMO conjugation involves the covalent attachment of a SUMO peptide to the side chain of specific Lys residues, through isopeptide bonds, that often lie within the SUMO consensus motif.Citation4,Citation8 It occurs through a cascade of enzymatic activities that is analogous to ubiquitination: activation by an E1 class enzyme and target conjugation by E2 enzymes, which can be facilitated by E3 SUMO ligases that enhance sumoylation and target Lys specificity.Citation8,Citation19,Citation20 Although many proteins are sumoylated, typically only a small fraction (< 5%) of polypeptides of each target protein is modified at any one time, which is partly the result of the constitutive activity of SUMO proteases, including the SENP family of isopeptidases in human cells.Citation21 In yeast, there are two SUMO proteases, Ulp1, which is essential for viability, and Ulp2, which prevents accumulation of polysumoylate chains that form when conjugated SUMO peptides themselves become sumoylated.Citation22–24 In sharp contrast with ubiquitination, the number of SUMO conjugation and deconjugation enzymes is small, with both yeast and mammals harboring only a single E2 conjugase, Ubc9. As such, eukaryotic cells can simultaneously control the sumoylation level of hundreds of substrate proteins by regulating the activity of just one SUMO conjugating or deconjugating enzyme at a time. For example, exposure of yeast to ethanol triggers the nucleolar sequestration of Ulp1, which results in a global increase in protein sumoylation within minutes.Citation25
Such rapid increases in global levels of SUMO conjugation occur in response to a variety of stress conditions, and are collectively referred to as the SUMO stress response (SSR).Citation18,Citation26–28 Exposure of mammalian cells to heat, oxidative, osmotic, or ethanol stress results in a spike in sumoylation levels caused by increased conjugation of numerous proteins with SUMO isoforms SUMO2 and/or SUMO3, specifically.Citation29–31 As with sequestration of Ulp1 in yeast upon exposure to ethanol, the SSR that occurs with heat shock in human cells is at least partly caused by inactivation of multiple SENPs, suggesting that constitutive desumoylation is required for maintaining steady state levels of sumoylation.Citation32 In addition to ethanol exposure, yeast also respond with dramatically elevated SUMO conjugation levels in response to oxidative and osmotic conditions.Citation27,Citation33 Inhibitors of transcription virtually eliminate the SSR in yeast, implying that elevated sumoylation is largely a consequence of stress-induced transcriptional reprogramming and is not necessarily linked to the stress itself.Citation27 However, SUMO2/3 isoforms are required for human cells to survive heat shock, and elevated levels of sumoylation appear to have a protective effect on neuronal tissue during exposure to reduced oxygen and glucose.Citation8,Citation29,Citation34–36 This suggests that elevated sumoylation itself can serve a role in the adaptation of cells to unfavorable conditions, but the physiological effects of modulated cellular sumoylation levels are largely unknown.
To explore this, here we examined how reduced sumoylation affects cell fitness and transcription patterns in yeast. Our data show that most SUMO modifications are not needed for normal growth under nonstress and some stress conditions, but that active SUMO conjugation by Ubc9 is critical when temperatures rise. Moreover, we find that a mild heat shock triggers elevated sumoylation levels at least partly by reducing the abundance of Ulp1. Mutant yeast with dramatically reduced sumoylation levels display widespread changes to RNA polymerase II (RNAPII) occupancy across protein-coding genes in normal growth, and the heat-shock triggered redistribution of RNAPII is greatly exacerbated in this strain. Taken together, our results suggest that, whereas some level of constitutive sumoylation is required to maintain normal transcription patterns, sumoylation and desumoylation systems play largely preemptive roles in preparing yeast for potential exposure to elevated temperature.
RESULTS
Cellular sumoylation levels are modulated under different conditions
Proteomics studies have identified nearly 600 SUMO conjugates in budding yeast,Citation1,Citation3,Citation5 and many of these can be detected by SUMO immunoblot analysis. For example, cell lysates from normally growing, unstressed yeast of the common lab strain W303a were analyzed by immunoblot with the y-84 SUMO antibody, which showed multiple bands as well as unconjugated (“free”) SUMO, as shown in . The detection level of SUMO conjugation was reduced when the SUMO protease inhibitor N-ethylmaleimide (NEM) was excluded from lysate preparations, and sumoylation patterns were shifted upward on the immunoblot when a strain expressing SUMO with an 8xHIS N-terminal epitope tag was used, confirming that the antibody recognizes SUMO and SUMO conjugates, specifically. Intriguingly, the antibody also strongly detected a sumoylated species migrating at ∼30 kDa, which most likely corresponds to Ubc9 modified by SUMO (“Ubc9-S” in diagrams) since (i) it comigrates with Ubc9-SUMO as detected in a Ubc9 immunoblot (, right), (ii) its migration in the 8HIS-Smt3 strain is retarded to a similar degree in the SUMO and Ubc9 blots, and (iii) fusion of polypeptide sequences to Ubc9 results in a corresponding upward shift in migration of this species (see below). Specifically, this species likely derives from Ubc9 modified with SUMO through a Lys-linked isopeptide bond, not through a thioester bond to Cys, since it is resistant to highly reducing conditions (). Altogether, this analysis confirms that we can detect high levels of protein sumoylation in normally growing yeast in addition to unconjugated SUMO and sumoylated Ubc9.
FIG 1 Many proteins are dynamically sumoylated in nonstressed yeast. (A) W303a and 8HIS-Smt3 yeast strains were grown in SC medium at the standard temperature (30 °C) and cell lysates were prepared under nondenaturing conditions, in the presence of NEM, except where indicated. Lysates were analyzed by SUMO, GAPDH, and Ubc9 immunoblots under conditions that facilitate detection of smaller proteins (see Experimental Procedures). The positions of SUMO conjugates, Ubc9-SUMO (“Ubc9-S”), unconjugated SUMO (“Free SUMO”), and unmodified Ubc9 are indicated. (B) Lysates from W303a and 8HIS-Smt3 strains were prepared in sample buffer containing the indicated concentrations of β-mercaptoethanol (β-me), then analyzed by PAGE and SUMO and GAPDH immunoblots. (C) Aliquots of a W303a culture grown in SC medium at 30 °C were collected at six intervals over 15 h and protein extracts, approximately normalized by cell numbers, were prepared and analyzed by immunoblot with antibodies for SUMO, GAPDH, ribosomal protein RPL3, and RNAPII subunit Rpb3. The culture density (cells/mL) at each time point is indicated at top and depicted in the curve below. The inset at the right of the SUMO immunoblot (lane 7; “1.0 norm.”) is from lysate of a culture grown to ∼1.0 × 107 cells/mL in which the exposure (signal intensity) of the SUMO immunoblot was adjusted to approximately match the signal level of the sample in lane 6. (D) The indicated common lab yeast strains, including haploid and diploid W303 (“W303a” and “W303 dip.,” respectively), were grown in either SC or YPD medium to exponential phase, then protein extracts were generated and analyzed by SUMO, Ubc9, and GAPDH immunoblots.
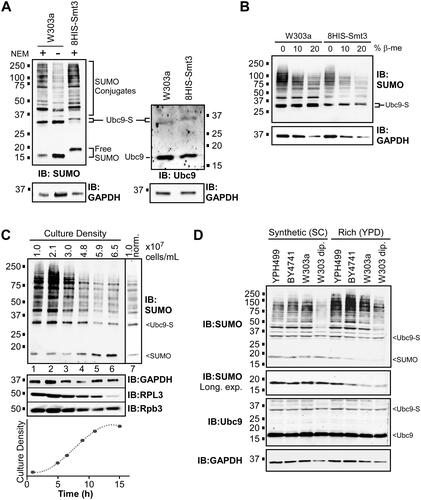
Next, we tested whether factors other than the previously reported stress conditions can affect SUMO conjugation levels. The W303a strain was grown in liquid culture and protein extracts were generated from aliquots taken from the culture at various time points and examined by SUMO immunoblot. Remarkably, the level of sumoylation dropped dramatically as the culture density increased (). This suggests that the physiological changes that accompany the approach to culture saturation as nutrients are depleted, like the diauxic shift, result in diminished SUMO conjugation levels (). As translation rates are reduced at high culture densities (e.g., see Rpb3 in ),Citation37 the effect may also be due to reduced synthesis of some SUMO targets, like the ribosomal protein RPL3 (.Citation5 We compared the overall pattern of sumoylation in high and low culture densities by normalizing the intensity of the SUMO immunoblot signal derived from a culture at 1.0 × 107 cells/mL to that of a culture at 6.5 × 107 cells/mL (; compare lanes 6 and 7 on SUMO immunoblot). The patterns are nearly identical implying that it is mostly the same set of targets that are sumoylated at all densities, but that fewer polypeptides of each target are modified at higher densities. Notably, this comparison also rules out elevated polysumoylation as the major source of increased SUMO signals at low culture densities, since this would appear as an upward shift compared to the signal at the higher density.Citation22 Although levels of sumoylation in dense cultures are reduced, they are not abolished, suggesting that any SUMO conjugation events that are essential for maintaining viability persist as cultures approach saturation. Finally, we compared sumoylation levels in different common lab strains of yeast grown in either synthetic (SC) or nutrient-rich (YPD) media. Minor differences were seen between the YPH499, BY4741, and W303a haploid yeast strains, but significantly less sumoylation was detected in a diploid yeast strain (W303 diploid), and approximately two times more sumoylation was detected when strains were grown in YPD versus SC medium (). Together, our results indicate that yeast cells modulate cellular sumoylation levels in response to culture density, nutrient availability (i.e., medium “richness”), ploidy, and to a lesser extent, strain genetic background.
Yeast tolerate dramatically reduced sumoylation levels with no apparent growth defect
To explore the physiological consequences of reduced global sumoylation, we applied the anchor away methodology to Ubc9.Citation38 By this system, Ubc9 is fused with the FRB domain of human mTOR (“Ubc9-AA”) in a yeast strain that also expresses the ribosomal protein Rpl13A fused to human FKBP12, which binds FRB in the presence of rapamycin. Exposing the Ubc9-AA strain to rapamycin causes nuclear Ubc9-AA to bind Rpl13A-FKB12, which is then rapidly translocated to the cytoplasm, effectively depleting the nucleus of Ubc9 and de novo sumoylation. As most sumoylated proteins are nuclear and associated with chromatin,Citation11 this is expected to cause a significant reduction in overall cellular sumoylation levels. The anchor away parent strain, Parent-AA, harbors a mutation in the TOR1 gene such that exposure to rapamycin has no effect on growth or sumoylation levels (see , and ). We compared sumoylation levels in the Parent-AA and Ubc9-AA strains, which are isogenic except for the C-terminal FRB fusion on Ubc9 in the latter. Notably, even in the absence of rapamycin, about 75% less sumoylation was detected in the Ubc9-AA strain, indicating that the FRB fusion itself hampers Ubc9 activity (). This might reflect a lower abundance of Ubc9-AA compared to normal Ubc9, as detected in a Ubc9 immunoblot (), but we cannot rule out that the fusion inhibits binding of the Ubc9 antibody, which recognizes the C-terminus of Ubc9. Many chromatin-associated proteins are sumoylated in the Ubc9-AA strain (Citation11), and it responds normally to ethanol, oxidative, and osmotic stress with increased SUMO conjugation (Citation27,Citation33), implying that Ubc9-AA maintains many of its normal nuclear functions, but at reduced levels. Addition of rapamycin for 30 min during growth caused a further dramatic reduction in overall sumoylation levels (to ∼3% of parental levels) in the Ubc9-AA strain, which is consistent with the exclusion of Ubc9-AA from the nucleus or further inhibition of Ubc9-AA activity because of its interaction with rapamycin and Rpl13A (). The Ubc9-AA system, therefore, allows us to examine the effects of constitutively low SUMO conjugation levels which can be further reduced by exposure to rapamycin.
FIG 2 Yeast display very high tolerance for reduced sumoylation levels in nonstress growth conditions. (A) Cultures of the Ubc9 anchor away strain, Ubc9-AA, and its parent, Parent-AA, were left untreated or treated with rapamycin (“+Rap.”) for 30 min, then extracts were prepared and analyzed by SUMO and GAPDH immunoblots. SUMO conjugation levels were quantified by densitometry and the average and standard deviations (error bars) from three experiments are plotted at right. (B) Lysates were prepared from cultures of W303a, Parent-AA, Ubc9-AA, or from an unrelated control anchor away strain, Rpb1-AA, and analyzed by immunoblots with antibodies that recognize Ubc9 or a subunit of RNA Polymerase II, Rpb3, which serves as a loading control. (C) Chromatin fractionation was performed with the Ubc9-AA strain, and whole cell extract (WCE), soluble (i.e., nonchromatin), and chromatin fractions (Chrom.) were analyzed by immunoblots with antibodies for SUMO, GAPDH, which is predominantly cytoplasmic, and H3, which is chromatin-bound. (D) The Ubc9-AA strain was grown in the indicated conditions (see Experimental Procedures for details), then lysates were prepared and analyzed by SUMO and GAPDH immunoblot. I A spot assay was performed with the Ubc9-AA and Parent-AA strains, on either SC medium, or SC supplemented with rapamycin. Plates were imaged after 2 days of growth. (F) Cultures of the Ubc9-AA and Parent-AA strains were prepared in SC medium, with or without rapamycin, at an absorbance (595 nm) of ∼0.2, then grown for 20 h while absorbance (culture density) measurements were made every 15 min. The two Parent-AA curves are virtually indistinguishable and are therefore marked with “overlap.” Triplicate cultures were grown for each sample, and the average values were plotted to create the growth curves shown. Values for average absorbance measurements and standard deviations for each triplicate set are listed in Table S5.
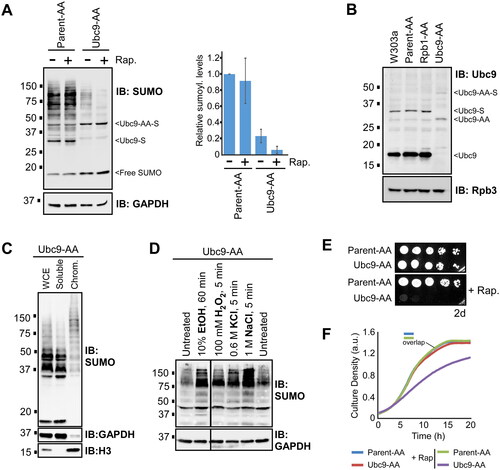
We then compared growth rates of the Parent-AA and Ubc9-AA strains in the presence or absence of rapamycin. Strikingly, despite the ∼75% reduction in sumoylation levels in Ubc9-AA, the strains grew equally well in untreated SC medium, indicating that normal levels of SUMO conjugation are not needed for growth in standard conditions on solid medium () or during exponential growth in liquid culture (). Further reducing sumoylation levels by the addition of rapamycin, however, was lethal to the Ubc9-AA strain, as indicated by spot assay (). This effect is supported by the liquid growth assay that showed significantly slowed escape from lag phase and attenuated exponential growth for Ubc9-AA in the presence of rapamycin, which had no effect on the Parent-AA strain (). As also shown in experiments described below, we note that small differences in our growth curves, specifically at the level of cell saturation, generally relate to much more dramatic growth defects in spot assays. In any case, our data indicate that yeast growth is surprisingly unaffected by a dramatic reduction in cellular sumoylation levels, although levels of ∼3% are lethal.
Yeast with constitutively low Ubc9 activity show high temperature sensitivity
To determine whether the high tolerance for reduced sumoylation can be observed in other contexts, we examined the ubc9-6 and ubc9-1 mutant strains and their respective isogenic parental strains (Fig. S1ACitation11,Citation39) The ubc9-6 and ubc9-1 strains harbor a point mutation that partly inactivates Ubc9, and like Ubc9-AA, they show constitutively low levels of sumoylation, with ubc9-6 showing the most dramatic reduction (∼5%) compared to its parent, W303a (). Consistent with this, spot assays and liquid growth curves showed that only ubc9-6 shows a growth defect, albeit modest, in standard growth conditions, which supports the idea that only a very low threshold level of sumoylation is needed for normal growth ( and ).
FIG 3 Yeast strains with constitutively low levels of Ubc9 activity cannot survive elevated temperatures. (A) Protein extracts were prepared from cultures of the indicated strains grown in untreated SC medium and analyzed by SUMO and GAPDH immunoblots. (B) Spot assays were performed with the indicated strains, onto either SC medium, or SC supplemented with the indicated stressors (see Experimental Procedures) and incubated at the standard growth temperature of 30 °C, or at the elevated temperature of 37 °C. Lower panel shows spot assay for a series of mutant strains, grouped with their respective wild-type parent strains, as positive controls for the various stress conditions. (C) Growth curves were generated, as in , for the indicated strains and conditions. Curves that overlap significantly and are indistinguishable are marked with “overlap.” Individual graphs include two curves except for the Parent-AA and Ubc9-AA set at 37 °C, which includes untreated and rapamycin-treated samples for both strains. Graph data are in Table S5.
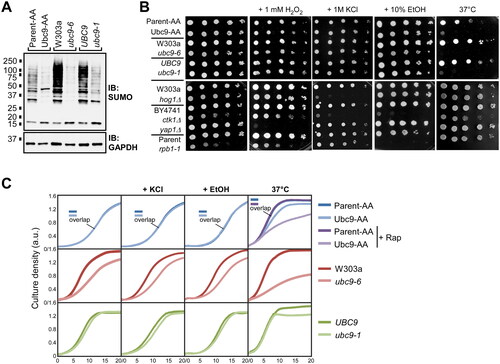
Why do cells maintain levels of sumoylation that are significantly higher than needed for normal growth? Considering that sumoylation levels increase in response to oxidative, osmotic, ethanol, and temperature stresses,Citation26–28,Citation33 we wished to determine whether normal sumoylation levels might play a role in preparing cells for possible exposure to these conditions. Surprisingly, the Ubc9-AA, ubc9-6, and ubc9-1 strains grew as well on medium containing H2O2, KCl, or ethanol as they did on standard medium, relative to their parental strains (). This was also observed in liquid growth assays in media containing KCl or ethanol (), and it indicates that normal levels of sumoylation are not required for survival, or even optimal growth, through these stress conditions. In sharp contrast, all three mutant strains showed a strong growth defect when grown at the elevated temperature of 37 °C on solid medium (), and significantly impaired growth in the liquid culture assay, with ubc9-6 showing the most severe defect (). The ubc9-6 and ubc9-1 strains were previously characterized as temperature-sensitive,Citation11,Citation39 and it was suggested that this sensitivity is due to instability of mutant Ubc9 at elevated temperature (see below). However, also considering the temperature-sensitivity of the Ubc9-AA strain, one intriguing possibility is that these strains are unable to grow well at 37 °C because sumoylation is required to survive when temperatures increase.
Yeast with induced loss of Ubc9 also grow normally except at high temperatures
To examine the effects of reduced cellular sumoylation levels using an additional approach, we employed the Ubc9-TO (“Tet-Off”) strain, in which the UBC9 gene is under the control of a tetracycline repressible promoter.Citation40 The strain was grown for several hours in medium containing varying concentrations of the tetracycline analog doxycycline, then lysates were prepared and analyzed by SUMO and Ubc9 immunoblots. Even at a low concentration of doxycycline (0.5 µg/mL), both Ubc9 and SUMO conjugation showed a dramatic reduction to less than 5% of their normal levels (). We then examined how well the Ubc9-TO strain tolerates low sumoylation levels, and whether reduced sumoylation also confers temperature sensitivity in this strain. Despite the dramatic reduction in sumoylation levels, treatment with doxycycline had no effect on exponential growth of the Ubc9-TO strain at 30 °C, which is again consistent with our finding that yeast cells show high tolerance for low levels of sumoylation when grown at optimal temperatures (). When the analysis was performed at elevated temperatures, however, Ubc9-TO cells treated with doxycycline displayed a growth defect which was more pronounced at 39.5 °C than at 37 °C, whereas the drug had no effect on the isogenic Parent-TO strain at any temperature (). Using this different, inducible system, these observations support our finding that many SUMO conjugation events are dispensable under optimal growth conditions but are needed to prepare cells for elevated temperatures.
FIG 4 Induced reduction of Ubc9 levels also sensitizes yeast to high temperatures. (A) Cultures of a strain expressing the UBC9 gene from a tetracycline-repressible promoter, Ubc9-TO, and its isogenic parent strain, Parent-TO, were exposed to the indicated concentrations of the tetracycline analog doxycycline for several hours, then lysates were prepared and examined by SUMO, Ubc9, and H3 immunoblots. (B) Growth curves were generated, as in , for the Ubc9-TO and Parent-TO strains grown in the absence or presence of doxycycline at the indicated temperatures. (C) Spot assays were performed with the Ubc9-TO and Parent-TO strains, onto either SC medium, or SC supplemented with doxycycline. Plates were incubated at the indicated temperatures and were imaged after 2 days of growth. As none of the strains grow in the presence of doxycycline at 39.5 °C on solid medium, a temperature of 38.5 °C was used as the highest temperature for this experiment. (D) A spot assay was performed comparing growth of the indicated background and parental yeast strains at 30 °C, 37 °C, or 39.5 °C, for the indicated number of days.
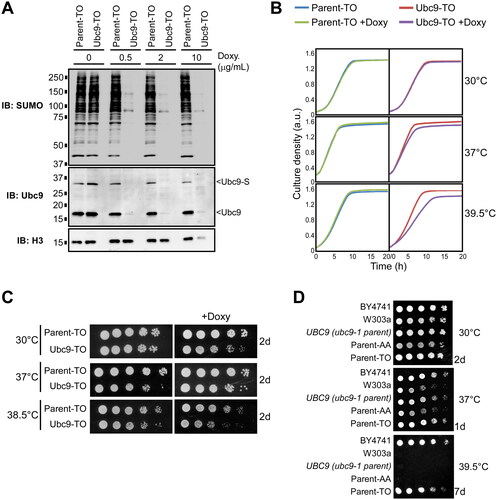
We also examined how reduced sumoylation affects growth of the Ubc9-TO strain on solid medium using spot assays. Parent-TO and Ubc9-TO strains were grown in culture for several hours, either untreated or in the presence of doxycycline, then spotted onto the same medium in solid form. Compared to the Parent-TO strain, a modest growth defect was observed in the Ubc9-TO strain when grown in the presence of doxycycline at 30 °C, suggesting that the dramatic reduction in sumoylation has some impact on growth on solid medium (). The growth defect was more apparent when samples were incubated at 38.5 °C, which was the highest temperature at which either strain could grow on doxycycline-containing solid medium (). As the Ubc9-TO and Parent-TO strains derive from a different background yeast strain (BY4741) than the Ubc9-AA and Parent-AA strains (W303a), we explored whether yeast strains of different origins show differential sensitivity to high temperatures. Indeed, as shown in , BY4741, and its derived strain Parent-TO, show strikingly higher thermotolerance than W303a-derived strains, including the Parent-AA strain and the parental strain for ubc9-1, UBC9. Although this inherent high tolerance for elevated temperatures may obscure temperature sensitivity to reduced sumoylation levels in the Ubc9-TO strain when spot assays are performed, the liquid growth assays shown in support our finding that yeast cells with reduced SUMO conjugation levels show growth defects at high temperatures. These data also imply, however, that W303a-derived strains are more dependent on sumoylation than BY4741-derived strains for growth at high temperatures.
Mild heat shock elevates global sumoylation levels and reduces Ulp1 protein abundance
Previous studies showed that sumoylation levels increase in response to heat shock in human cells and to a high-temperature heat shock (42 °C) in yeast.Citation28,Citation29,Citation32 To examine this in further detail, wild-type W303a yeast cultures were exposed to a mild heat shock (37 °C) for increasing durations, lysates were prepared, then examined by SUMO immunoblot. Indeed, high molecular-weight SUMO conjugation levels increased through the time-course and peaked at 30 min with about 50% more sumoylation (). In contrast, no increase in sumoylation was observed in the Ubc9-AA, ubc9-6, or ubc9-1 strains after a 30-min heat shock at 37 °C (). Potentially explaining this, Ubc9 protein levels decreased in the ubc9-6 and ubc9-1 strains during the heat shock, and Ubc9 levels in the Ubc9-AA strain are constitutively very low ( and ). Although the further reduction in Ubc9 levels in the ubc9-6 and ubc9-1 strains during heat shock may contribute to the temperature-sensitivity of these strains, very low Ubc9 abundance on its own is not sufficient to trigger growth defects, as we observed with the Ubc9-TO strain treated with doxycycline in which Ubc9 levels are nearly undetectable (). In any case, these data demonstrate that even a mild heat shock triggers elevated sumoylation in yeast, and suggest that active sumoylation, from functional levels of Ubc9, is needed for the increase in SUMO conjugation levels.
FIG 5 Mild heat shock elevates global sumoylation levels and reduces Ulp1 protein abundance. (A) Lysates were prepared from W303a yeast cultures that were heat shocked at 37 °C for the indicated durations, followed by SUMO and GAPDH immunoblots. Quantification of sumoylation levels of three independent analyses was performed, with average sumoylation levels graphed and standard deviations shown. Student’s t test analysis indicated that the 30- and 60-min time-point samples are significantly elevated compared to the 0-min control (P < 0.05; asterisks). (B) Cultures of the Ubc9-AA, ubc9-6, and ubc9-1 strains, and their respective parental strains, were heat-shocked at 37 °C for 30 min (+) or untreated (−). Lysates were prepared and analyzed by SUMO, Ubc9, and GAPDH immunoblots. (C) Spot assays were performed with the ULP1 and ulp1-mt strains on SC medium at 30 °C or 37 °C, or under the indicate stress conditions, and imaged after 4 days of growth. (D) Cultures of the ulp1-mt strain, its isogenic parental strain (ULP1), and the wild-type lab strain BY4741 were heat-shocked or untreated, then analyzed by SUMO and GAPDH immunoblots. The analysis was also performed using MG132-treated ulp1-mt cultures (inset; see Experimental Procedures). Quantification is shown at right with average relative sumoylation levels graphed with standard deviations. Student’s t test analysis indicates significantly elevated sumoylation levels in the wild-type strains after heat shock, and a significant but modest reduction in the ulp1-mt strain (asterisks). (E) Cultures of strains expressing HA-tagged versions of Ulp1 or Ulp2, or their parental untagged strain, YPH499, were heat-shocked or untreated. A strain expressing an HA-tagged version of an unrelated protein, Sko1, was used as a control. Lysates were analyzed by HA and GAPDH immunoblots. Quantification of HA-tagged protein levels is shown at right. Student’s t tests support that heat shock significantly reduces Ulp1-HA levels (asterisk) but does not affect Ulp2-HA or Sko1-HA levels. (F) HA and GAPDH immunoblots of extracts prepared using denaturing conditions (TCA precipitation; see Experimental Procedures) from the Ulp1-HA-expressing strain. (G) Cultures of the Ulp1-HA strain were treated with MG132 (+) or DMSO (−), as indicated in Experimental Procedures, then heat-shocked or left at 30 °C. Lysates were prepared and analyzed by HA and GAPDH immunoblots. (H) Cultures of the ULP1 or ulp1-mt strains were grown and aliquots were collected at the indicated culture densities. Lysates were prepared and analyzed by SUMO and GAPDH immunoblots. (I) Aliquots of a Ulp1-HA culture were collected at the indicated culture densities, then lysates were prepared and analyzed by HA and GAPDH immunoblots.
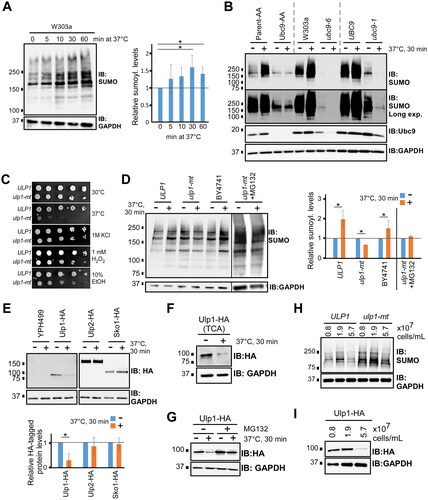
Ethanol stress triggers a marked increase in SUMO conjugation levels through nucleolar sequestration of the primary yeast SUMO protease, Ulp1.Citation25 To explore whether reduced desumoylation may also contribute to the increase in sumoylation levels during heat shock, we examined whether impairment of Ulp1 activity affects the heat shock-mediated increase in SUMO conjugation. The ulp1-mt strain harbors a point mutation (I615N) that reduces the function of Ulp1 and renders the strain temperature-sensitive.Citation41 Through spot assays, we confirmed that ulp1-mt does not grow at 37 °C and found that it is somewhat sensitive to ethanol, but not osmotic or oxidative conditions, which is consistent with a role for Ulp1 in the temperature and alcohol stress responses, specifically (). Cultures of ulp1-mt, its isogenic parent strain (ULP1), and the wild-type lab strain BY4741 were heat shocked, then lysates were prepared and analyzed by SUMO immunoblot (). As expected, the ulp1-mt strain showed higher sumoylation levels than its parent at the normal temperature. Whereas both wild-type strains showed the expected increase in sumoylation levels after heat shock, the ulp1-mt strain instead showed a modest decrease in SUMO conjugation levels. This slight reduction in sumoylation was not observed in yeast treated with the 26S proteasomal inhibitor MG132, indicating that it is likely due to degradation of sumoylated proteins as viability decreases in the heat-shocked ulp1-mt strain (). In any case, the absence of increased sumoylation in ulp1-mt at 37 °C might reflect the already-elevated constitutive levels of SUMO conjugation in that strain. Our data, therefore, suggest that functional levels of both Ulp1 and Ubc9 are needed for the heat shock-triggered increase in SUMO conjugation levels as well as for viability at elevated temperatures.
Protein levels and activities of human SUMO proteases (SENPs) were previously found to be reduced by heat shock.Citation32 We therefore examined whether Ulp1 protein levels are also affected by elevated temperature. Cultures of strains expressing C-terminally 6xHA tagged Ulp1, its untagged parental strain (YPH499), and control strains expressing 6xHA-tagged Ulp2 or the unrelated transcription factor Sko1, were grown and heat shocked at 37 °C for 30 min. Lysates were prepared and examined by HA immunoblot which showed that the elevated temperature did not affect Ulp2-HA or Sko1-HA abundance but, remarkably, it caused a marked reduction in Ulp1-HA levels (). The drop in Ulp1 levels is not likely caused by reduced ULP1 transcription since heat shock does not significantly affect its mRNA levels, as determined by a previous RNA-seq study,Citation42 or the level of RNAPII associated with the ULP1 gene, as determined by our ChIP-seq analysis described below (see Table S1). The heat shock-induced reduction in Ulp1-HA levels was also detected when extracts were prepared using denaturing conditions (trichloroacetic acid; see Experimental Procedures), indicating that the loss of Ulp1-HA signal is not due to reduced solubility of the protein at elevated temperatures (). Notably, however, treatment of cultures with MG132 effectively diminished the reduction of Ulp1-HA at 37 °C, indicating that the loss of Ulp1 at high temperature is due largely to proteolysis (). In addition, we examined whether the reduction in overall sumoylation levels observed at high culture densities (see ) could be the result of elevated Ulp1 activity, but the effect was still observed in the ulp1-mt strain, and Ulp1-HA levels actually diminished at high cell density, ruling out the possibility that Ulp1-mediated desumoylation is responsible for reduced SUMO conjugation as cultures approach saturation (). Together, these data indicate that increased temperature causes elevated SUMO conjugation through active sumoylation, which requires a threshold level of functional Ubc9, and by reduced desumoylation through reduced abundance/stability of Ulp1. If Ulp1 function is already impaired, as in the ulp1-mt strain, the increase in sumoylation during heat shock is not observed.
Yeast with constitutively low sumoylation levels display altered transcription and gene expression patterns
Proteins involved in transcription, and more generally in gene expression, are among the most frequently identified SUMO conjugates across species.Citation1,Citation3–5,Citation9 To explore whether constitutively reduced levels of sumoylation affect transcription and gene expression, we carried out RNA polymerase II (RNAPII) ChIP-seq and RNA-seq analyses in the ubc9-6 strain. Since promoter-proximal pausing is rare in yeast, the level of RNAPII occupancy on gene bodies can be a reasonable measure of transcriptional activity.Citation43 We analyzed data from our previously reported RNAPII ChIP-seq study in which independent duplicate experiments were performed in the ubc9-6 and W303a parental (WT) strains at the normal growth temperature, and after a 12-min heat shock at 37 °C (; Table S1).Citation11 Comparison of RNAPII occupancy levels across all protein-coding genes revealed that, at the normal growth temperature, 736 genes showed significantly increased RNAPII occupancy whereas 846 genes had reduced RNAPII occupancy in ubc9-6 compared to its parent (; Table S1). Sample gene alignments and ChIP-qPCR validations of five genes are shown in Fig. S1C and D, respectively. Gene ontology (GO) term analysis showed that some biological processes are enriched in the subsets of genes that have elevated or reduced RNAPII occupancy in the ubc9-6 strain (; Table S2). For example, as we further discuss below, many genes involved in protein synthesis, including most ribosomal protein genes (RPGs), show significantly elevated RNAPII occupancy when sumoylation levels are low (; ; Table S1; see below).
FIG 6 Constitutively reduced sumoylation alters gene expression patterns and expands the heat shock-induced genomic redistribution of RNAPII. (A) Scatterplot comparing RNAPII levels at all protein-coding genes in WT and ubc9-6 strains, as determined by our previously reported ChIP-seqCitation11 (see Table S1). (B) GO term analysis was performed on genes showing elevated or reduced RNAPII levels across their ORFs in ubc9-6 (see Experimental Procedures and Table S2 for details). (C) Comparison of global steady-state mRNA levels in ubc9-6 and WT strains as determined by RNA-seq (see and S3). (D) Scatterplot comparing ubc9-6-mediated changes in mRNA abundance and RNAPII occupancy for all protein-coding genes based on the RNA-seq and RNAPII ChIP-seq analyses. Genes showing significantly altered levels of both RNAPII occupancy and mRNA abundance are shown in red or blue, as indicated (see Table S4). The four quadrants (Qd1 to Qd4) refer to subsets of genes that show correlated or anticorrelated effects in the RNA-seq and RNAPII ChIP-seq studies (see Table S4). (E) Scatterplots comparing heat shock-induced changes to RNAPII occupancy levels across protein-coding genes in WT and ubc9-6 strains. RNAPII ChIP-seq was performed in both strains in untreated (NT) or heat-shocked cultures (HS; 37 °C for 12 min) as previously described.Citation11 Genes showing significantly elevated or reduced RNAPII occupancy in the heat shock samples are shown in red or blue, respectively (see Table S1). (F) Doughnut plots showing the proportion of protein-coding genes that display altered RNAPII levels after heat shock in WT and ubc9-6 strains. (G) Venn diagram comparing the number of genes showing significantly reduced or elevated RNAPII levels after heat shock in the WT and ubc9-6 strains. The number of genes in each set is shown above the Venn diagram, whereas the numbers of genes that overlap different sets is indicated on the diagram itself.
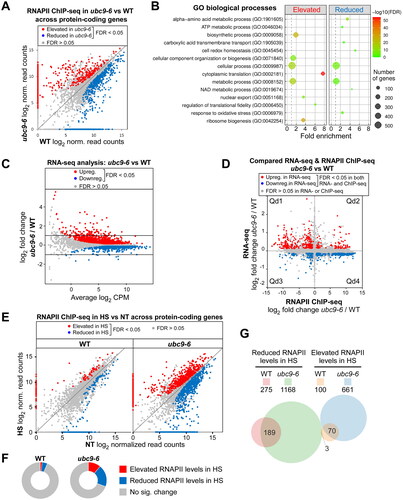
To determine whether changes to genome-wide transcription in the ubc9-6 strain result in corresponding altered steady-state mRNA levels, independent duplicate RNA-seq analyses were performed (; and Table S3). Differential expression analysis revealed that many genes showed a small, significant difference in mRNA levels in ubc9-6 and wild-type yeast, with 700 genes upregulated and 517 genes downregulated in ubc9-6 (; Table S3). GO term analysis identified certain biological processes that are enriched among these groups (; Table S2). We compared our data with a previously reported RNA-seq analysis, which was performed with the ubc9-1 strain and its parent grown in rich medium (YPD; whereas our experiments used synthetic medium, SC).Citation44 As shown in , even though different mutant strains and growth conditions were used, there is substantial overlap in the datasets, supporting the idea that expression of many genes is controlled by cellular sumoylation levels (Table S3). Sample gene alignments from our RNA-seq analysis and RT-qPCR validation of a subset of six genes are shown in and , respectively.
Although approximately one-fifth of all protein-coding genes showed altered expression in ubc9-6 in our RNA-seq analysis, the majority displayed only a minor change (< 2-fold; ). This suggests that the many changes to transcription that occur when sumoylation levels are constitutively low do not necessarily translate to corresponding changes in steady-state mRNA levels. To explore this more directly, we compared our RNAPII ChIP-seq and RNA-seq data, examining genes that are significantly altered in both datasets (referred to as “sumoylation-regulated genes”; Table S4). As shown in , no clear positive correlation was observed, suggesting that sumoylation can regulate transcription and steady-state mRNA levels independently of each other. We examined the genes that lie within each of the four quadrants shown in , representing subsets of sumoylation-regulated genes that are correlated or anticorrelated in the ChIP- and RNA-seq datasets, and GO term analysis identified biological processes enriched in each subset except Qd1 (; Table S2). Intriguingly, most sumoylation-regulated genes showed anticorrelated effects in the two studies (i.e., genes with elevated RNAPII occupancy showed reduced mRNA, and vice versa). This suggests that a mechanism might be in place for compensating abnormal transcription of many genes with a reciprocal change in the stability of their mRNAs to maintain homeostasis of protein expression.
Genes expressing translation-related proteins, including most RPGs, showed the most significant increase in RNAPII occupancy in the ubc9-6 strain, but RPG mRNA levels were unaffected or modestly reduced in ubc9-6 ( and ), similar to what was previously reported for RPG mRNAs in the ubc9-1 strain.Citation44 Consistent with minimal changes to mRNA levels, we found that levels of proteins expressed by two RPGs, RPL5 and RPL20, are unaffected in the ubc9-6 strain (). To explore why increased transcription of RPGs does not lead to increased RPG mRNA levels, we first examined whether high rates of transcription of RPGs are countered with elevated degradation of RPG mRNAs. However, after blocking transcription with the inhibitor thiolutin, RNA levels of RPL5 and RPS20 did not show elevated rates of decay in the ubc9-6 strain (). We then considered the possibility that the increased transcription of RPGs in the ubc9-6 strain is not productive, generating many transcripts that fail to be processed into mRNAs. To address this possibility, we examined two intron-containing RPGs, RPL25 and RPS27B, both of which showed highly elevated RNAPII occupancy in ubc9-6 (; top). Processed (polyA-containing) mRNA levels for the two genes were approximately equal in the WT and ubc9-6 strains, as determined by our RNA-seq analysis (; bottom). However, when we examined the level of precursor mRNAs from the two genes, by qRT-PCR with primers specific for intron-containing transcripts, we observed significantly higher levels in the ubc9-6 strain (; middle). Together, these data indicate that, when sumoylation levels are reduced, RPG transcription increases, but many of the excess transcripts that are produced fail to be processed into functional mRNAs.
The heat shock-induced genomic redistribution of RNAPII is greatly expanded when sumoylation levels are low
Heat shock in yeast is accompanied by rapid changes to gene expression patterns.Citation42,Citation45–47 Considering that increased temperature also triggers elevated sumoylation, we examined whether constitutively low sumoylation levels affect the transcriptional response to heat shock. Occupancy levels of RNAPII across protein-coding gene bodies were compared in wild-type and ubc9-6 strains after a 12-min heat shock at 37 °C. In the wild-type strain, a small number of genes (∼6%) showed significantly increased or decreased RNAPII occupancy after the heat shock ( and ; Table S1). This is less than the reported ∼10–15% of genes that show altered mRNA levels after heat shock,Citation47 which might reflect that the expression of many heat shock-regulated mRNAs can occur posttranscriptionally (e.g., by regulating their stability), or that our differential binding analysis comparing the untreated and heat shocked RNAPII ChIP-seq datasets was highly stringent. Strikingly, however, under the same growth and analysis conditions as for the wild-type strain, the ubc9-6 strain displayed a far more dramatic change to RNAPII occupancy levels across gene bodies ( and ; ; Table S1). Indeed, ∼30% of all protein coding genes showed elevated or decreased RNAPII levels after heat shock in ubc9-6. This includes most of the genes that show heat shock-altered RNAPII levels in the wild-type strain plus a large number of additional genes (). This analysis strongly suggests that normal levels of Ubc9 activity and/or sumoylation are important for restraining the redistribution of RNAPII that occurs in response to elevated temperatures.
DISCUSSION
A variety of environmental stressors have been shown to trigger rapid increases in SUMO conjugation levels in yeast, plants, and mammalian systems, leading to the concept of the SSR.Citation26,Citation48,Citation49 In yeast, these include exposure to oxidative, osmotic, and ethanol stresses, high-temperature heat shock, nitrogen starvation, and DNA damaging agents.Citation25,Citation27,Citation28,Citation33 We have now identified nonstress conditions that result in dramatically modulated sumoylation levels. Specifically, we found that diploid yeast harbors less sumoylation than their haploid counterparts, and that nutrient availability, or the enhancement in growth that it enables, has a positive effect on levels of SUMO conjugation. This was apparent by comparing SUMO conjugation levels in yeast grown in rich versus synthetic medium, and by monitoring sumoylation during growth in liquid culture. Sumoylation levels were greatest during early exponential growth, but dropped dramatically with the diauxic shift, which occurs as glucose becomes depleted. Many proteins that become differentially sumoylated during the SSR have been identified, leading to speculation that certain processes, in particular transcription by RNAPII or RNAPIII, are influenced by altered sumoylation levels during stress.Citation27,Citation28,Citation33 However, Lewicki et al. found that impairing transcription prior to exposing yeast to osmotic stress prevented a surge in SUMO conjugation levels, leading the investigators to speculate that the SSR is a consequence of altered transcription programs that occur during stress exposure and are not directly part of the stress response itself.Citation27 Likewise, the elevated sumoylation that we detected in yeast during nutrient-rich and exponential growth conditions may simply be a consequence of increased rates of certain physiological processes that involve sumoylated proteins. At the same time, elevated sumoylation itself might facilitate these processes and/or high rates of growth.
Arguing against a role for high levels of sumoylation in facilitating growth, however, is our finding that yeast with dramatically reduced sumoylation levels shows no apparent growth defect, implying that cellular processes that are normally affected by sumoylation remain functional. Remarkably, on both solid media and in liquid culture, strains with low Ubc9 activity, either through mutation (ubc9-1) or reduced Ubc9 expression (Ubc9-AA, Ubc9-TO in presence of doxycycline), have normal growth rates, even when sumoylation levels are ∼75% lower than in wild-type counterparts. This strongly implies that most SUMO conjugation events are dispensable under normal growth conditions, even during exponential growth where sumoylation levels normally rise. Consistent with the finding that multiple components of the sumoylation pathway are essential, including Ubc9 and Ulp1, we found that a low threshold level of sumoylation, of greater than ∼3% of wild-type levels, is needed for normal growth, however.Citation23,Citation50 Indeed, the ubc9-6 strain, which shows a more dramatic reduction in sumoylation than the ubc9-1 or untreated Ubc9-AA strains, has a modest growth defect, and the Ubc9-AA strain treated with rapamycin, which nearly abolishes sumoylation, cannot grow. This might reflect that there are essential roles for the modification in some processes, such as progress thorough mitosis and the cell-cycle.Citation23,Citation50,Citation51 The bulk of sumoylation, however, is not needed and might be a consequence of physiological processes that involve sumoylation but do not require much sumoylation. Alternatively, as we propose below, constitutive Ubc9 activity might be required preemptively for protecting yeast from some, but not all, anticipated stress conditions.
Our examination of the effects of reduced sumoylation on transcription showed that about a quarter of all protein-coding genes showed significantly elevated or reduced RNAPII occupancy levels in the ubc9-6 strain compared to its wild-type parent. This indicates that reduced sumoylation has a widespread effect on transcription by RNAPII and implies that normal levels of sumoylation are needed to maintain global transcriptional patterns. Transcriptome analyses in human cells in which SUMO1 or Ubc9 expression levels were reduced by RNA interference showed that steady-state mRNA levels of a variety of gene classes were upregulated or downregulated when sumoylation levels dropped.Citation14,Citation15 Our RNA-seq analysis in the ubc9-6 strain, however, showed only modest changes to the mRNA levels of a number of genes compared to wild-type, which is consistent with a previous study using the ubc9-1 mutant which found that the transcriptome was barely affected by the mutation.Citation44 In yeast with constitutively low sumoylation, then, it is possible that an adaptive mechanism is in place to buffer changes in transcription, perhaps by controlling RNA degradation rates correspondingly, such that the transcriptome is mostly unaffected. Supporting this, we find that most of the small but significant changes to mRNA levels in ubc9-6 anticorrelate with changes to RNAPII occupancy on the corresponding genes, as though cells are working to elevate mRNA stability for genes with abnormally reduced transcription levels, and reduce stability for mRNAs of genes with abnormally elevated transcription. Indeed, systems for buffering changes in mRNA synthesis, due to perturbation of transcription, with reciprocal changes to mRNA degradation have been described.Citation52,Citation53 In contrast, as we showed for RPGs, for genes that show elevated RNAPII occupancy but little or no change to mRNA levels in the ubc9-6 strain, it is possible that the excess transcripts that are produced are not efficiently processed (i.e., spliced and polyadenylated) into functional mRNAs that can be detected by RNA-seq that is specific to polyA + mRNAs. Supporting this idea, sumoylation has been previously implicated in the efficient splicing of pre-mRNAs.Citation54
The rise in SUMO conjugation that occurs as part of the SSR after exposure to a variety of stress conditions suggests that sumoylation plays a protective role or facilitates the stress response.Citation18,Citation26 It is surprising then, that we observed no growth defect in yeast with low Ubc9 activity and reduced sumoylation levels that were exposed to oxidative, osmotic, or ethanol stress. Our observations align with the proposal that the SSR is primarily a consequence of altered transcription genome-wide, which involves a wave of transcription-mediated SUMO conjugation, and suggest that high levels of overall sumoylation do not play a protective role during these particular stresses.Citation17,Citation27 In contrast, all tested strains with reduced Ubc9 activity, and consequently low sumoylation levels, showed growth defects when incubated at elevated temperatures, with most showing severe growth impairment. This is consistent with the finding that sumoylation facilitates heat tolerance in human cells and plants.Citation29,Citation55,Citation56 For example, siRNA-mediated depletion of SUMO1 and SUMO2 levels resulted in a 7-fold reduction in survival after heat shock in human U2OS cells.Citation29 Heat shock in human cells leads to increased polysumoylation of proteins involved in multiple processes, including transcription, and it triggers altered sumoylation of promoter- and enhancer-bound proteins, specifically.Citation18,Citation29 These findings suggest that the elevated sumoylation that occurs with high temperature can function, at least in part, to adjust transcriptional programs as needed for surviving heat stress.
We explored this by examining how reduced constitutive sumoylation levels affect the transcriptional response to heat shock. In yeast, 10–15% of genes show upregulated or downregulated mRNA levels in response to heat shock.Citation45–47 Although heat shock can change steady-state mRNA levels through regulation of mRNA stabilities, much of the heat shock-altered transcriptome is likely due to changes at the transcriptional level.Citation57 Indeed, significant reorganization of multiple transcription-related factors on chromatin has been observed in quick response to elevated temperature.Citation47 In our analysis, ∼6% of protein-coding genes showed altered RNAPII occupancy levels in wild-type cells after heat shock, but this number increased dramatically to ∼30% in the ubc9-6 strain. This strongly implicates Ubc9 and sumoylation in controlling or restraining the transcriptional response to elevated temperatures. More specifically, we believe that a rise in SUMO conjugation levels, as opposed to merely a threshold constitutive level of sumoylation, is needed to endure high temperatures, at least partly by regulating transcription. Supporting this, we observed that active sumoylation (i.e., functional levels of Ubc9) is required, but also that Ulp1 protein levels drop significantly after heat shock, implying that reduced desumoylation plays a major, evolutionarily conserved role in elevating sumoylation levels when temperature rises.Citation32 Taken together, our results demonstrate that SUMO conjugations, although plentiful, are largely dispensable for normal growth in many stress and nonstress conditions. However, we find that active sumoylation and desumoylation systems are critical for enduring elevated temperatures where they function, at least in part, to temper the transcriptional response to heat shock. Further study in this area will be invaluable for uncovering the molecular mechanisms by which sumoylation facilitates heat tolerance in eukaryotes.
EXPERIMENTAL PROCEDURES
Yeast media and growth
Yeast strains used in this study are listed in . Previously unreported strains were generated by transformation with PCR-amplified DNA fragments that were incorporated at specific genomic loci through homologous recombination.Citation61 Unless otherwise noted, yeast were grown at 30 °C in synthetic complete (SC) medium. Overnight cultures were diluted to an absorbance (595 nm) of ∼0.2 in a volume of 10–50 mL, then grown to exponential phase (absorbance <1.0) unless otherwise noted, treated as indicated below if appropriate, then harvested by centrifugation at 3000 g for 5 min. Where appropriate, the following were added to media (final concentrations indicated): 0.6 M KCl for 5 min for liquid cultures, 1 M KCl on solid media; 1 M NaCl; 100 mM H2O2 for 5 min for liquid cultures, 1 mM H2O2 for solid media; 10% ethanol for 60 min for liquid cultures, 10% ethanol for solid media; 1 µg/mL rapamycin for 30 min, or as indicated; doxycycline, 10 µg/mL, or as indicated, for duration of culture growth. For inhibition of the 26S proteasome, 75 µM of MG132 was added to cultures for 1 h prior to further treatment or harvesting. To increase permeability of the drug, the method of Liu et al. was used.Citation62 Where rapamycin or MG132 were added, the control or “untreated” sample was supplemented with the same volume of their solvent, DMSO. Chromatin fractionation was performed as previously described.Citation63
TABLE 1 Yeast strains used in this study
Protein lysates and extracts
Protein samples were generated under nondenaturing conditions (“lysates”) or through precipitation with trichloroacetic acid (TCA; “protein extracts”). For lysate preparation, pellets were washed with ice-chilled IP buffer (50 mM Tris-HCl, pH8; 150 mM NaCl; 0.1% Nonidet P-40 (NP40); supplemented with a protease inhibitor cocktail, 1 mM phenylmethylsulphonyl fluoride (PMSF), and 2.5 mg/mL N-ethylmaleimide (NEM)) at a volume equal to the culture volume, then resuspended in 500 µL of IP buffer. Acid-washed glass beads (0.25 g) were then added, and samples were vortexed at 4 °C for 30 min, with a 5-min ice incubation after the first 15 min. Lysates were transferred to fresh microfuge tubes and clarified by two rounds of 5-min 4 °C centrifugation at maximum microfuge speed. Prior to analysis by immunoblot, an equal volume of 2X sample buffer (140 mM Tris-HCl, pH 6.8; 4% SDS; 20% glycerol; 0.02% bromophenol blue; supplemented with 10% 2-mercaptoethanol prior to use) was added, and samples were boiled for 4 min.
For protein extract preparation through TCA precipitation, volumes equal to 2.5 absorbance units (595 nm) of the exponentially growing yeast cultures, treated if necessary, were centrifuged at 3000 g, then resuspended in 1 mL of ice-cold water. 150 µL of freshly prepared extraction buffer (1.85 NaOH; 7.5% 2-mercaptoethanol) was added, and the mixed samples were incubated on ice for 10 min. Then, 150 µL of cold 50% TCA was added, samples were mixed, then incubated again on ice for 10 min before they were centrifuged at 4 °C at maximum microfuge speed. Supernatants were removed and the protein pellets were resuspended in 100 µL of 2X sample buffer, with 5–10 µL of Tris-HCl, pH 8 added if the sample was yellow in color, to raise the pH and restore the blue color. Samples were boiled for 4 min, centrifuged for 3 min at maximum microfuge speed to separate debris, then used for immunoblot analysis.
Immunoblots and quantifications
Protein samples were analyzed by standard polyacrylamide gel electrophoresis (PAGE) techniques. For preferential detection of high-molecular weight SUMO conjugates, PAGE was performed with lysates followed by wet transfer (100 V for 1 h) in a buffer consisting of 0.1% SDS, 10% methanol, 50 mM Tris-HCl, and 380 mM glycine. For preferential detection of free SUMO and lower molecular-weight SUMO conjugates, TCA extracts were analyzed by PAGE, followed by semi-dry transfer using a Power Blotter system (Thermo Fisher). Chemiluminescence-based imaging was performed using the MicroChemi imager (DNR). For quantification of signals, TIFF-format images were analyzed using ImageJ software (version 1.52a; NIH). Specifically for determining sumoylation levels, all SUMO signals (above the Ubc9-SUMO band, if it was present) were considered SUMO conjugates, and normalization was made to the corresponding signal from GAPDH immunoblots. Antibodies used for immunoblot analyses were: 1:500–1:1000 SUMO/Smt3 (y-84; Santa Cruz, sc-28649); 1:3000 GAPDH (Sigma, G9545); 1:500 Ubc9 (Santa Cruz, sc-6721); 1:3000 histone H3 (Abcam, ab1791); 1:1000 Myc epitope tag (Sigma, 05-724); 1:1000 Rpb3 (Abcam, ab202893); 1:5000 HA (12CA5; Sigma, 11583816001). The RPL3 antibody, which we used at a dilution of 1:200, was deposited to the DSHB by Warner, J. R. (DSHB Hybridoma Product ScRPL3 supernatant).
Spot assays and liquid growth assays
Yeast strain fitness was examined by spot assays, as previously described,Citation64 or liquid growth analysis. To generate liquid growth curves, growing cultures were diluted to an absorbance of 0.2–0.5 in appropriate medium and treated if needed. Each sample was transferred, in triplicate, to wells in a 96-well microtiter dish, and inserted into an accuSkan™ absorbance microplate reader (Fisherbrand™, Thermo Fisher), which shook and incubated samples at the appropriate temperature while taking absorbance measurements (595 nm) at 15-min intervals over a course of 20 h. Background absorbance readings were made from wells containing uninoculated medium, and the background-subtracted average absorbance readings, and standard deviations, are listed in Table S5. Curves shown in figures were generated using average values without standard deviations indicated since they are minimal and would otherwise obscure curves on the diagrams. Because the light path distance in the accuSkan™ is less than 1 cm, absorbance readings are indicated as arbitrary units (a.u.) and are not directly comparable to values obtained through a standard spectrophotometer.
RNA-seq and RNAPII ChIP-seq analysis
RNA preparation and ChIP were performed as previously described.Citation11 Details of the ubc9-6 RNA-seq experiment are presented in . RNAPII ChIP-seq analysis was previously reported,Citation11 with the heat shock-treated samples (37 °C for 12 min) analyzed in the same manner. Bioinformatics analysis of the previously published ubc9-1 RNA-seq datasetCitation44 was also performed as indicated in . Read counts and differential expression analysis results for the RNA-seq analyses are in Table S3, and RNAPII ChIP-seq analysis results are in Table S1. GO term analysis and visualization were performed as previously describedCitation65 with data shown in Table S2. Validation qRT-PCRs, and the analyses in and , were performed as previously described using primers indicated in .Citation11
TABLE 2 Analysis details for RNA-seq in ubc9-6 vs WT
TABLE 3 Sequences of oligonucleotides used in this study
Supplemental Material
Download Zip (6 MB)SUPPLEMENTAL MATERIAL
Supplemental data for this article can be accessed online at https://doi.org/10.1080/10985549.2023.2166320.
DISCLOSURE STATEMENT
No potential conflict of interest was reported by the authors.
DATA AVAILABILITY STATEMENT
Data for the RNA-seq analysis with WT and ubc9-6 strains were deposited in the Gene Expression Omnibus (GEO) repository with accession number GSE167427 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167427). The GEO accession number for the previously reported RNAPII ChIP-seq with the same strains is GSE167424 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167424).
Additional information
Funding
REFERENCES
- Albuquerque CP, Yeung E, Ma S, Fu T, Corbett KD, Zhou H. A chemical and enzymatic approach to study site-specific sumoylation. PLoS One. 2015;10:e0143810. doi:10.1371/journal.pone.0143810.
- Boulanger M, Chakraborty M, Tempé D, Piechaczyk M, Bossis G. SUMO and transcriptional regulation: the lessons of large-scale proteomic, modifomic and genomic studies. Molecules. 2021;26:828.
- Esteras M, Liu I-C, Snijders AP, Jarmuz A, Aragon L. Identification of SUMO conjugation sites in the budding yeast proteome. Microb Cell. 2017;4:331–341. doi:10.15698/mic2017.10.593.
- Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal ACO, Nielsen ML. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325–336. doi:10.1038/nsmb.3366.
- Makhnevych T, Sydorskyy Y, Xin X, Srikumar T, Vizeacoumar FJ, Jeram SM, Li Z, Bahr S, Andrews BJ, Boone C, et al. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol Cell. 2009;33:124–135. doi:10.1016/j.molcel.2008.12.025.
- Zhao X. SUMO-mediated regulation of nuclear functions and signaling processes. Mol Cell. 2018;71:409–418. doi:10.1016/j.molcel.2018.07.027.
- Chymkowitch P, Nguéa P A, Enserink JM. SUMO-regulated transcription: challenging the dogma. Bioessays. 2015b;37:1095–1105. doi:10.1002/bies.201500065.
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi:10.1146/annurev-biochem-061909-093311.
- Rosonina E, Akhter A, Dou Y, Babu J, Sri Theivakadadcham VS. Regulation of transcription factors by sumoylation. Transcription. 2017;8:220–231. doi:10.1080/21541264.2017.1311829.
- Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–1392. doi:10.1042/BST0351389.
- Baig MS, Dou Y, Bergey BG, Bahar R, Burgener JM, Moallem M, McNeil JB, Akhter A, Burke GL, Sri Theivakadadcham VS, et al. Dynamic sumoylation of promoter-bound general transcription factors facilitates transcription by RNA polymerase II. PloS Genet. 2021;17:e1009828. doi:10.1371/journal.pgen.1009828.
- Chang P-C, Cheng C-Y, Campbell M, Yang Y-C, Hsu H-W, Chang T-Y, Chu C-H, Lee Y-W, Hung C-L, Lai S-M, et al. The chromatin modification by SUMO-2/3 but not SUMO-1 prevents the epigenetic activation of key immune-related genes during Kaposi’s sarcoma associated herpesvirus reactivation. BMC Genomics. 2013;14:824. doi:10.1186/1471-2164-14-824.
- Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol. 2005;25:4272–4282. doi:10.1128/MCB.25.10.4272-4282.2005.
- Liu HW, Zhang J, Heine GF, Arora M, Gulcin Ozer H, Onti-Srinivasan R, Huang K, Parvin JD. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 2012;40:10172–10186. doi:10.1093/nar/gks819.
- Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi:10.1101/gr.154872.113.
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100:13225–13230. doi:10.1073/pnas.1735528100.
- Enserink JM. The SUMO stress response in transcriptional regulation: causal relationships or secondary bystander effects? BioEssays. 2022;44:2200065. doi:10.1002/bies.202200065.
- Niskanen EA, Palvimo JJ. Chromatin SUMOylation in heat stress: to protect, pause and organise?: SUMO stress response on chromatin. BioEssays. 2017;39:1–10.
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi:10.1006/jmbi.1998.1839.
- Sheng W, Liao X. Solution structure of a yeast ubiquitin-like protein Smt3: the role of structurally less defined sequences in protein-protein recognitions. Protein Sci. 2002;11:1482–1491. doi:10.1110/ps.0201602.
- Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi:10.1038/nrm3478.
- Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi:10.1074/jbc.M308357200.
- Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi:10.1038/18457.
- Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–2377. doi:10.1128/MCB.20.7.2367-2377.2000.
- Sydorskyy Y, Srikumar T, Jeram SM, Wheaton S, Vizeacoumar FJ, Makhnevych T, Chong YT, Gingras A-C, Raught B. A novel mechanism for SUMO system control: regulated Ulp1 nucleolar sequestration. Mol Cell Biol. 2010;30:4452–4462. doi:10.1128/MCB.00335-10.
- Enserink JM. Sumo and the cellular stress response. Cell Div. 2015;10:4. doi:10.1186/s13008-015-0010-1.
- Lewicki MC, Srikumar T, Johnson E, Raught B. The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J Proteomics. 2015;118:39–48. doi:10.1016/j.jprot.2014.11.012.
- Nguéa P A, Robertson J, Herrera MC, Chymkowitch P, Enserink JM. De-sumoylation of RNA polymerase III lies at the core of the Sumo stress response in yeast. J Biol Chem. 2019;294:18784–18795. doi:10.1074/jbc.RA119.009721.
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi:10.1126/scisignal.2000282.
- Hendriks IA, D’Souza RCJ, Yang B, Verlaan-de Vries M, Mann M, Vertegaal ACO. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21:927–936. doi:10.1038/nsmb.2890.
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi:10.1074/jbc.275.9.6252.
- Pinto MP, Carvalho AF, Grou CP, Rodríguez-Borges JE, Sá-Miranda C, Azevedo JE. Heat shock induces a massive but differential inactivation of SUMO-specific proteases. Biochim Biophys Acta. 2012;1823:1958–1966. doi:10.1016/j.bbamcr.2012.07.010.
- Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–32268. doi:10.1074/jbc.M404173200.
- Lee YJ, Castri P, Bembry J, Maric D, Auh S, Hallenbeck JM. SUMOylation participates in induction of ischemic tolerance. J Neurochem. 2009;109:257–267. doi:10.1111/j.1471-4159.2009.05957.x.
- Guo C, Henley JM. Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life. 2014;66:71–77. doi:10.1002/iub.1244.
- Lee Y, Mou Y, Maric D, Klimanis D, Auh S, Hallenbeck JM. Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PloS ONE. 2011;6:e25852. doi:10.1371/journal.pone.0025852.
- Fuge EK, Braun EL, Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J Bacteriol. 1994;176:5802–5813. doi:10.1128/jb.176.18.5802-5813.1994.
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi:10.1016/j.molcel.2008.07.020.
- Betting J, Seufert W. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J Biol Chem. 1996;271:25790–25796. doi:10.1074/jbc.271.42.25790.
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi:10.1016/j.cell.2004.06.013.
- Soustelle C, Vernis L, Freon K, Reynaud-Angelin A, Chanet R, Fabre F, Heude M. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol Cell Biol. 2004;24:5130–5143. doi:10.1128/MCB.24.12.5130-5143.2004.
- Scholes AN, Lewis JA. Comparison of RNA isolation methods on RNA-Seq: implications for differential expression and meta-analyses. BMC Genomics. 2020;21:249. doi:10.1186/s12864-020-6673-2.
- Tan K, Wong KH. RNA polymerase II ChIP-seq-a powerful and highly affordable method for studying fungal genomics and physiology. Biophys Rev. 2019;11:79–82. doi:10.1007/s12551-018-00497-9.
- Chymkowitch P, Nguéa AP, Aanes H, Koehler CJ, Thiede B, Lorenz S, Meza-Zepeda LA, Klungland A, Enserink JM. Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res. 2015a;25:897–906. doi:10.1101/gr.185793.114.
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi:10.1091/mbc.12.2.323.
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi:10.1091/mbc.11.12.4241.
- Vinayachandran V, Reja R, Rossi MJ, Park B, Rieber L, Mittal C, Mahony S, Pugh BF. Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res. 2018;28:357–366. doi:10.1101/gr.226761.117.
- Augustine RC, Vierstra RD. SUMOylation: re-wiring the plant nucleus during stress and development. Curr Opin Plant Biol. 2018;45:143–154. doi:10.1016/j.pbi.2018.06.006.
- Niskanen EA, Malinen M, Sutinen P, Toropainen S, Paakinaho V, Vihervaara A, Joutsen J, Kaikkonen MU, Sistonen L, Palvimo JJ. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi:10.1186/s13059-015-0717-y.
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi:10.1038/373078a0.
- Dieckhoff P, Bolte M, Sancak Y, Braus GH, Irniger S. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol Microbiol. 2004;51:1375–1387. doi:10.1046/j.1365-2958.2003.03910.x.
- Rodríguez-Molina JB, Tseng SC, Simonett SP, Taunton J, Ansari AZ. Engineered covalent inactivation of TFIIH-kinase reveals an elongation checkpoint and results in widespread mRNA stabilization. Mol Cell. 2016;63:433–444. doi:10.1016/j.molcel.2016.06.036.
- Timmers HTM, Tora L. Transcript buffering: a balancing act between mRNA synthesis and mRNA degradation. Mol Cell. 2018;72:10–17. doi:10.1016/j.molcel.2018.08.023.
- Pozzi B, Mammi P, Bragado L, Giono LE, Srebrow A. When SUMO met splicing. RNA Biol. 2018;15:689–695. doi:10.1080/15476286.2018.1457936.
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun D-J, Bressan RA, Hasegawa PM. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;142:1548–1558. doi:10.1104/pp.106.088831.
- Zhang S, Wang S, Lv J, Liu Z, Wang Y, Ma N, Meng Q. SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato. Plant Cell Physiol. 2018;59:58–71. doi:10.1093/pcp/pcx160.
- Castells-Roca L, García-Martínez J, Moreno J, Herrero E, Bellí G, Pérez-Ortín JE. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PloS One. 2011;6:e17272. doi:10.1371/journal.pone.0017272.
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR, Yates 3rd JR. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–45668. doi:10.1074/jbc.M409203200.
- Fan X, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3’-end formation. Proc Natl Acad Sci U S A. 2010;107:17945–17950. doi:10.1073/pnas.1012674107.
- McNeil JB, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 1998;12:2510–2521. doi:10.1101/gad.12.16.2510.
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi:10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W.
- Liu C, Apodaca J, Davis LE, Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques. 2007;42:158,160,162.
- Svejstrup JQ, Petrakis TG, Fellows J. Purification of elongating RNA polymerase II and other factors from yeast chromatin. Methods Enzymol. 2003;371:491–498. doi:10.1016/S0076-6879(03)71036-2.
- Sri Theivakadadcham VS, Bergey BG, Rosonina E. Sumoylation of DNA-bound transcription factor Sko1 prevents its association with nontarget promoters. PloS Genet. 2019;15:e1007991. doi:10.1371/journal.pgen.1007991.
- Bonnot T, Gillard M, Nagel D. A simple protocol for informative visualization of enriched gene ontology terms. BIO-PROTOCOL. 2019;9:e3429. doi:10.21769/BioProtoc.3429.
