Abstract
The European snow vole (Chionomys nivalis) is a microtine rodent with a highly fragmented distribution range, mostly associated with the main mountain systems from southern Europe to Turkmenistan. In this paper we confirm the occurrence of the snow vole in Portugal, based on morphological characteristics, biometrics and genetic analysis of two individuals captured in the Montesinho Mountain range (northeastern Portugal). Both mitochondrial and nuclear genetic markers were used to confirm the species identity. The analysis of cytochrome b supports previous conclusions on the phylogeographic structure of the species, revealing the existence of several distinct lineages. Moreover, it shows that the Portuguese specimens are closely related to the other Iberian populations. This finding is of great interest as it adds new information regarding the spatial distribution of the snow vole, by redefining the southwestern limits of the species’ range, and it highlights the need for accurate assessment of regional small mammal population trends and conservation status.
Introduction
The European snow vole Chionomys nivalis (Martins, 1842) is a rock-dwelling microtine that lives in mountain regions (1000–4700 m; Amori Citation1999; Luque-Larena et al. Citation2002; Pérez-Aranda et al. Citation2007b). The species inhabits stable rocky formations with open slopes and patches of mixed shrubs and grass meadows (LeLouarn & Janeau Citation1975; Jones & Carter Citation1980; Janeau & Aulagnier Citation1997; Luque-Larena et al. Citation2002), and displays some morphological and behavioural characteristics, such as excellent climbing ability, long tail (nearly half the body length), long vibrissae (up to 60 mm) and well-developed hind feet, which are considered to be an adaptation to those habitats (; Janeau & Aulagnier Citation1997; Luque-Larena et al. Citation2002; Darvish et al. Citation2005).
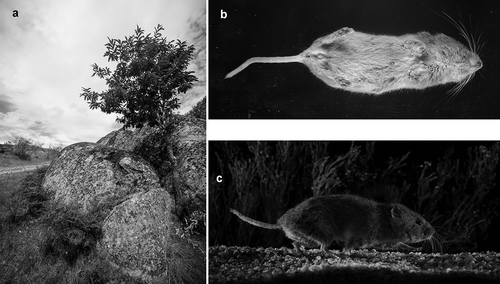
Figure 1. (a) Habitat features of the sampling site; (b) the juvenile female snow vole captured; (c) the adult male snow vole captured (this picture was obtained through infrared beam cells).
The distribution of the European snow vole is wide, ranging from southwestern Europe through southeastern Europe and the Caucasus, to Iran and Turkmenistan (Wilson & Reeder Citation2005). However, due to the specificity of its ecological requirements, the species distribution is highly fragmented and restricted to mountainous regions including the Pyrenees, the Alps, the Apennines, the Tatra Mountains, the Carpathians and the Balkan Peninsula (Nadachowski Citation1991; Nappi Citation2002; Darvish et al. Citation2005; Pérez-Aranda et al. Citation2007a). Although considered a widespread species with stable populations and an International Union for Conservation of Nature (IUCN) conservation status of least concern (LC), the snow vole is regionally protected, being included in the Mammal Red Books of Spain, Italy and Ukraine (Pérez-Aranda et al. Citation2007b; Kryštufek & Amori Citation2008; Bertolino et al. Citation2014). Indeed, in a recent study that defined a ranking system for small mammal conservation priorities, the snow vole in Italy was identified as one of those with the highest score, due to its ecological specialisation and potential vulnerability to habitat changes (Bertolino et al. Citation2014).
Several studies have focused on the evolutionary history of the species through the analysis of mitochondrial and nuclear DNA (Castiglia et al. Citation2009; Yannic et al. Citation2012; Bannikova et al. Citation2013). The first of these, based on mitochondrial DNA, suggested a European origin of the species; however, the later studies, which sampled a wider range of individuals and analysed both mitochondrial and nuclear markers, supported a Middle Eastern origin (Yannic et al. Citation2012; Bannikova et al. Citation2013). Nonetheless, they all demonstrate the existence of a mitochondrial phylogeographic structure with several lineages that occupy geographically congruent ranges, mostly associated with previously described subspecies (Castiglia et al. Citation2009; Yannic et al. Citation2012; Bannikova et al. Citation2013). The Iberian Peninsula represents the southwestern limit of the snow vole distribution range and is occupied by one of the lineages that were highly supported in all of the studies. This lineage includes sequences from two populations (the Pyrenees and Central System) of the five main core areas that have been identified in Spain (Pérez-Aranda et al. Citation2007b). The Iberian populations, particularly those from southern areas, are already considered of conservation concern due to their highly fragmented distribution and low densities (Pérez-Aranda et al. Citation2007b). There are no historical or recent records of the species’ occurrence in Portugal, despite the presence of suitable habitat in both the northern (Serra do Gerês and Serra de Montesinho) and central (Serra da Estrela) regions. This study describes the first record of Chionomys nivalis in Portugal, which is confirmed by morphologic characteristics, biometrics and genetic analysis, and discusses the importance of this new distributional data in terms of the species’ genetic structure and conservation.
Materials and methods
In October 2014, live trapping was performed during six consecutive days in Lama Grande, Montesinho Mountain (41°59′00″N, 6°47′24″W; Bragança, northeastern Portugal) at an elevation of 1370 m. The habitat is heterogeneous and includes stable and extensive patches of large granite rocks, together with shrubs and low herbaceous cover (; Carvalho Citation2005). A total of 27 Sherman traps (23 × 7.5 × 8.5 cm) were set, baited with fresh apples and carrots, and with hydrophobous cotton as bedding material. The two snow voles captured were weighed and sexed, and the reproductive status and standard biometric measurements were recorded (Barnett Citation1992; Blanco Citation1998; Gurnell & Flowerdew Citation2006). The animals were released immediately at the trapping location.
Tissue samples from the snow voles captured (1 mm ear punch) were collected to confirm the species identity by molecular analysis. DNA was extracted using an Easy Spin® Genomic DNA Minipreps Tissue Kit (Citomed, Lisbon, Portugal) following the manufacturer’s instructions. The amplification and sequencing of one mitochondrial gene [cytochrome b (cyt-b)] and one nuclear gene [interphotoreceptor retinoid-binding protein (IRBP)] were then performed, according to the procedures described in Barbosa et al. (Citation2013). The sequences obtained were visually inspected and assembled in BioEdit version 7.2.4 (Hall Citation1999) and were subsequently matched to the reference data set compiled by Barbosa et al. (Citation2013) for species identification. In addition, 41 cyt-b snow vole sequences and three IRBP sequences were retrieved from GenBank () and aligned with the sequences obtained from the specimens captured, using the CLUSTAL W algorithm (Thompson et al. Citation1994) as implemented in BioEdit (Hall Citation1999). The cyt-b 855-bp alignment was reduced into haplotypes with DnaSP 5.10 (Librado & Rozas Citation2009). A median-joining network was then constructed for the cyt-b alignment, to represent the evolutionary relationships among haplotypes, with the algorithm implemented in the software Network 4.6.1.3 (Bandelt et al. Citation1999; http://fluxus-engineering.com).
Table I. Locality and accession numbers of the Chionomys nivalis cyt-b and interphotoreceptor retinoid-binding protein (IRBP) sequences retrieved from GenBank and used in the analysis. Number of sequences is given in parentheses.
A Bayesian coalescent genealogy of the C. nivalis cyt-b sequences was also inferred using BEAST version 1.8.0, inputting through BEAUti version 1.8.0 (Drummond & Rambaut Citation2007). A two-partition (first and second codon position linked; third position separate) HKY (Hasegawa, Kishino and Yano) (+Γ) nucleotide substitution model was used for this, as recommended for protein coding sequence data (Shapiro et al. Citation2006). Three replicate runs of 100 million generations were performed using a constant-size population model and with a sampling frequency that provided a total of 10,000 samples for each run. Tracer version 1.5 (Rambaut & Drummond Citation2007) was used to assess convergence of the runs. The results were then combined, after the removal of 10% burn-in, and a representative genealogy was identified from the posterior sample of trees.
Results and discussion
The two snow voles captured during the trapping session, one juvenile female and one adult male, exhibit morphological characteristics () and biometric measurements within the range described for the species (; Luque-Larena et al. Citation2004; Darvish et al. Citation2005; Pérez-Aranda et al. Citation2007b; Metcheva et al. Citation2008). Although the comparison with previous studies of snow voles from Iran and Bulgaria shows that the tail of the captured male is relatively long, the length is compatible with the range of tail size that has been found in Spanish populations ().
Table II. Age, sex, number of specimens analysed in each study (N) and biometric measures of Chionomys nivalis (standard errors are given in parentheses when available). According to body weight and hair colour, two age classes, juveniles and adults (body mass > 35 g), were defined (LeLouarn & Janeau Citation1975; Luque-Larena et al. Citation2002; Pérez-Aranda et al. Citation2007b).
Genetic analysis of both mitochondrial and nuclear markers confirmed the species identity. The 1143-bp sequences obtained for cyt-b corresponded to the same haplotype (GenBank accession number KT175903) and showed 11 nucleotide substitutions (10 transitions and one transversion) from the most closely related haplotype, which came from Sierra de Gredos in Spain (AM392367). Two different 1040-bp sequences were obtained for the IRBP gene (GenBank accession numbers KT175904 and KT175905), and these had three polymorphisms in relation to the published data (two transitions and one transversion; JX457671-JX457673, Barbosa et al. Citation2013).
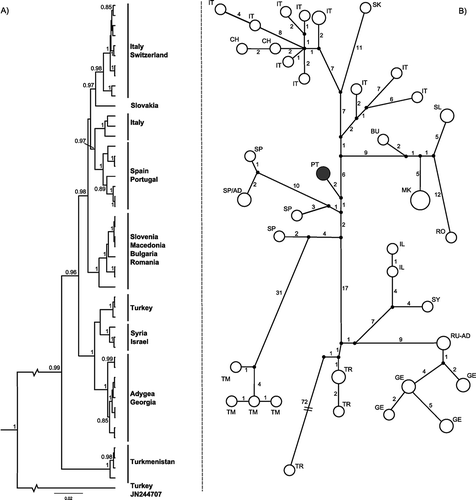
The analysis of the evolutionary relationships of cyt-b haplotypes revealed several geographic clusters, in accordance with previous studies on the species’ phylogeography (; Castiglia et al. Citation2009; Yannic et al. Citation2012; Bannikova et al. Citation2013). In the phylogenetic analysis three main clades were retrieved with high support (), concordant with the results obtained by Yannic et al. (Citation2012). Moreover, there is a haplotype from Turkey that does not cluster with any of the main groups (JN244707; Bannikova et al. Citation2013), showing a divergence of about 9% to these clades. This particular specimen has already been reported by the previous authors as divergent both at the mitochondrial and nuclear levels (Bannikova et al. Citation2013). Considering the three main clades, the most divergent includes the specimens from Turkmenistan (C. nivalis dementievi) and has an uncorrected p-distance of 3.5 to 4.0% from the other clades. The other two clades, which correspond to the Near East and western European specimens, are more closely related (uncorrected p-distance of 1.8% ± 0.34) and each includes several highly supported lineages [Near East: Turkey, Israel, Syria and Georgia; western Europe: Iberian Peninsula, Italy (two lineages), Slovakia, Slovenia, Macedonia, Bulgaria and Romania; see ]. The haplotype from Lama Grande clustered with the remaining Iberian haplotypes with high support (). Moreover, although the records available for the IRBP gene are limited, as they only include a small number of specimens from the Iberian Peninsula (Sorpe and Andorra; Barbosa et al. Citation2013), there is some evidence of differentiation between the western Iberian and Pyrenean populations. This overall pattern is consistent with a Middle Eastern origin of the species, as proposed by other authors (Yannic et al. Citation2012; Bannikova et al. Citation2013), and suggests that there is a certain degree of divergence between fragmented populations.
Figure 2. Phylogenetic analysis of the Chionomys nivalis cyt-b gene 855-bp, considering the sequences retrieved from GenBank (see ) and the sequences obtained from the captured individuals. (A) Maximum clade credibility tree based on Bayesian coalescence genealogy of the 49 C. nivalis cyt-b sequences with BEAST (). Values on branches indicate posterior probability support for the nodes (≥ 0.8). (B) Median-joining network of the C. nivalis cyt-b haplotypes. Each circle represents one haplotype, and circle size is proportional to the number of sequences of each haplotype. The number of nucleotide differences between haplotypes is indicated on branches. Letters indicate country codes: Adygea (RU-AD), Andorra (AD), Bulgaria (BG), Georgia (GE), Israel (IL), Italy (IT), Macedonia (MK), Portugal (PT; in dark grey), Romania (RO), Slovakia (SK), Slovenia (SI), Spain (ES), Syria (SY), Switzerland (CH), Turkey (TR) and Turkmenistan (TM).
This newly detected population in the highest areas of Montesinho Mountain may be spatially connected with the populations already recorded in the contiguous region of Sanabria (Spain). Although this new finding might be seen as evidence for the recent expansion of the species’ distribution range in the northern part of the Iberian Peninsula, it is highly likely that it actually represents the discovery of an existing but previously undetected population in northeastern Portugal. In fact, although information about the distribution of small mammals in Portugal has been gradually updated (e.g. Cruz et al. Citation2002; Vale-Gonçalves & Cabral Citation2014), the available records of species distributions are insufficient to support a consistent national mammal atlas, and the spatial gaps subsist for the range of several species (Vale-Gonçalves & Cabral Citation2014). Therefore, this new record of snow vole occurrence highlights the need for accurate regional surveys of small mammal distribution and population trends to facilitate the definition of species conservation status, as previously emphasised by Bertolino et al. (Citation2014) for Italy. In Portugal, additional sampling effort is needed to delimit the species’ distribution and determine its conservation status. We recommend as the Portuguese common name the designation of ‘Rato-das-neves’, which is a strict translation of the English name.
Overall, our finding contributes new information regarding the southwestern limit of the snow vole distribution in particular, and regarding Portuguese small mammal diversity as a whole, while emphasising the need for further studies on the genetic structure of the species. Our analysis of both mitochondrial and nuclear data, including that from this newly detected population in Portugal, reinforces previous conclusions on the species’ phylogeographic structure (Castiglia et al. Citation2009; Yannic et al. Citation2012; Bannikova et al. Citation2013), and highlights the existence of several divergent and geographically distinct lineages. Although the divergence between these lineages is within the limit of intraspecific differentiation (5% as suggested by Baker & Bradley Citation2006), it may indicate that these constitute potential management units for conservation, due to the level of current fragmentation and isolation and their expected vulnerability to habitat change (Bertolino et al. Citation2014). However, the combination of a more comprehensive sampling design, throughout the species range, with additional genetic analysis, using molecular markers that are widespread in the genome (e.g. microsatellites or SNPs (Single Nucleotide Polymorphisms)), will be crucial to accurately define its distribution and to assess the level of gene flow between the lineages and clarify the level of divergence and isolation of these populations.
Ethical standards
Animals were trapped and ethically handled according to Portuguese law and under permit no. 587/2014/CAPT.
Acknowledgements
This study was supported by funding from several ecological monitoring projects of the Laboratory of Applied Ecology (University of Trás-os-Montes and Alto Douro, UTAD), including the grants BGCT/LEA/402/UTAD/2014 (Hélia Marisa Vale-Gonçalves) and BGCT/LEA/403/UTAD/2014 (Paulo Barros), and by the Portuguese Foundation for Science and Technology (FCT) through the project PEst-OE/AGR/UI4033/2014. Joana Paupério has a postdoctoral grant funded by the project “Genomics and Evolutionary Biology” co-financed by the North Portugal Regional Operational Programme 2007/2013 (ON.2 – O Novo Norte), under the National Strategic Reference Framework, through the European Regional Development Fund. We are grateful to C. M. Ferreira and F. M. S. Martins for collaboration in the genetic analysis, and to P. C. Alves for early discussions of the manuscript. We also thank J. S. Herman for reviewing the English, and the reviewers for valuable comments.
References
- Amori G. 1999. Chionomys nivalis (Martins 1842). In: Mitchell-Jones AJ, Amori G, Bogdanowicz W, Krystufek B, Reijnders PJH, Spitzenberger F, Stubbe M, Thissen JBM, VohralõÂka V, Zima J, editors. Atlas of European mammals. London: Academic Press. pp. 256–257.
- Baker RJ, Bradley RD. 2006. Speciation in mammals and the genetic concept. Journal of Mammalogy 87:643–662. doi:10.1644/06-MAMM-F-038R2.1.
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16:37–48. doi:10.1093/oxfordjournals.molbev.a026036.
- Bannikova AA, Sighazeva AM, MAlikov VG, Golenishev FN, Dzuev RI. 2013. Genetic diversity of Chionomys genus (Mammalia, Arvicolinae) and comparative phylogeography of snow voles. Animal Genetics 49:561–575. doi:10.1134/S1022795413050025.
- Barbosa S, Paupério J, Searle JB, Alves PC. 2013. Genetic identification of Iberian rodent species using both mitochondrial and nuclear loci: Application to noninvasive sampling. Molecular Ecology Resources 13:43–56. doi:10.1111/1755-0998.12024.
- Barnett A. 1992. Small mammals, expedition field techniques. London: Expedition Advisory Centre, Royal Geographical Society.
- Bertolino S, Girardello M, Amori G. 2014. Identifying conservation priorities when data are scanty: A case study with small mammals in Italy. Mammalian Biology 79:349–356. doi:10.1016/j.mambio.2014.06.006.
- Blanco J. 1998. Mamíferos de España. Barcelona: GeoPlaneta.
- Carvalho AMP. 2005. Etnobotánica del Parque Natural de Montesinho. Plantas, tradición y saber popular en un territorio del Nordeste de Portugal. Tesis Doctoral, Universidad Autónoma de Madrid.
- Castiglia R, Annesi F, Kryštufek B, Filippucci MG, Amori G. 2009. The evolutionary history of a mammal species with a highly fragment range: The phylogeography of the European snow vole. Journal of Zoology 279:243–250. doi:10.1111/j.1469-7998.2009.00612.x.
- Cruz R, Santos S, Mira A, Monteiro A, Queirós F, Mathias M. 2002. First record of the common vole Microtus arvalis (Pallas, 1778) for Portugal. Mammalia 66:606–609. doi:10.1515/mamm.2002.66.4.599.
- Darvish J, Siahsarvie R, Javidkar M, Mirshamsi O. 2005. New records of the snow vole Chionomys nivalis (Rodentia: Arvicolinae) from the Binaloud and Elburz Mountains of Iran. Acta Zoologica Cracoviensia 48:67–70. doi:10.3409/173491505783995716.
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7:214. doi:10.1186/1471-2148-7-214.
- Fink S, Fischer MC, Excoffier L, Heckel G. 2010. Genomic scans support repetitive continental colonization events during the rapid radiation of voles (Rodentia: Microtus): The utility of AFLPs versus mitochondrial and nuclear sequence markers. Systematic Biology 59:548–572. doi:10.1093/sysbio/syq042.
- Galewski T, Tilak M, Sanchez S, Chevret P, Paradis E, Douzery EJP. 2006. The evolutionary radiation of Arvicolinae rodents (voles and lemmings): Relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evolutionary Biology 6:80. doi:10.1186/1471-2148-6-80.
- Gurnell J, Flowerdew JR. 2006. Live trapping small mammals: A practical guide, 4th ed. London: The Mammal Society.
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98.
- Jaarola M, Martínková N, Gündüz I, Brunhoff C, Zima J, Nadachowski A, Amori G, Bulatova NS, Chondropoulos B, Fraguedakis-Tsolis S, González-Esteban J, López-Fuster MJ, Kandaurov AS, Kefelioglu H, Luz Mathias M, Villate I, Searle JB. 2004. Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Molecular Phylogenetics and Evolution 33:647–663. doi:10.1016/j.ympev.2004.07.015.
- Janeau G, Aulagnier S. 1997. Snow vole, Chionomys nivalis (Martins, 1842). IBEX J.M.E 4:1–11.
- Jones JK, Carter DC. 1980. The snow vole, Microtus nivalis, in the lowlands of western Yugoslavia. Journal of Mammalogy 61:572. doi:10.2307/1379860.
- Kryštufek B, Amori G 2008. Chionomys nivalis. The IUCN Red List of Threatened Species, Version 2014.2. Available: www.iucnredlist.org. Accessed Oct 2014 30.
- LeLouarn H, Janeau G. 1975. Répartition et biologie du campagnol des neiges Microtus nivalis Martins dans la région de Briançon. Mammalia 39:589–604. doi:10.1515/mamm.1980.44.1.1.
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi:10.1093/bioinformatics/btp187.
- Luque-Larena JJ, López P, Gosálbez J. 2002. Microhabitat use by the snow vole Chionomys nivalis in alpine environments reflects rock-dwelling preferences. Canadian Journal of Zoology 80:36–41. doi:10.1139/z01-197.
- Luque-Larena JJ, López P, Gosálbez J. 2004. Spacing behaviour and morphology predict promiscuous mating strategies in the rock-dwelling snow vole, Chionomys nivalis. Canadian Journal of Zoology 82:1051–1060. doi:10.1139/z04-083.
- Metcheva R, Beltcheva M, Chassovnikarova T. 2008. The snow vole (Chionomys nivalis) as an appropriate environmental bioindicator in alpine ecosystems. Science of the Total Environment 391:278–283. doi:10.1016/j.scitotenv.2007.10.007.
- Nadachowski A. 1991. Systematics, geographic variation, and evolution of snow voles (Chionomys) based on dental characters. Acta Theriologica 36:1–45. doi:10.4098/0001-7051.
- Nappi A. 2002. Vertical distribution of the snow vole Chionomys nivalis (Martins, 1842) (Rodentia, Arvicolidae) in Italy. Hystrix 13:45–52. doi:10.4404/hystrix-13.1-2-4185.
- Pérez-Aranda D, Carro F, Garrido JA, Cano J, Castillo A, Granados JE, Suárez F, Soriguer RC. 2007a. Nuevos datos sobre la distribución del topillo nival Chionomys nivalis (Martins, 1842) en Sierra Nevada (Andalucía, España). Galemys 19:17–24. ISSN: 1137-8700.
- Pérez-Aranda D, Garrido García JA, Soriguer R. 2007b. Chionomys nivalis (Martins, 1842). Ficha Libro Rojo. In: Palomo LJ, Gisbert J, Blanco JC, editors. Atlas y Libro Rojo de los Mamíferos terrestres de España. Madrid: Dirección General de Conservación de la Biodiversidad-SECEM-SECEMU. pp. 410–414.
- Rambaut A, Drummond AJ. 2007. Tracer v1.5. Available: http://beast.biod.ed.ac.uk/Tracer. Accessed Dec 2013
- Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Molecular Biology and Evolution 23:7–9. doi:10.1093/molbev/msj021.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22:4673–4680. doi:10.1093/nar/22.22.4673.
- Vale-Gonçalves HM, Cabral JA. 2014. New records on the distribution of three rodent species in NE Portugal from barn owl (Tyto alba) diet analysis. Galemys 26:100–104. doi:10.7325/Galemys.2014.N3.
- Wilson DE, Reeder DM. 2005. Mammal Species of the World. A Taxonomic and Geographic Reference, 3rd ed. Baltimore, Maryland: Johns Hopkins University Press.
- Yannic G, Burri R, Malikov VG, Vogel P. 2012. Systematics of snow voles (Chionomys, Arvicolinae) revisited. Molecular Phylogenetics and Evolution 62:806–815. doi:10.1016/j.ympev.2011.12.004.
