Abstract
CircRNAs have been implicated in the development of resistance in triple-negative breast cancer (TNBC). However, the association between circRNA_0044556 and paclitaxel (PTX) resistance in TNBC is still limited. Therefore, the purpose of this study was to investigate the effect of circRNA_0044556 on biological function and PTX resistance in TNBC cells. PTX-resistant TNBC cells (MDA-MB-231/PTX) were obtained by continuously exposing MDA-MB-231 cells to increasing paclitaxel levels. The expression levels of circRNA_0044556 and miR-665 were measured by qRT–PCR. The regulatory relationship between miR-665 and circRNA_0044556 was verified by biological information website analysis and double-luciferase reporter gene detection experiments. MTT assay, clone assay, flow cytometry and Western blot analysis were used to evaluate the influence of cell biological function. Elevated circRNA_0044556 was observed in TNBC, and paclitaxel increased the expression of circRNA_0044556 in TNBC cells. In TNBC, circRNA_0044556 acted as a ceRNA for miR-665. In addition, low expression of circRNA_0044556 combined with miR-665 inhibited the proliferation of TNBC cells and paclitaxel-resistant TNBC cells while inducing cell death. Our study demonstrated that the downregulation of circRNA_0044556 inhibits the malignant progression of TNBC cells and paclitaxel resistance via miR-665. Thus, circRNA_0044556 may be a potential therapeutic target for PTX-resistance TNBC.
Introduction
Breast cancer (BC) is the most common cancer among women, accounting for about 15% to 20% of cases worldwide and poses a serious threat to the health of women worldwide [Citation1,Citation2]. Based on molecular markers, including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), BC is divided into three main subtypes: hormone receptor (HR) positive, HER2-positive, and triple-negative breast cancer (TNBC). TNBC accounts for about 15%–20% of all breast cancers [Citation3,Citation4]. TNBC has a worse prognosis than HR-positive BCs. More than 50% of patients relapse within the first 3–5 years after diagnosis [Citation5], and the median OS currently treated is 10.2 months [Citation6]. Due to the lack of relevant receptor markers, patients with TNBC do not benefit from established endocrine or HER2-targeting drugs [Citation7]. Therefore, chemotherapy still remains as the main standard treatment for the non-surgical TNBC approach. TNBC responds best to standard chemotherapy regimens, such as paclitaxel or anthracyclines [Citation8]. However, less than 30% of TNBC patients achieve complete remission, and recurrence and mortality rates remain higher than non-TNBC subtypes [Citation9]. Therefore, we urgently need to further explore the molecular mechanism of TNBC occurrence and development and PTX chemotherapy resistance, and explore new therapeutic targets of TNBC, to provide a new theoretical basis for clinical PTX chemotherapy resistance targeted therapy for TNBC.
As a unique group of non-coding RNAs, circRNAs are derived from the reverse splicing of a single pre-mRNA and have a covalent closed-loop structure [Citation5,Citation6]. Due to their remarkable stability and high abundance, CircRNAs have the potential to be ideal biomarkers for the diagnosis of diseases, including cancer [Citation7]. Many studies have demonstrated that abnormal expression of circRNAs is inextricably linked to tumorigenesis and chemical resistance in TNBC [Citation8,Citation9]. For example, Zheng et al. reported that CircGFRA1 affects the sensitivity of TNBC cells to paclitaxel [Citation9]. Similarly, Li et al. showed that silencing hsa_circ_0000199 enhanced the chemical sensitivity of TNBC [Citation10]. This evidence strongly demonstrates the importance of circRNAs in the treatment of chemotherapy-resistant TNBC. CircRNA_0044556 (circCOL1A1) is a newly discovered circRNA. CircBase database analysis revealed that circRNA_0044556 originated from the exon of the COL1A1 gene on chromosome 17q21.33, and the length of the spliced mature sequence was 699 bp. This research group previously confirmed that circRNA_0044556 is upregulated in TNBC tissues [Citation11]. However, we have yet to elucidate the effect of circRNA_0044556 on PTX resistance in TNBC cells.
CircRNAs interact with miRNAs and play an indispensable role as miRNA sponges in tumor progression and chemical resistance [Citation12]. miRNAs have been confirmed to be related to TNBC progression, prognosis and chemotherapy resistance [Citation13,Citation14]. Among these, miR-665 has been shown to be associated with the progression of multiple cancers and chemotherapy resistance. More importantly, miR-665 was found to be under-expressed in TNBC [Citation15]. In a previous study, the prediction results of circRNA_0044556 targeting miRNAs showed that miR-665 has a targeting relationship with circRNA_0044556 [Citation16]. However, the exact role of circRNA_0044556 in sensitivity to paclitaxel chemotherapy in TNBC by regulating miR-665 expression is unclear.
Based on this background, we hypothesized that circRNA_0044556 promotes the malignant behavior of TNBC cells and PTX resistance by targeting miR-665 expression. Here, we investigated the expression, action and mechanism of circRNA_0044556 activity in PTX resistance in TNBC. These results may provide a new perspective on the antitumor mechanism of PTX in TNBC.
Materials and methods
Cell culture
The TNBC cell line (MDA-MB-231) and non-TNBC cell line (MCF-10A) were purchased from the Type Culture Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were removed from the −80 °C freezer and immediately placed in a water bath. Then, the cells were added to a DMEM medium containing 10% fetal bovine serum (FBS, Excell Bio, China) and 1% penicillin-streptomycin (MACKLIN, China) (Hyclone, Cat.No.SH30809.18), and the cells were suspended. The suspension is centrifuged at 37 °C at ∼300xg for 5–10 min. The supernatant was discarded and the cells were transferred to a culture bottle and cultured at 37 °C in 5% CO2.
Construction of PTX-resistant cells
MDA-MB-231 cells become resistant to PTX (HY-B0015, MCE, USA) through continuous exposure to increased PTX levels. In short, 1 × 105/well MDA-MB-231 cells were exposed to 5 nM PTX medium for a long period of time to maintain PTX resistance, and then PTX concentration was increased to 15 nM. After 4 weeks, when the cells were maintained at 15 nM PTX, the concentration was increased to 18 nM. After 2 weeks, the concentration was increased to 20 nM. After 8 weeks of PTX treatment, PTX concentration reached 20 nM and PTX-resistant TNBC cell line (MDA-MB-231/PTX) was induced. Finally, the IC50 value of PTX in MDA-MB-231 cells was used to determine the drug resistance of MDA-MB-231.
Experimental grouping
The experiment was divided into 8 groups: sh-NC group, sh-circRNA_0044556 group, pcDNA 3.1 group, pcDNA 3.1-circRNA_0044556 group, miR-665 mimics group, NC mimics group, sh-circRNA_0044556 + in-NC group, and sh-circRNA_0044556 + in-miR-665 group. Short hairpin RNA and pcDNA 3.1 overexpression plasmid vectors targeting circRNA_0044556, miR-665 mimics, miR-665 inhibitor and negative control were purchased from GenePharma company and named sh-circRNA_0044556, sh-NC, pcDNA 3.1, pcDNA 3.1-circRNA_0044556, miR-665 mimics, NC mimics, in-miR-665 and in-NC. Each 125 μl diluted plasmid carrier was added to 125 μl diluted Lipofectamine 3000 reagent (Invitrogen) and incubated at room temperature for 15 min. MDA-MB-231 and MDA-MB-231/PTX cells at the logarithmic growth stage were selected, and the vector-lipid complex was added to the cells, gently shaken, and then returned to the incubator at 37 °C and 5% CO2 for further incubation and culture. After 48 h, the transfection efficiency was observed by an inverted fluorescence microscope and subsequent experiments were conducted.
RT-qPCR
The different groups of MDA-MB-231 and MDA-MB-231/PTX cells (1 × 105/well) were collected after each treatment. TRIzol (Beyotime) was used to extract total RNA from the MDA-MB-231 and MDA-MB-231/PTX cells. The total RNA concentration was among 200–400 ng/μL. The PrimeScript RT reagent kit (Vazyme) and miRNA 1st Strand cDNA Synthesis Kit (Vazyme) were used for the reverse transcription of circRNA and miRNA, respectively. RT-qPCR was performed using a SYBR Green kit (Vazyme). The U6 and GAPDH were selected as the internal reference genes for miRNA and circRNA, respectively. The primer concentrations were 100 μM, the sequences are shown in . The differences of the circRNA or miRNA levels were calculated through the 2−ΔΔCt method.
Table 1. Primer sets for real-time RT-PCR used in this study.
Western blot
The different groups of MDA-MB-231 and MDA-MB-231/PTX cells (1 × 105/well) were collected after each treatment. Proteins were extracted from the cells using a radioimmunoprecipitation (RIPA, Beyotime) buffer containing protease inhibitors. An equivalent amount of protein (20 μg) was loaded onto an SDS-PAGE gel and then transferred to a nitrocellulose membrane. After the membrane was sealed in 5% buttermilk for 1 h, it was diluted with TBST solution containing 5% BSA according to the manufacturer’s instructions (BAX, Proteintech, 50599-2-Ig, 1:1000; Bcl2, Proteintech, 68103-1-Ig, 1:1000; Ki67, Abcam, ab197547, 1:1000; GAPDH, bsm-33033M,1:1000), and incubated with the membrane overnight at 4 °C. The membrane was then incubated with a secondary antibody for 2 h. Tanon ECL was used for chemiluminescence detection. Images were captured using a Tanon 5200 chemiluminescence imager, and ImageJ software was used for analysis.
MTT assay
When the confluence of TNBC cells reached 70%–90%, the cells (1 × 103 cells/well) were inoculated into 96-well plates. Transfected cells were treated with 100 μl of sterile MTT dye (5 mg/ml; Beyotime, C0009S) and stained at 37 °C for 4 h. The medium was then removed, and 100 μl of formazan solution was added (Beyotime, C0009S). Absorbance was measured at 570 nm using an enzyme-labeled instrument.
Clone formation assay
Transfected cells were seeded into 6-well plates at a density of 1 × 103 cells/well. After seven days of culture in the incubator, the cells were fixed with 4% formaldehyde for 10 min and stained with 5% crystal violet for 10 min. Finally, the cell clone count was determined using a low-power microscope.
Flow cytometry (FCM)
Cells in the logarithmic growth phase were seeded in a 24-well plate at a density of 1 × 105 cells/well, with a final volume of 500 μL. After transfection, cells were digested with 0.25% trypsin, washed with PBS, and centrifuged. The supernatant was discarded, and the density was adjusted by resuspension in 400 μL 1× annexin-binding buffer. Next, 100 μL was absorbed and transferred to a sterile centrifuge tube. Then, 5 μL Annexin V-FITC and 5 μL PI were added successively, mixed well, and incubated in the dark for 15 min. Analysis of the mixture was performed using FCM.
Luciferase activity assay
Lipofectamine 3000 reagent (Invitrogen) was used to cotransfect the pGL3-basic vector with the circRNA_0044556-wild type (WT)/mutant one (MUT) and miR-665 mimics or NC mimics into TNBC cells (1 × 105 cells/well). Luciferase activity was detected using a dual- luciferase detection system.
Data analysis
Data were analyzed and mapped using Graphpad Prism9 (Version9.5.0). Photoshop was used to organize the images. All maps are presented as the mean ± SD, and significant differences between groups were tested using the t-test. A p value less than .05 was considered significant (*p < .05, **p < .01, ***p < .001).
Results
CircRNA_0044556 was highly expressed in TNBC, while miR-665 was low expressed
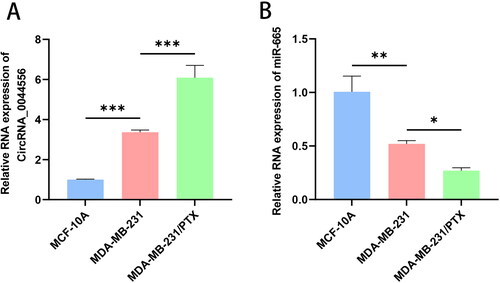
First, we investigated the levels of circRNA_0044556 and miR-665 in TNBC cells using RT–qPCR analysis. We found that the level of circRNA_0044556 was higher in MDA-MB-231 cells than in MCF-10A cells (p < .001), whereas miR-665 expression was downregulated (p < .01) (). To further investigate their relationship with paclitaxel resistance in TNBC, we cultured PTX-resistant cells and named them MDA-MB-231/PTX. MTT results showed that the average IC50 value of PTX in MDA-MB-231/PTX was 40.54, which was significantly higher than the average IC50 value of MDA-MB-231 (6.52), indicating that PTX-resistant cell lines were successfully established (p < .05) (Figure S2(A)). In addition, MDA-MB-231/PTX showed irregular character changes, larger size and irregular edges compared with parental cells under the microscope (Figure S2(B)), further supporting the successful construction of drug-resistant strains. Subsequently, we detected the expression levels of circRNA_0044556 and miR-665 in PTX-resistance MDA-MB-231 cells by RT-qPCR. Compared to normal TNBC cells, the expression of circRNA_0044556 significantly increased (p < .001), whereas the expression of miR-665 was significantly decreased in PTX resistance MDA-MB-231 cells (p < .05) ().
Figure 1. CircRNA_0044556 was highly expressed in TNBC, while miR-665 was low expressed. (A) The level of circRNA_0044556 in cells was analyzed by RT–qPCR. (B) The level of miR-665 in cells was analyzed by RT–qPCR. **p < .01, ***p < .001 vs. MCF-10A, N = 3. *p < .05, ***p < .001 vs. MDA-MB-231, N = 3.
Downregulated circRNA_0044556 inhibited the malignant phenotype and reduced paclitaxel resistance of TNBC cells
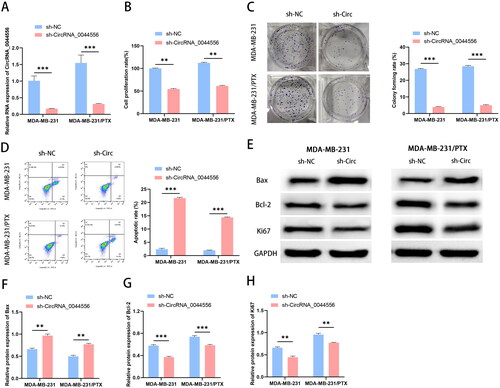
Next, to study the influence of circRNA_0044556 on the biological function of TNBC cells and the sensitivity to paclitaxel, we conducted a functional knockout experiment. Transfection efficiency was confirmed by RT-qPCR, and as shown in , the level of circRNA_0044556 was inhibited after sh-circRNA_0044556 treatment. The results of MTT and clonogenesis experiments showed that knockdown of circRNA_0044556 reduced cell viability and clonal colony number, suggesting that inhibition of circRNA_0044556 reduced the PTX-resistance in PTX resistant MDA-MB-231 cells (). FCM analysis showed that circRNA_0044556 knockdown increased the apoptosis rate of the cells (). Bax (apoptotic protein), Bcl-2 (anti-apoptotic protein), and Ki67 (proliferative protein) are some of the most important indicators of apoptosis and proliferation and are closely related to the occurrence, development, and prognosis of tumors. WB analysis confirmed that the knockdown of circRNA_0044556 depressed the expression levels of Ki-67 and Bcl-2 proteins in cells, but enhanced the expression levels of Bax protein (). The above data suggested that the downregulation of circRNA_0044556 suppressed the malignant phenotype and paclitaxel resistance of TNBC cells.
Figure 2. Down-regulated circRNA_0044556 inhibited the malignant phenotype and paclitaxel resistance of TNBC cells. (A) RT–qPCR was used to assess the endogenous expression of circRNA_0044556 after transfection of cells. (B) MTT assays were used to assess cell proliferation. (C) Clonal formation assays were used to assess cell proliferation. (D) Apoptosis was measured by FCM. (E–H): WB analysis (E) of the expression levels of Bax (F), Bcl-2 (G) and Ki67 (H) proteins. **p < .01, ***p < .001 vs. sh-NC, N = 3.
CircRNA_0044556 competitively adsorbs miR-665
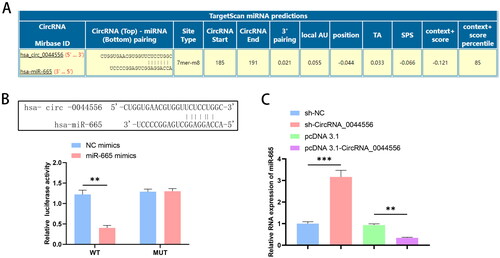
Next, we explored the relationship between circRNA_0044556 and miR-665 expression. The bioinformatics website found that miR-665 has a targeted binding site for circRNA_0044556 (). The circRNA_0044556-WT/MUT luciferase reporter vector was designed based on the predicted binding sites. Dual luciferase reporter assays showed that circRNA_0044556 targeted miR-665 (). Subsequently, we analyzed the effect of circRNA_0044556 on miR-665 expression. As shown in , up-regulation or down-regulation of circRNA_0044556 in MDA-MB-231 cells decreased and increased miR-665 expression, respectively. These data indicate that miR-665 is the downstream miRNA of circRNA_0044556 in TNBC and that the effect of circRNA_0044556 on TNBC paclitaxel resistance may be related to miR-665.
Figure 3. CircRNA_0044556 competitively adsorbs miR-665. (A) Predicted binding sites of circRNA_0044556 and miR-665. (B) Dual luciferase reporting experiment verified the targeting relationship between circRNA_0044556 and miR-665. (C) The expression of miR-665 in MDA-MB-231 cells with circRNA_0044556 knockdown or overexpression was analyzed by RT–qPCR. **p < .01, vs. NC mimics, N = 3. ***p < .001, vs. sh-NC, N = 3. **p < .01, vs. pcDNA 3.1, N = 3.
CircRNA_0044556 affects the malignant phenotype and paclitaxel resistance of TNBC cells by regulating miR-665 expression
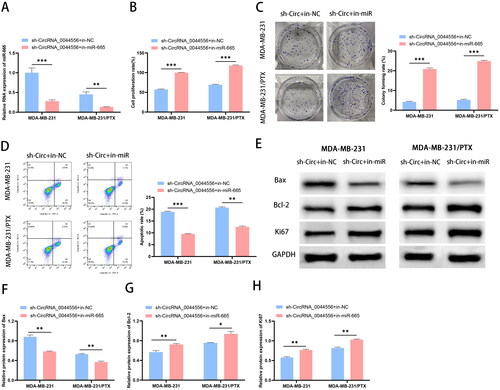
To observe the modulatory mechanism of circRNA_0044556 and miR-665 in TNBC, we co-transfected sh-circRNA_0044556 with in-NC or in-miR-665. RT-qPCR analysis showed that transfection with sh-circRNA_0044556 upregulated miR-665 expression, whereas co-transfection with miR-665 inhibitors significantly eliminated this reaction (). The results of biological function experiments showed that compared to cells transfected with sh-circRNA_0044556, cells co-transfected with sh-circRNA_0044556 and in-miR-665 showed increased cell proliferation and decreased apoptosis (, p < .01). WB analysis further confirmed these results (). In summary, circRNA_0044556 inhibits the malignant phenotype and paclitaxel resistance of TNBC cells, and these biological functions are achieved by sponging miR-665.
Figure 4. CircRNA_0044556 affects the malignant phenotype and paclitaxel resistance of TNBC cells by regulating miR-665 expression. (A) RT–qPCR was used to assess the endogenous expression of circRNA_0044556 after transfection of cells. (B) MTT assays were used to assess cell proliferation. (C) Clonal formation assays were used to assess cell proliferation. (D) Apoptosis was measured by FCM. (E–H) WB analysis (E) of the expression levels of Bax (F), Bcl-2 (G) and Ki67 (H) proteins. *p < .05, **p < .01, ***p < .001 vs. sh-circRNA_0044556 + in-NC, N = 3.
Discussion
Paclitaxel is a promising chemotherapeutic drug for the treatment of advanced TNBC [Citation17]. However, most advanced TNBC patients only respond to paclitaxel treatment in the initial stages, and acquired paclitaxel resistance occurs frequently in the later stage of treatment, resulting in no significant improvement in the 5-year survival of TNBC patients treated with paclitaxel [Citation18]. Therefore, acquired resistance to paclitaxel is undoubtedly the biggest obstacle to improving the overall response and survival of patients with TNBC. In recent years, owing to the stability and abundance of circRNAs, an increasing number of circRNAs have been identified as abnormally expressed in TNBC; as important regulators of TNBC cancer progression, circRNAs are of great significance for the resistance of TNBC [Citation8,Citation19]. However, research on circRNAs associated with paclitaxel resistance in TNBC remains limited. Therefore, we investigated the expression, action, and mechanism of circRNA_0044556 activity in PTX resistance in TNBC. The results showed that circRNA_0044556 promoted the malignant behavior of TNBC cells and PTX resistance by sponging miR-665.
In this study, we first verified the expression pattern of CircRNA_0044556 in TNBC. Compared to normal cells, the expression of CircRNA_0044556 was upregulated in TNBC cells. Subsequently, we constructed PTX-resistant TNBC cells and measured the levels of circRNA_0044556. PTX increased circRNA_0044556 expression in MDA-MB-231 cells, suggesting that circRNA_0044556 may be related to paclitaxel resistance. As a circRNA encoded by COL1A1, previous studies have shown that circRNA_0044556 plays a vital role in the occurrence and development of tumors, including colorectal cancer [Citation20,Citation21], gastric cancer [Citation22] and TNBC [Citation11]. Infinite malignant proliferation of tumor cells and inhibition of apoptosis are core factors in tumor development and change [Citation23,Citation24]. Here, we found that low expression of circRNA_0044556 restricted the proliferation of TNBC cells and induced cell apoptosis. In previous studies, we confirmed that circRNA_0044556 reduces the sensitivity of triple-negative breast cancer cells to adriamycin by acting as a sponge for miR-145 and regulating NRAS [Citation11]. However, the effect of circRNA_0044556 on PTX sensitivity in TNBC is unclear. Therefore, we further observed the effect of circRNA_0044556 knockdown on PTX-resistant TNBC cells and found that low expression of circRNA_0044556 restricted the cell proliferation of PTX-resistant TNBC cells in vitro and promoted cell apoptosis. In summary, circRNA_0044556 can be used as a therapeutic target for TNBC, and downregulation of circRNA_0044556 has a beneficial effect on slowing the progression of TNBC and PTX resistance by reducing cell proliferation and promoting cell apoptosis.
The development of circRNAs as competing ceRNA-binding miRNAs to regulate TNBC has been demonstrated in several studies [Citation25]. Previous studies have confirmed that the main regulatory mechanism of circRNA_0044556 is that ceRNA, which acts as a downstream miRNA, plays a role in tumors [Citation22]. Therefore, we screened miRNAs significantly enriched with circRNA_0044556 through the bioinformatics website and found that circRNA_0044556 showed highly specific enrichment of miR-665. To further verify the relationship between circRNA_0044556 and miR-665, we confirmed that circRNA_0044556 acts as a ceRNA for miR-665 in TNBC through luciferase reporter gene analysis. In addition, circRNA_0044556 actively regulated miR-665 expression in TNBC, leading us to propose a mechanism by which circRNA_0044556 may contribute to blocking TNBC progression by sponging miR-665.
Previous studies have confirmed that miRNAs play an important role in many biological processes, such as proliferation, apoptosis and drug resistance [Citation26]. In recent years, a number of studies have been conducted on the role of miR-665 in tumorigenesis and development, but the results vary, and different studies have reached different conclusions regarding the role of miR-665 as a tumor suppressor or tumor promoter, which seems to be determined by the cell type and the localization of miR-665. Currently, miR-665 has been demonstrated as a tumor suppressor in some cancer cells, such as bladder cancer [Citation27], hepatocellular carcinoma [Citation28], and gastric cancer [Citation29], whereas miR-665 has been described as a tumor promoter in lung cancer [Citation28] and ovarian cancer [Citation30]. Importantly, recent studies have found that miR-665 is significantly downregulated in TNBC, and the upregulation of miR-665 is positively correlated with the survival time of patients with TNBC [Citation15]. In addition, miR-665 is inextricably associated with paclitaxel resistance in cervical cancer cells [Citation31]. However, the mechanism of miR-665 resistance to paclitaxel in TNBC remains unknown. Therefore, in this study, sh-circRNA_0044556 and IN-NC or In-miR-665 were transfected into MDA-MB-231 and MDA-MB-231/PTX cells for the functional salvage experiments. The results showed that the knockdown of circRNA_0044556 increased the cell viability and colony number of MDA-MB-231 and MDA-MB-231/PTX cells and induced cell apoptosis, but these effects were reversed by an miR-665 inhibitor. We can also explain that circRNA_0044556 depletion attenuated the effects of low miR-624-3p levels in TNBC and paclitaxel-resistant cells.
In summary, our study confirmed our hypothesis that circRNA_0044556 inhibits TNBC cell resistance to PTX by regulating miR-665 to reduce cell activity and proliferation capacity, and induce cell apoptosis. By identifying a novel regulatory mechanism for TNBC, we aimed to broaden our current understanding of the molecular mechanisms associated with TNBC, which could help in the development of personalized treatment approaches. But also provides a new target for the clinical treatment of paclitaxel-resistant TNBC. Nevertheless, this study had some limitations that should be explored further. For example, mechanistic studies have been conducted in cells, and studies on other cell lines need to be further explored. Second, further studies are needed to determine which targets and signaling pathways miR-665 regulates TNBC progression and PTX resistance. Future multicenter trials and animal studies are required to verify the diagnostic and therapeutic effects of circRNA_0044556.
Conclusion
Our results suggest that circRNA_0044556 is an ‘oncogene’ in TNBC. Highly stable circRNA_0044556 promotes the malignant phenotype and PTX resistance of TNBC cells through miR-655.
Authors contributions
Jingjing Chen, Peng Shi: Conceptualization, Methodology, Experiments, manuscript reviewing and Editing; Zhichao Cui, Nan Jiang, Jie Ma: Data collection and analysis. All authors have written and approved the final manuscript.
Supplemental Material
Download MS Word (1.4 MB)Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Lemesle M, Geoffroy M, Alpy F, et al. CLDN1 sensitizes triple-negative breast cancer cells to chemotherapy. Cancers. 2022;14(20):5026. doi: 10.3390/cancers14205026.
- Shen L, O'Shea JM, Kaadige MR, et al. Metabolic reprogramming in triple-negative breast cancer through myc suppression of TXNIP. Proc Natl Acad Sci U S A. 2015;112(17):5425–5430. doi: 10.1073/pnas.1501555112.
- Green-Tripp G, Nattress C, Halldén G. Targeting triple negative breast cancer with oncolytic adenoviruses. Front Mol Biosci. 2022;9:901392. doi: 10.3389/fmolb.2022.901392.
- Ferrari P. Molecular mechanisms, biomarkers and emerging therapies for chemotherapy resistant TNBC. Int J Mol Sci. 2022;23(3):1665.
- Asano T. Drug resistance in cancer therapy and the role of epigenetics. J Nippon Med Sch. 2020;87(5):244–251. doi: 10.1272/jnms.JNMS.2020_87-508.
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361.
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi: 10.1080/15476286.2015.1122162.
- Weidle UH, Birzele F. Triple-negative breast cancer: identification of circRNAs with efficacy in preclinical in vivo models. Cancer Genomics Proteomics. 2023;20(2):117–131. doi: 10.21873/cgp.20368.
- Zheng S-R, Huang Q-D, Zheng Z-H, et al. circGFRA1 affects the sensitivity of triple-negative breast cancer cells to paclitaxel via the miR-361-5p/TLR4 pathway. J Biochem. 2021;169(5):601–611. doi: 10.1093/jb/mvaa148.
- Li H, Xu W, Xia Z, et al. Hsa_circ_0000199 facilitates chemo-tolerance of triple-negative breast cancer by interfering with miR-206/613-led PI3K/akt/mTOR signaling. Aging. 2021;13(3):4522–4551. doi: 10.18632/aging.202415.
- Chen J, Shi P, Zhang J, et al. CircRNA_0044556 diminishes the sensitivity of triple‑negative breast cancer cells to adriamycin by sponging miR‑145 and regulating NRAS. Mol Med Rep. 2022;25(2):1–12. doi: 10.3892/mmr.2021.12567.
- Afzal S, Hassan M, Ullah S, et al. Breast cancer; discovery of novel diagnostic biomarkers, drug resistance, and therapeutic implications. Front Mol Biosci. 2022;9:783450. doi: 10.3389/fmolb.2022.783450.
- Ding L, Gu H, Xiong X, et al. MicroRNAs involved in carcinogenesis, prognosis, therapeutic resistance and applications in human triple-negative breast cancer. Cells. 2019;8(12):1492. doi: 10.3390/cells8121492.
- Li J, Lu M, Jin J, et al. miR-449a suppresses tamoxifen resistance in human breast cancer cells by targeting ADAM22. Cell Physiol Biochem. 2018;50(1):136–149. doi: 10.1159/000493964.
- Zhao X-G, Hu J-Y, Tang J, et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019;10(7):479. doi: 10.1038/s41419-019-1705-z.
- Ma X, Deng C. Circ_0044556 promotes the progression of colorectal cancer via the miR-665-Dependent expression regulation of diaphanous homolog 1. Dig Dis Sci. 2022;67(9):4458–4470. doi: 10.1007/s10620-021-07310-w.
- Xu K, Zhu W, Xu A, et al. Inhibition of FOXO1‑mediated autophagy promotes paclitaxel‑induced apoptosis of MDA‑MB‑231 cells. Mol Med Rep. 2022;25(2):1–11. doi: 10.3892/mmr.2022.12588.
- Yen W-C, Corpuz MR, Prudente RY, et al. A selective retinoid X receptor agonist bexarotene (targretin) prevents and overcomes acquired paclitaxel (taxol) resistance in human non-small cell lung cancer. Clin Cancer Res. 2004;10(24):8656–8664. doi: 10.1158/1078-0432.CCR-04-0979.
- Wang Z, Li Y, Yang J, et al. Circ-TRIO promotes TNBC progression by regulating the miR-432-5p/CCDC58 axis. Cell Death Dis. 2022;13(9):776. doi: 10.1038/s41419-022-05216-7.
- Hossain MT, Li S, Reza MS, et al. Identification of circRNA biomarker for gastric cancer through integrated analysis. Front Mol Biosci. 2022;9:857320. doi: 10.3389/fmolb.2022.857320.
- Jing L, Wu J, Tang X, et al. Identification of circular RNA hsa_circ_0044556 and its effect on the progression of colorectal cancer. Cancer Cell Int. 2020;20(1):427. doi: 10.1186/s12935-020-01523-1.
- Ma Y, Ren Y, Wen H, et al. circCOL1A1 promotes the progression of gastric cancer cells through sponging miR-145 to enhance RABL3 expression. J Immunol Res. 2021;2021:6724854–6724820. doi: 10.1155/2021/6724854.
- Ye M, Song Y, Pan S, et al. Evolving roles of lysyl oxidase family in tumorigenesis and cancer therapy. Pharmacol Ther. 2020;215:107633. doi: 10.1016/j.pharmthera.2020.107633.
- Fu L, Huo J, Fitrat H, et al. CircNRIP1 exerts oncogenic functions in papillary thyroid carcinoma by sponging miR-653-5p and regulating PBX3 expression. J Oncol. 2022;2022:2081501–2081512. doi: 10.1155/2022/2081501.
- Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–5021. doi: 10.7150/jca.30828.
- Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–76. doi: 10.1016/j.cub.2014.06.043.
- Wang W, Ying Y, Xie H, et al. miR-665 inhibits epithelial-to-mesenchymal transition in bladder cancer via the SMAD3/SNAIL axis. Cell Cycle. 2021;20(13):1242–1252. doi: 10.1080/15384101.2021.1929677.
- Bai N, Peng E, Xia F, et al. CircABCC2 regulates hepatocellular cancer progression by decoying MiR-665. J Cancer. 2019;10(17):3893–3898. doi: 10.7150/jca.31362.
- Wu K-Z, Zhang C-D, Zhang C, et al. miR-665 suppresses the Epithelial-Mesenchymal transition and progression of gastric cancer by targeting CRIM1. Cancer Manag Res. 2020;12:3489–3501. doi: 10.2147/CMAR.S241795.
- Zhou P, Xiong T, Yao L, et al. MicroRNA-665 promotes the proliferation of ovarian cancer cells by targeting SRCIN1. Exp Ther Med. 2020;19(2):1112–1120. doi: 10.3892/etm.2019.8293.
- Dong M, Li P, Xie Y, et al. CircMYBL2 regulates the resistance of cervical cancer cells to paclitaxel via miR-665-dependent regulation of EGFR. Drug Dev Res. 2021;82(8):1193–1205. doi: 10.1002/ddr.21834.
