Abstract
The genus Pseudoplatystoma includes catfish species distributed throughout the fresh waters of South America. These species are important fisheries resources and play a significant ecological role due to their piscivorous and migratory habits. The taxonomy of this genus is still debated: traditionally, only three species have been recognised, but recently, this number was raised to eight. The validity of these eight morphospecies, however, was not confirmed by two subsequent molecular phylogenetic studies, which identified either five or four main clades. In this study, we focused on the two morphospecies restricted to the Orinoco basin, P. metaense and P. orinocoense, which have been assigned to either the same or different clades in previous studies. We carried out cytogenetic analyses to describe their unknown karyotypes and to look for cytotaxonomic markers. We also analysed their mitochondrial sequences in order to assign the sampled specimens to the previously identified molecular clades. The two presumptive species show similar karyotypes (2n = 56, 42 biarmed and 14 uniarmed chromosomes) and cytogenetic features in terms of the constitutive heterochromatin distribution and the number and location of minor and major ribosomal genes. Thus, no species-specific chromosome markers could be identified. The analysis of cytochrome b and cytochrome oxidase I mitochondrial genes (carried out by retrieving all the mtDNA Pseudoplatystoma sequences available in GenBank) distributed the sampled specimens into two distinct molecular clades and confirmed the need to re-evaluate, by parallel morphological and molecular analyses, the monophyly of some lineages.
Keywords:
Introduction
Pimelodidae (Siluriformes) is an extremely diversified group of catfishes distributed in the fresh waters of South America and in the lower Isthmian regions. The family has undergone multiple taxonomic revisions, as is reflected by the different number of taxa reported by different authors. Nelson (Citation2006) reported approximately 31 genera and at least 85 species, Ferraris (Citation2007) noted 29 genera with approximately 93 species, and Eschmeyer and Fong (Citation2013) recognised 109 valid species.
Among the Pimelodidae, Pseudoplatystoma species are boldly striped or spotted catfishes that are distributed over the major river basins in South America (Burgess Citation1989). These fish species represent very important fisheries resources and play a significant ecological role due to their piscivorous and migratory habits. The taxonomy of the genus has been debated. Three species, P. fasciatum, P. corruscans and P. tigrinum, were recognised (Burgess Citation1989; Lundberg & Littmann Citation2003) until the morphological revision by Buitrago-Suárez and Burr (Citation2007). These authors recognised the validity of two formerly described species, P. punctifer and P. reticulatum, and described three new species, P. orinocoense (formerly P. fasciatum), P. metaense (formerly P. tigrinum) and P. magdaleniatum (formerly P. fasciatum), raising the total number of species in the genus to eight.
This morphology-based classification (Buitrago-Suárez & Burr Citation2007) was tested by Torrico et al. (Citation2009) with mitochondrial molecular markers [cytochrome b (cyt b); control region (CR)]. The molecular analysis only partially supported the morphological taxonomy and identified five main clades, four of which correspond to P. corruscans, P. magdaleniatum, P. reticulatum and P. tigrinum. The remaining clade, identified through the CR only, includes the two taxa endemic to the Orinoco basin, P. metaense and P. orinocoense. More recently, Carvalho-Costa et al. (Citation2011) carried out a comprehensive molecular phylogenetic analysis of the genus based on nuclear (Rag1 and S7int1) and mitochondrial (cyt b) DNA markers. Their results demonstrated that molecular markers are not able to identify the eight putative morphospecies (Buitrago-Suárez & Burr Citation2007) and that only four clades (i.e., distinct species), slightly different from those identified by Torrico et al. (Citation2009), can be identified: P. corruscans, P. magdaleniatum, P. tigrinum sensu lato (including P. tigrinum and P. metaense) and P. fasciatum sensu lato (including P. fasciatum, P. punctifer, P. reticulatum and P. orinocoense).
The number of valid species for the genus currently reported in FishBase (Froese & Pauly version 05/2013) is seven, i.e., all of the morphospecies identified by Buitrago-Suárez and Burr (Citation2007), with the exclusion of P. punctifer. Thus, the systematics of the genus still remains to be clarified.
In this context, we performed a cytogenetic analysis of presumptive P. metaense and P. orinocoense specimens, the two morphospecies restricted to the Orinoco basin (Buitrago-Suárez & Burr Citation2007), whose karyotypes and cytogenetic features (distribution of constitutive heterochromatin and location of minor and major ribosomal genes) are still undescribed. This study also aimed to identify species-specific cytotaxonomic markers. This cytogenetic analysis allowed for a cytotaxonomic comparison with the previously described four Pseudoplatystoma species (), which share the same diploid number (2n = 56) but also show a certain degree of intra- and interspecific karyotype diversity.
Table I. Chromosome data for the genus Pseudoplatystoma
Meanwhile, in order to assign the sampled specimens to the previously identified molecular clades (Torrico et al. Citation2009; Carvalho-Costa et al. Citation2011), we sequenced the mitochondrial cyt b gene in a subset of specimens. In these same specimens, we analysed a fragment of the mitochondrial cytochrome oxidase I (COI) gene and, for both markers, we carried out a phylogenetic analysis by retrieving all the Pseudoplatystoma sequences available in GenBank.
Materials and methods
Twelve Pseudoplatystoma metaense specimens and 14 P. orinocoense specimens were collected from a flood plain lagoon, Laguna de Castillero in Caicara del Orinoco, Bolívar State, Venezuela (N 07° 37′ 49.8″, W 66° 09′ 21.3″).
The voucher specimens were deposited at the Ichthyology Collection of the Escuela de Ciencias Aplicadas del Mar, Universidad de Oriente (ECAM 852–859). As the individuals were juveniles, it was not possible to determine their sex. The specimens were morphologically identified as described by Buitrago-Suárez and Burr (Citation2007).
Forty-eight hours before the chromosome preparations, the fishes were injected intramuscularly with a yeast glucose solution (Lee & Elder Citation1980) to stimulate mitosis. The mitotic chromosome preparations were obtained from kidney cells according to the technique described by Foresti et al. (Citation1993). Nucleolus organiser regions (NORs) were revealed by silver (Ag) staining (Howell & Black Citation1980), and C-banding was performed as described by Sumner (Citation1972).
The 45S rDNA, which corresponds to the NOR, and the 5S rDNA clusters were mapped by fluorescence in situ hybridization (FISH) following the method of Pinkel et al. (Citation1986). The Drosophila melanogaster 45S rDNA probe containing the 18S-5.8S-28S genes plus an intergenic spacer was labelled by random priming with biotin using the BioPrime Labelling system (Invitrogen). The 5S rDNA repeat probe was generated by polymerase chain reaction (PCR) with the primers 5SA (5′TAC GCC CGA TCT CGT CCG ATC3′) and 5SB (5′CAG GCT GGT ATG GCC GTA AGC3′) (Martins & Galetti Citation1999) from the genomic DNA of Squalius lucumonis and directly labelled with biotin-14-dATP. The post-hybridisation washes were performed at a low stringency (37°C, 10 min each). The signals were detected and amplified by a two-round application of Avidin-FITC/biotinylated Anti-avidin (Vector). The metaphases were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and propidium iodide and examined under a Zeiss Axiophot epifluorescence microscope equipped with the appropriate selective filters. The images were acquired using a SenSys-1400 CCD camera and then artificially coloured and edited with Photoshop CS3 (Adobe Systems, Inc).
For a subset of 10 specimens (five P. metaense and five P. orinocoense), fragments of the cyt b and COI mitochondrial genes were amplified. DNA was isolated from white muscle preserved in 95% ethanol and amplified through polymerase chain reaction using primers GluFor (Milana et al. Citation2008) and H15913 (Minegishi et al. Citation2005) for cyt b, and FishF1 and FishR2 (Ward et al. Citation2005) for COI following the procedures reported by Milana et al. (Citation2008). The sequences obtained were deposited into the GenBank database (Acc. No. JQ733550–JQ733556). BLAST (Basic Local Alignment Search Tool) software was used for similarity searching of each sequence in GanBank and, for COI, also in the Barcode of Life Data Systems (BOLD).
For the phylogenetic analysis, all the cyt b and COI Pseudoplatystoma sequences available in GenBank (consulted 13 May 2013) were included (). Zungaro zungaro (DQ119459) and Pimelodus pictus (AP012019) were used as outgroups in the cyt b and COI analyses, respectively. Two types of phylogenetic analyses were carried out: neighbor-joining (NJ) and maximum-likelihood (ML). The NJ analysis (1000 bootstrap replicates) was performed using MEGA4 (Tamura et al. Citation2007); the ML tree reconstructions and likelihood calculations (1000 bootstrap replicates) were conducted in PhyML v. 2.4.4 (Guindon & Gascuel Citation2003). Modeltest v. 3.7 (Posada & Crandall Citation1998) was used to choose the model of nucleotide substitution for the ML analysis. According to the Akaike Information Criterion (AIC), TrN + I + G was the evolutionary model selected for the cyt b data and TrN + G for the COI data.
Table II. Cyt b and COI sequences available in GenBank for Pseudoplatystoma species. N: number of sequences, and (h): number of haplotypes; AN: GenBank Accession Number
Results
In both species, the counts of diploid metaphase cells revealed a modal diploid number of 2n = 56 chromosomes. P. metaense () and P. orinocoense () have a similar chromosome formula, with a karyotype characterised by 21 biarmed chromosome pairs (24 M + 18 SM) and seven uniarmed chromosome pairs (14 ST/A), with a fundamental number of arms (FN) of 98. The short arms of one of the largest submetacentric chromosome pairs, classified as number 13 in both species, appear often as decondensed and heteromorphic in size.
Figure 1. Giemsa-stained karyotypes of (a) Pseudoplatystoma metaense and (d) P. orinocoense. M indicates metacentric, SM submetacentric, ST subtelocentric and A acrocentric chromosomes. Selected samples of chromosome pair 13 for each species are shown in the insets: (b, e) Giemsa-stained above, Ag-stained below; (c, f) Giemsa-stained above, C-banded below.
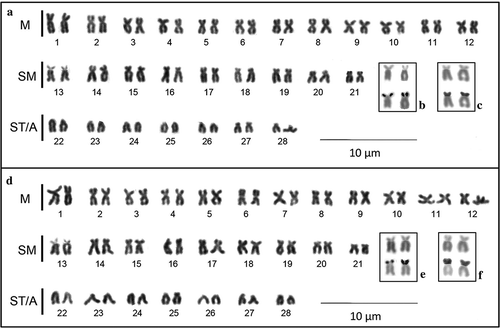
As expected, the sequential silver staining (, e) produced Ag-positive signals on the short arms of this chromosome pair. No additional Ag-positive sites were observed.
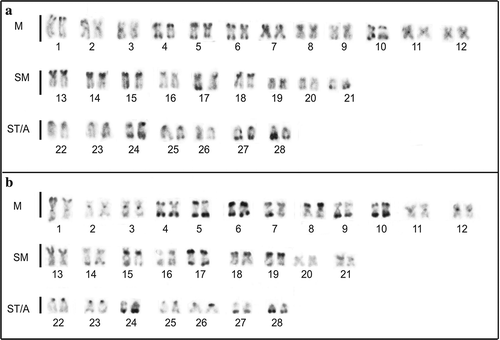
C-banding (–b) also revealed a generally similar pattern of heterochromatin distribution in the two species. C-bands can be seen in the terminal regions of several chromosomes, sometimes on both chromosome arms of biarmed chromosomes. Constitutive heterochromatin is evident in the pericentromeric region of several chromosome pairs, especially in the metacentric chromosomes. In both species, chromosome pair number 13 presents C-positive short arms (, f).
Figure 2. C-banded karyotypes of (a) Pseudoplatystoma metaense and (b) P. orinocoense. M indicates metacentric, SM submetacentric, ST subtelocentric and A acrocentric chromosomes.
In both species, FISH using the 45S rDNA probe (–b) produced two bright signals on the same short-arm locations of the Ag-positive sites on chromosome pair number 13; no additional major ribosomal genes clusters were identified.
Figure 3. Metaphases of Pseudoplatystoma metaense (left) and P. orinocoense (right) after FISH with 45S rDNA (a, b) and with 5S rDNA (c, d). Arrows indicate the NOR-bearing chromosomes. Arrowheads indicate the 5S rDNA-bearing chromosomes.
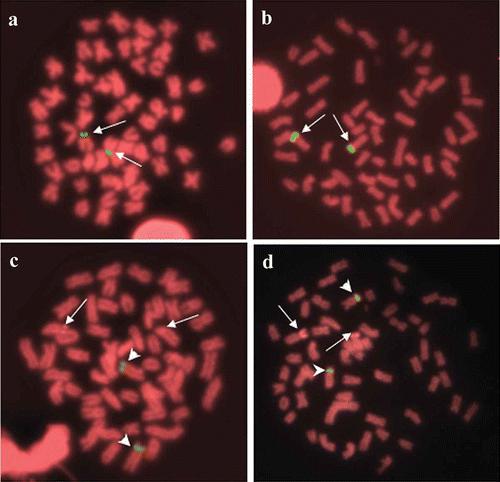
FISH using the 5S rDNA probe (–d) produced signals located in a paracentromeric position on the short arms of a large submetacentric chromosome pair, different from the NOR-bearing one. In fact, after FISH using the 5S rDNA probe, the NORs (arrows in –d) can be identified as an indirect effect of FISH procedures involving propidium iodide staining (Rab et al. Citation1996). Thus, the co-localisation of major and minor ribosomal genes can be excluded.
The almost complete sequence of the cyt b (1050 bp) and a fragment (432 bp) of the COI genes were obtained from the five putative P. metaense and the five putative P. orinocoense specimens. The P. metaense individuals showed two haplotypes for cyt b and one for COI, while the P. orinocoense specimens showed three haplotypes for cyt b and one for COI.
The cyt b sequence similarity was 99.9% among the five P. metaense and 99.8% among the five P. orinocoense specimens; there was a 96.8% similarity between the two species. When BLASTed in GenBank, our P. metaense sequences showed 100% similarity with haplotype H54 found by Carvalho-Costa et al. (Citation2011) in 18 specimens of P. metaense from the Orinoco basin and in 10 specimens of P. tigrinum from the Amazon basin. The remaining top nine matches indicated a 99% similarity with P. tigrinum. After BLASTing our P. orinocoense sequences, the top 10 matches showed 99–100% similarity with P. orinocoense (top four) and 97–98% similarity with both P. reticulatum and P. punctifer.
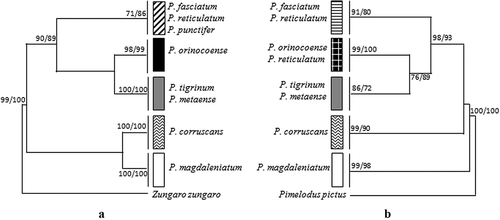
For all (167) of the Pseudoplatystoma cyt b gene sequences available in GenBank (details in ), it was possible to align a fragment of 402 bp; 79 variable sites and 52 haplotypes were detected. This fragment was used for gene genealogy analysis by NJ and ML methods that generated the same tree topology () showing five clades. Our P. metaense and P. orinocoense specimens fall in two separate clades, respectively, within the P. tigrinum-P. metaense and the P. orinocoense clades identified by Carvalho-Costa et al. (Citation2011) using the same molecular marker.
Figure 4. Phylogenetic relationships within the Pseudoplatystoma genus based on (a) cyt b and (b) COI genes. Only the maximum-likelihood (ML) tree topology is shown. The neighbor-joining (NJ) analysis generates the same tree topology. Bootstrap values above 70% for NJ and ML are indicated at each node.
Regarding the COI sequences, the sequence similarity was 100% among the five P. metaense specimens, 100% among the five P. orinocoense specimens, and 97.9% between the two species. We did not find COI sequences of the two Orinoco basin putative taxa in GenBank or in the BOLD database. After BLASTing our COI sequences both in GenBank and in the BOLD system, the top 10 matches showed 100% similarity between our P. metaense and P. tigrinum specimens and 99–100% similarity between our P. orinocoense and P. reticulatum specimens.
For all (315) of the Pseudoplatystoma COI gene sequences available in GenBank (details in ), it was possible to align a fragment of 411 bp; 63 variable sites and 40 haplotypes were detected. Also for COI, the NJ and ML gene genealogy trees show a similar topology (). Five clades were identified, three of which correspond to those identified by cyt b. The fourth clade includes P. orinocoense and nine sequences of P. reticulatum (out of the 34 included in the analysis, Acc. No. GU570871-62). The last clade includes P. fasciatum and P. reticulatum, consistent with these species clustering into the same cyt b clade. However, in the cyt b phylogeny, P. punctifer, represented by over 70 sequences and 23 haplotypes, was also included in this clade. Unfortunately, no COI sequences are available for P. punctifer; thus, a complete comparison between the two gene trees cannot be made.
Discussion
The molecular analysis (see below) distributed the examined specimens, morphologically classified as P. metaense and P. orinocoense, into two distinct Pseudoplatystoma molecular clades corresponding to those identified in the Orinoco basin by Carvalho-Costa et al. (Citation2011). More precisely, our P. metaense sequences fall within the P. tigrinum sensu lato clade, while those of P. orinocoense cluster with the discrete cyt b clade exclusively including the P. orinocoense haplotypes.
Regarding cytogenetics, the two Pseudoplatystoma species from the Orinoco basin show a high degree of similarity both in chromosome number and formulae and in all the considered cytogenetic features, i.e., the constitutive heterochromatin distribution and location of the minor and major ribosomal genes. These results appear to be inconsistent with the picture of the intra- and interspecific karyological diversity, in terms of karyotype formula, fundamental number (FN) and NOR-bearing chromosomes, reported in the genus (). Nevertheless, this variation is probably overestimated, as in such small chromosomes the short arms of submetacentric and subtelocentric chromosomes gradually decrease in size and, depending on the condensation level, it can be quite difficult to define a clear-cut distinction/classification between these chromosomes. It is therefore likely that many of the reported differences are due to technical artefacts and/or subjectivity in classification. Another source of apparent variation between the species is in the FN, which is calculated differently by different authors, as some count subtelocentric as uniarmed and others as biarmed chromosomes (respectively, the first and second number in the FN column of ).
By specifically considering the two Orinoco species reported here in the light of their still-unclear systematic position, it is worthwhile to compare our P. metaense (P. tigrinum sensu lato group, according to Carvalho-Costa et al. Citation2011) data with those previously obtained in nominal P. tigrinum specimens from the Solimões River, Amazon basin (Fenocchio & Bertollo Citation1992). Despite the difference in the reported chromosome formulae (), by examining the original figures of Fenocchio and Bertollo (Citation1992), the chromosome morphology, constitutive heterochromatin distribution and Ag-NOR location demonstrate a karyotype similarity between the analysed specimens of P. tigrinum (Fenocchio & Bertollo Citation1992) and P. metaense (this study).
Regarding P. orinocoense, its cytogenetic features can be compared with those of P. fasciatum (from the Amazon basin, Fenocchio & Bertollo Citation1992; from the Paraná-Paraguay system, Porto-Foresti et al. Citation2000) and P. reticulatum (from the Paraguay River, Prado Citation2010; Neto et al. Citation2011) within the P. fasciatum sensu lato group (Carvalho-Costa et al. Citation2011). Again, under a parsimonious criterion and examining the original figures of the quoted papers, all the considered cytogenetic features appear similar in P. orinocoense and in P. fasciatum, as well as in P. reticulatum. For P. reticulatum, data are also available for the 5S rDNA gene clusters (Prado Citation2010; Neto et al. Citation2011). The number and location of these clusters on a submetacentric chromosome pair, different from the chromosome bearing NORs, suggest that this chromosome pair is likely the same observed in this study in P. orinocoense.
Thus, karyotypic data do not disclose important macrostructural species-specific chromosome markers that can be useful in discriminating the number of valid taxa within the genus.
As mentioned above, the sequence similarity and tree topology of both cyt b and COI genes assign the analysed specimens, identified as belonging to the two Orinoco basin morphospecies, to two distinct Pseudoplatystoma molecular clades. These results, obtained by retrieving all the available sequences from GenBank, therefore, do not support the existence in this basin of a single clade including both P. orinocoense and P. metaense, as was detected by Torrico et al. (Citation2009). Nevertheless, Torrico et al. (Citation2009) themselves hypothesised that their result was surprising and could be due to either a mtDNA introgression between the two species or to a misidentification of the samples, as they used CR sequences retrieved from GenBank.
The cyt b gene genealogy obtained using our haplotypes along with all the haplotypes presently available on GenBank strengthens the cyt b tree obtained by Carvalho-Costa et al. (Citation2011) using a smaller number of sequences.
The lack of COI data from P. punctifer and the fact that the sequences for the cyt b and COI genes were obtained from different specimens/studies do not allow for a straightforward comparison of the two gene phylogenies. Nevertheless, the number and composition of the identified clades are the same in the two gene trees, with the exception of P. reticulatum. For this species, in the cyt b tree, all the available sequences cluster into the same clade with P. punctifer and P. fasciatum, while in the COI tree, these sequences are split in two well-supported clades, one including P. fasciatum and the other including P. orinocoense.
Most of the retrieved P. reticulatum sequences (details in ) were obtained from specimens collected in the southern and more extended area of the discontinuous species range (Buitrago-Suárez & Burr Citation2007), i.e., the Paranà and Paraguay basins, or were purchased in supermarkets in this area. All of these sequences clustered with those of P. fasciatum for both genes. The specimens collected in the northern and more limited area of species distribution, i.e., the Negro River, Amazon basin, were only sequenced for COI gene, and these sequences (Acc. No. GU570862-71) clustered with those of P. orinocoense obtained in this study. As the Negro River is also linked to the Orinoco basin through the Casiquiare River, mtDNA introgression between the two species is the most likely explanation, although a misidentification cannot be excluded.
The phylogenies obtained using all of the available sequences of the Pseudoplatystoma cyt b and COI genes confirm the results of Carvalho-Costa et al. (Citation2011), who concluded that P. metaense cannot be identified as a clade distinct from P. tigrinum. Furthermore, although in the Carvalho-Costa et al. (Citation2011) species tree P. orinocoense falls within the P. fasciatum sensu lato clade, its mitochondrial lineages can still be identified among the others, likely disclosing a historical component of the present genetic diversity distribution.
The additional sampling/sequencing of more reliably identified specimens, and of established geographic origin, are undoubtedly needed. Nevertheless, the available molecular data highlight the need to complement the traditional taxonomic data (morphology-based species identification) with molecular tools (DNA-based species identification) (Teletchea Citation2010) to more precisely define species boundaries and distributions both for conservation and fishery purposes.
Milana_et_al._Table_SI.doc
Download MS Word (40 KB)Acknowledgements
Financial support was provided by the Consejo de Investigación, Universidad de Oriente, Venezuela, and by the Sapienza University, Rome, Italy. We are grateful to Angelo Libertini [National Research Council (CNR), Venice], a dear colleague who will be sadly missed, for kindly providing us with 45S rDNA probe. We declare that any experiments comply with the current laws of the country in which they were performed.
Notes
1. During processing of this manuscript, a new paper, by García-Dávila et al. (2013), Genetica 141:347--358, reported microsatellite data and COI sequences suggesting the existence of two distinct taxa within P. punctifer in the Peruvian Amazon basin.
References
- Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E. 2010. DNA barcoding for conservation and management of Amazonian commercial fish. Biological Conservation 143:1438–1443.
- Bigoni APV, Almeida-Toledo LF, Toledo Filho SA. 1992. Estudos citogenéticos em Pseudoplatystoma coruscans (Pimelodidae, Sorubiminae) do Rio Mogi-Guaçu. In: Resumos do IV Simpósio de Citogenética Evolutiva Aplicada em Peixes Neotropicais. Rio de Janeiro. 32 pp.
- Buitrago-Suárez UA, Burr BM. 2007. Taxonomy of the catfish genus Pseudoplatysoma Bleeker (Siluriformes: Pimelodidae) with recognition of eight species. Zootaxa 1512:1–38.
- Burgess WE. 1989. An atlas of freshwater and marine catfishes: A preliminary survey of the Siluriformes. Neptune City: TFH Publications.
- Carvalho DC, Neto DA, Brasil BS, Oliveira DA. 2011a. DNA barcoding unveils a high rate of mislabeling in a commercial freshwater catfish from Brazil. Mitochondrial DNA 22:97–105.
- Carvalho DC, Oliveira DA, Beheregaray LB, Torres RA. 2012. Hidden genetic diversity and distinct evolutionarily significant units in an commercially important Neotropical apex predator, the catfish Pseudoplatystoma corruscans. Conservation Genetics 13:1671–1675.
- Carvalho DC, Oliveira DA, Pompeu PS, Leal CG, Oliveira C, Hanner R. 2011b. Deep barcode divergence in Brazilian freshwater fishes: The case of the Sao Francisco River basin. Mitochondrial DNA 22(suppl.1):80–86.
- Carvalho-Costa LF, Piorski NM, Willis SC, Galetti PM, Ortí G. 2011. Molecular systematics of the neotropical shovelnose catfish genus Pseudoplatystoma Bleeker 1862 based on nuclear and mtDNA markers. Molecular Phylogenetics and Evolution 59:177–194.
- Eschmeyer WN, Fong JD. 2013. Species of fishes by family/subfamily. Available: http://research.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp. Accessed May 2013 24.
- Fenocchio AS, Bertollo LAC. 1992. Karyotype similarities among Pimelodidae (Pisces, Siluriformes) from the Brazilian Amazon región. Cytobios 69:41–46.
- Ferraris CJ Jr. 2007. Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa 1418:1–628.
- Foresti F, Oliveira C, Almeida-Toledo LF. 1993. A method for chromosome preparations from large specimens of fishes using in vitro short treatment with colchicine. Experientia 49:810–813.
- Froese R, Pauly D. 2013. FishBase. Available: http://www.fishbase.org. Accessed May 2013 24.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52:696–704.
- Howell W, Black D. 1980. Controlled silver staining of Nucleolus Organizer Regions with a protective colloidal developer: A 1-step method. Experientia 36:1014–1015.
- Lee MR, Elder FFB. 1980. Yeast stimulation of bone marrow mitosis for cytogenetic investigations. Cytogenetics and Cell Genetics 26:36–40.
- Lundberg JG, Littmann MW. 2003. Family Pimelodidae (Long–whiskered catfishes). In: Reis R, Kullander SO, Ferraris Jr CJ, editors. Check list of the freshwater fishes of South and Central America. Porto Alegre: Edipucrs. pp. 432–455.
- Lundberg JG, Sullivan JP, Hardman M. 2011. Phylogenetics of the South American catfish family Pimelodidae (Teleostei: Siluriformes) using nuclear and mitochondrial gene sequences. Proceedings of the Academy of National Science, Philadelphia 161:153–189.
- Martins C, Galetti PM Jr. 1999. Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Research 7:363–367.
- Martins-Santos IC, Julio HF Jr, Burin I. 1996. Karyotypic studies of four species of the Sorubiminae subfamily (Pisces, Siluriformes). Caryologia 49:73–80.
- Milana V, Sola L, Congiu L, Rossi AR. 2008. Mitochondrial DNA in Atherina (Teleostei: Atheriniformes): Differential distribution of an intergenic spacer in lagoon and marine forms of Atherina boyeri. Journal of Fish Biology 73:1216–1227.
- Minegishi Y, Aoyama J, Inoue JG, Miya M, Nishida M, Tsukamoto K. 2005. Molecular phylogeny and evolution of the freshwater eels genus Anguilla based on the whole mitochondrial genome sequences. Molecular Phylogenetics and Evolution 34:134–146.
- Nelson JS. 2006. Fishes of the world. 4th ed. Hoboken, NJ: John Wiley & Sons.
- Neto AM, Silva M, Matoso DA, Vicari MR, Almeida MC, Collares-Pereira MJ, Artoni RF. 2011. Karyotype variability in neotropical catfishes of the family Pimelodidae (Teleostei: Siluriformes). Neotropical Ichthyology 9:97–105.
- Pereira LH, Hanner R, Foresti F, Oliveira C. 2013. Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genetics 14:20.
- Pinkel D, Straume T, Gray JW. 1986. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences of the United States of America 83:2934–2938.
- Porto-Foresti F, Andreata AA, Oliveira C, Foresti F. 2000. The karyotype of Pseudoplatystoma fasciatum (Teleostei, Siluriformes) from the Rio Paraguay basin. Chromosome Science 4:99–102.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818.
- Prado FD. 2010. Caracterização citogenética e molecular das espécies pintado (Pseudoplatystoma corruscans), cachara (Pseudoplatystoma reticulatum) e seus híbridos utilizados na piscicultura brasileira. Master Thesis, Universidade Estadual Paulista, Botucatu.
- Rab P, Reedz KM, Ponce de Leon FA, Phillips RB. 1996. A new method for detecting nucleolus organizer regions in fish chromosomes using denaturation and Propidium Iodide staining. Biotechnic and Histochemistry 71:157–162.
- Souza AB, Fonseca CG, Pinheiro L-EL, Ribeiro LP. 1992. Estudos citogenéticos preliminares em Pseudoplatystoma corruscans (Siluriformes, Pimelodidae) da bacia do rio Paraguai. In: Resumos do IV Simpósio de Citogenética Evolutiva Aplicada a Peixes Neotropicais. Rio de Janeiro. 28 pp.
- Souza AB, Fonseca CG, Ribeiro LP, Pinheiro LEL. 1997. Análise cromossomica do surubim Pseudoplatystoma corruscans das bacias dos ríos Sao Francisco e Paraguai. In: Miranda MOT, editor. Belo Horizonte: IBAMA. pp. 57–68.
- Sumner AT. 1972. A simple technique for demonstrating centromeric heterocromatin. Experimental Cell Research 75:304–306.
- Swarça AC, Fenocchio AS, Cestari MM, Dias AL. 2005. Chromosomal divergence among populations of Pseudoplatystoma corruscans (Pisces, Pimelodidae): I. Evidences obtained from conventional Giemsa staining, C- and G- banding. Ichthyological Exploration of Freshwaters 16:325–330.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24:1596–1599.
- Teletchea F. 2010. After 7 years and 1000 citations: comparative assessment of the DNA barcoding and the DNA taxonomy proposals for taxonomists and non-taxonomists. Mitochondrial DNA 21:206–226.
- Torrico JP, Hubert N, Desmarais E, Duponchelle F, Nuñez Rodriguez J, Montoya-Burgos J, Garcia Davila C, Carvajal-Vallejos FM, Grajales AA, Bonhomme F, Renno J-F. 2009. Molecular phylogeny of the genus Pseudoplatystoma (Bleeker, 1862): biogeographic and evolutionary implications. Molecular Phylogenetics and Evolution 51:588–594.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. Barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences 360:1847–1857.