Abstract
In our previous studies, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15 have been shown to be antagonistic to Aspergillus flavus in stored rice grains. In this study, the biocontrol activities of these strains were evaluated against Aspergillus candidus, Aspergillus fumigatus, Penicillium fellutanum, and Penicillium islandicum, which are predominant in stored rice grains. In vitro and in vivo antifungal activities of the bacterial strains were evaluated against the fungi on media and rice grains, respectively. The antifungal activities of the volatiles produced by the strains against fungal development and population were also tested using I-plates. In in vitro tests, the strains produced secondary metabolites capable of reducing conidial germination, germ-tube elongation, and mycelial growth of all the tested fungi. In in vivo tests, the strains significantly inhibited the fungal growth in rice grains. Additionally, in I-plate tests, strains KU143 and AS15 produced volatiles that significantly inhibited not only mycelial growth, sporulation, and conidial germination of the fungi on media but also fungal populations on rice grains. GC-MS analysis of the volatiles by strains KU143 and AS15 identified 12 and 17 compounds, respectively. Among these, the antifungal compound, 5-methyl-2-phenyl-1H-indole, was produced by strain KU143 and the antimicrobial compounds, 2-butyl 1-octanal, dimethyl disulfide, 2-isopropyl-5-methyl-1-heptanol, and 4-trifluoroacetoxyhexadecane, were produced by strain AS15. These results suggest that the tested strains producing extracellular metabolites and/or volatiles may have a broad spectrum of antifungal activities against the grain fungi. In particular, B. megaterium KU143 and P. protegens AS15 may be potential biocontrol agents against Aspergillus and Penicillium spp. during rice grain storage.
1. Introduction
Stored rice grains contain various microbes such as bacteria and fungi, in which the microbial structure depends on not only the harvesting process but also the field and storage conditions. Deleterious fungal growth in stored rice grains leads to reduced milling quality and nutritional values, discoloration, and production of hazardous mycotoxins [Citation1]. During grain storage, low relative humidity promotes the growth of xerophilic fungi (i.e., dry condition-tolerant fungi) such as Aspergillus and Penicillium spp., which are predominant in rice grains [Citation2–4]. These fungi, along with Fusarium spp., are responsible for mycotoxin contamination of rice grains [Citation5]. Mycotoxins, as secondary metabolites of certain filamentous fungi, are known to elicit toxic biological responses in humans and animals [Citation4,Citation6].
Aspergillus flavus is the most deleterious fungus in stored rice grains, and it receives particular attention because of its ability to produce potent carcinogenic aflatoxins [Citation7]. Along with A. flavus, Aspergillus candidus, Aspergillus fumigatus, Penicillium fellutanum, and Penicillium islandicum are other major contaminants of stored rice grains [Citation2,Citation8–10]. First of all, A. candidus is a common fungus of the natural mycoflora in rice grains [Citation11]. This fungus was reported to represent a potential respiratory hazard and a strong immune-stimulating factor for grain workers [Citation12]. Moreover, A. candidus produces several phenolic compounds such as cytotoxic terphenyllin and xanthoascin (a xanthocillin analog), which are hepato- and cardio-toxic to animals [Citation13,Citation14]. A. fumigatus is the main causative agent of invasive aspergillosis, a life-threatening disease caused by inhaled conidia, particularly in immuno-compromised individuals [Citation15]. Several ergot alkaloids were found to be associated with A. fumigatus conidia in a respirable form at high levels. Ergot alkaloids are known to cause several health problems in humans and animals, including negative effects on the cardiovascular, nervous, and immune systems [Citation16]. In addition, P. islandicum, the main cause of “yellow rice” in Japan, can produce the toxic peptide, islanditoxin [Citation17]. P. fellutanum is also a mycotoxigenic fungus producing diketopiperazine alkaloids (fellutanines A–E), in addition to other alkaloids (agroclavine I and epoxyagroclavine I) [Citation18,Citation19]. Because of the toxic activities and health hazards associated with the fungal contamination of stored rice grains, several attempts have been made in search of environmentally safe, efficient, and sustainable control methods [Citation4,Citation6,Citation20–22].
Conventionally, synthetic agro-chemicals have been used for the management of seed-borne fungal diseases. Fungicides such as ammonia, benomyl, formaldehyde, iprodione, propionic and sorbic acid, and thiabendazole have been applied for controlling grain fungi during storage [Citation23–28]. However, fungicide application on stored grains has been limited because of their adverse effects on the sensory quality of grains, inefficiency in storage conditions, and their toxic potential [Citation29]. Moreover, increasing evidence and awareness of related health issues, as well as the occurrence of fungicide resistance, has highlighted the need for environmentally sound and efficient alternatives to chemical application [Citation29]. A promising approach for controlling deleterious fungi in stored grains may be the use of antagonistic microbes. In particular, microbes producing antifungal volatiles, which is one of the most significant biocontrol traits, may have a potential to control deleterious grain fungi [Citation30,Citation31]. For example, the volatile compound, 2-phenylethanol, produced by the biocontrol yeast Pichia anomala suppressed A. flavus growth and aflatoxin production [Citation32]. Additionally, antagonistic microbes can also serve as an untapped resource of bioactive metabolites [Citation6,Citation33].
In our previous study, three antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, were selected from 460 strains isolated from stored rice grains in Korea [Citation20,Citation34]. These strains exhibited significant biocontrol activities against A. flavus and aflatoxin production in stored rice grains [Citation20]. Moreover, volatiles from B. megaterium KU143 and P. protegens AS15 were found to inhibit A. flavus growth and aflatoxin production [Citation21]. Further examination of antifungal activities of the antagonistic strains against grain fungi such as A. candidus, A. fumigatus, P. fellutanum, and P. islandicum are needed to enable these strains to use as biocontrol agents during grain storage. Therefore, the objectives of this study were (1) to evaluate the antifungal activities of the bacterial strains against Aspergillus and Penicillium spp. predominant in stored rice grains, (2) to investigate the antifungal activities of the volatiles produced by the strains against the tested fungi, and (3) to identify bacterial volatile compounds using gas chromatography-mass spectrometry (GC-MS) analysis.
2. Materials and methods
2.1. Fungal and bacterial strains and rice grains
The antagonistic bacterial strains, B. megaterium KU143, M. testaceum KU313, and P. protegens AS15, were stored in nutrient broth (NB) containing 20% glycerol at −70 °C until used. To prepare bacterial cultures, the strains were streaked on nutrient agar (NA) and incubated at 28 °C for 2 d. Single colonies were transferred to NB and incubated in a rotary shaker (160 rpm) at 28 °C for 2 d. The cultured bacterial cells were harvested and washed twice by centrifugation at 5000×g for 15 min, and suspended in a 10 mM MgSO4 solution. The bacterial suspensions were adjusted to 108 cells/ml (OD600 = 0.5) with the MgSO4 solution, using a spectrophotometer.
The tested fungi, A. candidus AC317, A. fumigatus AF8, P. fellutanum KU53, and P. islandicum KU101, isolated from stored rice grains in Korea [Citation8–10], were stored on potato dextrose agar (PDA) at 4 °C until used. To prepare fungal inocula, the isolates were cultured on PDA for 5 d at 28 °C. Subsequently, conidia were harvested using sterile distilled water (SDW) containing 0.03% Tween 20. Harvested conidia were adjusted to 107 conidia/ml using a hemocytometer. Unhulled, stored rice grains (cv. “Ilpum”) from the Korea University Farm (Namyangju, Korea) were surface-sterilized using 1% sodium hypochlorite for 3 min and 70% ethanol for 5 min [Citation35]; then, washed with SDW three times and blotted on sterile Whatman (No. 1) filter papers.
2.2. In vitro antifungal activity of antagonistic bacterial strains and bacterial culture filtrates in media
Antifungal activity of antagonistic bacterial strains was examined against the tested grain fungi, A. candidus AC317, A. fumigatus AF8, P. fellutanum KU53, and P. islandicum KU101, in a dual-culture assay. The assay was based on the inhibition of the fungal mycelia on PDA plates (90 mm in diameter), streaked in the center with each tested bacterial strain, Sphingomonas aquatilis KU408 (bacterial negative control), or SDW (untreated control). Streaked plates were inoculated with a conidial suspension (2 µl) of each tested fungus, prepared as described above, to the opposite edges of the plates. The plates were sealed with Parafilm M (Sigma-Aldrich, St. Louis, MO), and incubated at 28 °C until mycelia in the untreated controls reached the center of the plates. Inhibition (mm) of mycelial growth of the tested fungi was determined and compared with the mycelial growth of the untreated control plates.
Culture filtrates of antagonistic strains were examined for their activity against conidial germination, germ-tube elongation, and mycelial growth of the tested fungi. The cell-free NB culture filtrates of the bacterial strains, prepared as described above, were obtained using 0.22 µm micro-filters (Merck Millipore, Darmstadt, Germany). One milliliter of the culture filtrate of each tested strain, S. aquatilis KU408 (bacterial negative control) or NB (untreated control) was added to 9 ml of potato dextrose broth (PDB) containing 105 conidia/ml. These samples were incubated at 28 °C for 10 h and conidial germination percentage was assessed by counting the number of germinated conidia among a total of 100 conidia observed. Subsequently, germ-tube lengths (µm) were measured using 20 germinated conidia. Another set of samples was incubated at 28 °C for 2 d in a rotary shaker (200 rpm) for assessing fungal growth. Mycelia of the tested fungi were harvested by filtration using Whatman (No. 1) filter papers, and mycelial dry weights (mg) were determined after drying the mycelia at 60 °C for 2 d.
2.3. In vivo antifungal activity of antagonistic bacterial strains in rice grains
Biocontrol activity of antagonistic bacterial strains against the tested fungi was evaluated according to the procedure described by Mannaa et al. [Citation20]. The surface-sterilized rice grains (2 g) were treated with the bacterial suspensions for 3 h. S. aquatilis KU408, 10 mM MgSO4 solution, and SDW were used as negative controls and a commercial fungicide, Benlate® (50% benomyl; Syngenta Korea, Seoul, Korea) at a concentration of 3 mg/g of rice grains was used as a positive control. Treated rice grains were inoculated with 200 µl of 107 conidia/ml, equivalent to 106 conidia/g dry weights of rice grains.
After 7 d of inoculation, fungal and bacterial populations of the treated rice grains were evaluated as followings. Rice grains were finely ground aseptically with an analytical mill (IKA A11 basic, IKA Works, Wilmington, NC). Grains (1 g) were suspended in 10 ml SDW and incubated in a rotary shaker (160 rpm) at 28 °C for 30 min. After incubation, rice suspensions were serially diluted 10-fold, and 200 µl of each dilution was smeared on 18% glycerol agar (DG18; Fluka 40587, Sigma-Aldrich, St. Louis, MO), followed by incubation at 28 °C for 4 d for the assessment of fungal populations [Citation36]. Another 200 µl of each dilution was smeared on NA supplemented with cycloheximide (50 mg/l) and incubated at 28 °C for 2 d to assess bacterial populations. Fungal and bacterial populations were expressed as colony-forming units (CFUs)/g dry weights of rice grains.
2.4. In vitro antifungal activity of bacterial volatiles on I-plates
In vitro antifungal activity of the volatiles produced by antagonistic bacterial strains against mycelial growth, sporulation, and conidial germination of the tested fungi was conducted using I-plates (plates containing a center partition; Fisher Scientific, Pittsburgh, PA) according to the method described by Mannaa et al. [Citation21]. Briefly, 10 ml of NA supplemented with 50 mg/l cycloheximide was applied to one side of the I-plate, and PDA was poured on the opposite side. Then, 100 µl of the bacterial suspension of each tested strain, S. aquatilis KU408 (bacterial negative control), or 10 mM MgSO4 solution (untreated control) was smeared onto the NA side of the I-plate. After 24 h of incubation at 28 °C, 2 µl of the conidial suspension (107 conidia/ml) of each tested fungus was drop-inoculated on the opposite (containing PDA) edges of the I-plates. These plates were sealed with Parafilm and incubated at 28 °C until mycelia approximately reached the center of the untreated control plates. Mycelial growth area (cm2) was determined using ImageJ software (NIH Image J system, Bethesda, MD) after photography [Citation37]. To evaluate fungal sporulation, conidia in the plates were harvested with SDW containing 0.03% Tween 20, vortexed for 1 min, and the number of conidia was counted using a hemocytometer. Fungal sporulation was expressed as the total number of conidia from each plate, divided by the mycelial growth area (cm−2) of each plate. The conidial germination percentage of the tested fungi was then determined using the harvested conidia as described above. The harvested conidia were filtered through two layers of sterile cheesecloth and transferred to PDB at a final concentration of 105 conidia/ml. After stationary incubation of the samples (three sub-replicates per sample) at 28 °C for 10 h, the number of germinated conidia was counted among a total of 100 conidia observed per sample.
2.5. In vivo antifungal activity of bacterial volatiles on I-plates
In vivo antifungal activity of the volatiles produced by antagonistic bacterial strains against the tested fungi was examined on unhulled rice grains on I-plates. A 100 µl aliquot of the bacterial suspension of each antagonistic strain, the negative control strain or 10 mM MgSO4 solution was smeared on I-plates containing NA as described above, and incubated at 28 °C. After 24 h of incubation, surface-sterilized grains (2 g) were placed on the opposite side of the I-plate, and inoculated with 200 µl of the conidial suspension of each tested fungus, as described above. These plates were sealed with Parafilm and incubated at 28 °C. After 7 d of incubation, the populations of the tested fungi in grains were evaluated as described above, and expressed as CFU/g dry weights of rice grains.
2.6. GC-MS analysis of bacterial volatiles
To identify the volatile compounds produced by B. megaterium KU143 and P. protegens AS15, GC-MS analysis was conducted using the method described by Sang et al. [Citation30]. Bacterial strains were streaked and grown on tryptic soy agar (TSA) and single colonies were pre-cultured in 2 ml of tryptic soy broth (TSB) as described for the bacterial preparation. These pre-cultured strains were further cultured in 1000 ml baffled Erlenmeyer flasks containing 100 ml of TSB. That is, flasks (three flasks per treatment) containing each tested strain or TSB (control) were sealed with two layers of Parafilm and incubated in a rotary shaker (160 rpm) at 28 °C. After 24 h of incubation, volatiles from the headspace of each flask were collected using a 50 ml gastight syringe (Hamilton Co., Reno, NV). Then, 10 ml of the collected volatiles was eluted in 500 µl of hexane and 2 µl of this solution was injected into a GC-MS system (Agilent 7890A for GC and 5975C MSD for MS; Agilent Technologies, Santa Clara, CA) equipped with a DB-5 MS column (307 m × 0.25 mm ×0.25 μm) (Agilent Technologies, Santa Clara, CA). The running conditions were: injector at 200 °C; detector at 210 °C; column oven at 30 °C for 3 min with programing at a rate of 10 °C/min up to 180 °C, then ramped up at a rate of 40 °C min−1 to 200 °C for 2 min. Helium was the carrier gas at a flow rate of 1 ml/min. The mass spectra of the obtained peaks were compared with the spectra corresponding to the best library match using Chemstation software (Hewlett Packard, Palo Alto, CA).
2.7. Statistical analysis
The experiments were designed to be completely randomized designs and were conducted twice with three replicates per treatment. Data from repeated experiments were combined after confirming the homogeneity of variances using Levene’s test [Citation38]. For analysis of fungal populations, the data were analyzed after log transformation. Analysis of variance (ANOVA) was conducted using the general linear model (GLM) procedures, and the means were separated using the least significant difference (LSD) test at p < .05. Statistical analysis of the data was performed using the Statistical Analysis Systems software (SAS Institute, Cary, NC).
3. Results
3.1. In vitro antifungal activity of antagonistic bacterial strains and bacterial culture filtrates against Aspergillus and Penicillium in media
When the antifungal activity of antagonistic bacterial strains was tested in a dual-culture assay, all the tested strains significantly (p < .05) inhibited mycelial growth of the tested fungi compared with the bacterial negative control strain, S. aquatilis KU408 (). Among the tested strains, P. protegens AS15 caused the most significant (p < .05) reduction in mycelial growth in all the tested fungi, followed by B. megaterium KU143 ().
Table 1. Antifungal activities of antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain, Sphingomonas aquatilis KU408, on mycelial growth of Aspergillus candidus AC317, Aspergillus fumigatus AF8, Penicillium fellutanum KU53, and Penicillium islandicum KU101 in dual-culture assays and the effects of cell-free culture filtrates of these strains on conidial germination, germ-tube lengths, and mycelial dry weights of the fungi in potato-dextrose broth tests.
Similarly, the cell-free culture filtrates of the antagonistic strains significantly (p < .05) inhibited conidial germination, germ-tube elongation, and mycelial dry weights of all the tested fungi, compared with untreated and negative controls (). S. aquatilis KU408 showed no antifungal activity against the tested fungi as detected in the untreated control. As observed in the dual-culture assays, P. protegens AS15 showed the highest antifungal activity against all the tested fungi ().
3.2. In vivo antifungal activity of antagonistic bacterial strains against Aspergillus and Penicillium in rice grains
When biocontrol activity of the antagonistic bacterial strains was examined against the grain fungi on unhulled rice grains, B. megaterium KU143, M. testaceum KU313, and P. protegens AS15 significantly (p < .05) inhibited populations of all the tested fungi when compared with SDW and MgSO4 (untreated controls) treatments (Supplementary Figure 1; ). Among these strains, P. protegens AS15 caused the largest reduction in fungal populations in the grains, regardless of the tested fungus. As expected, the commercial agrochemical Benlate® (positive control) caused the largest population reduction in the tested fungi among all the treatments. However, S. aquatilis KU408 (bacterial negative control), failed to reduce the fungal populations compared with the untreated controls in the tested fungi ().
Figure 1. Antifungal activities and populations of antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain Sphingomonas aquatilis KU408 against (A) Aspergillus candidus AC317, (B) Aspergillus fumigatus AF8, (C) Penicillium fellutanum KU53, and (D) Penicillium islandicum KU101, 7 d after treatment in unhulled rice grains. Sterile distilled water (SDW) and 10 mM MgSO4 solution were used as negative controls whereas a commercial fungicide, Benlate® was used as a positive control. Fungal populations were determined after 4 d of incubation at 28 °C on DG18 and total bacterial populations were evaluated after 2 d of incubation at 28 °C on nutrient agar. Different letters on the vertical bars with error bars (standard deviations, n = 6) indicate significant differences between treatments according to the least significant difference test at p < .05. CFU: colony-forming unit.
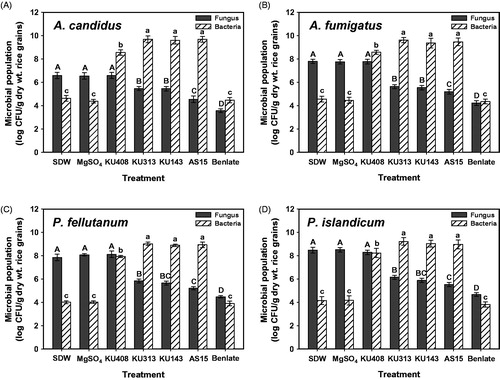
The tested bacterial strains significantly (p < .05) enhanced the bacterial populations in grains compared with the untreated controls, regardless of the tested fungus (). S. aquatilis KU408 also showed larger bacterial populations than the untreated controls for all the tested fungi. However, the tested antagonistic strains exhibited significantly (p < .05) higher bacterial populations than the negative control strain. Untreated and positive control treatments showed similar levels of bacterial populations in the grains for all the tested fungi ().
3.3. In vitro antifungal activity of bacterial volatiles against Aspergillus and Penicillium on I-plates
Volatiles produced by B. megaterium KU143 and P. protegens AS15 significantly (p < .05) inhibited the mycelial growth of all the tested fungi compared with M. testaceum KU313, S. aquatilis KU408 (bacterial negative control), and MgSO4 (untreated control) treatments ( and ). In addition, volatiles from M. testaceum KU313 significantly (p < .05) reduced the mycelial growth of A. fumigatus AF8 and P. islandicum KU101, compared with the negative and untreated controls. The volatiles also significantly (p < .05) reduced the mycelial growth of A. candidus AC317 and P. fellutanum KU53 compared with the untreated control, though these were not significant compared with the bacterial negative control. As expected, S. aquatilis KU408 failed to inhibit the mycelial growth of the tested fungi ( and ).
Figure 2. Photographs of antifungal activities of the volatiles produced by antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain Sphingomonas aquatilis KU408 against (A) Aspergillus candidus AC317, (B) Aspergillus fumigatus AF8, (C) Penicillium fellutanum KU53, and (D) Penicillium islandicum KU101 on the I-plates. Bacterial strains or 10 mM MgSO4 solution (untreated control) were smeared on one side (nutrient agar) of the I-plates and the other side (potato dextrose agar) was inoculated with a conidial suspension of the tested fungi 24 h after the bacterial treatment. Insets are photographs of the conidial germination of the fungi affected by the bacterial volatiles or MgSO4 solution. C: conidium; gt: germ tube. Scale bar, 20 µm.
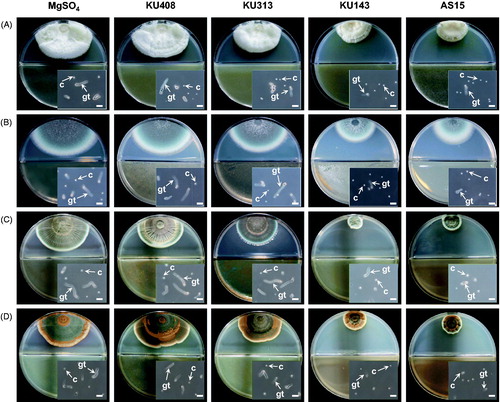
Table 2. Antifungal activities of the volatiles produced by antagonistic bacterial strains, Bacillus megaterium KU143, Microbacterium testaceum KU313, and Pseudomonas protegens AS15, and negative control strain, Sphingomonas aquatilis KU408, on mycelial growth, sporulation, and conidial germination of Aspergillus candidus AC317, Aspergillus fumigatus AF8, Penicillium fellutanum KU53, and Penicillium islandicum KU101 on I-plates.
In the case of bacterial volatile activity against fungal sporulation, volatiles from B. megaterium KU143 and P. protegens AS15 significantly (p < .05) inhibited fungal sporulation regardless of the tested fungus (except for the inhibition of A. candidus by strain AS15) compared with M. testaceum and negative and untreated controls (). Volatiles from M. testaceum KU313 did not inhibit the sporulation of the tested fungi except for A. fumigatus AF8 compared with negative and untreated controls. S. aquatilis KU408 did not inhibit the fungal sporulation ().
In the tests of bacterial volatile effects on conidial germination, volatiles from B. megaterium KU143 and P. protegens AS15 significantly (p < .05) inhibited the conidial germination of the tested fungi (except the inhibition of A. candidus by strain AS15) compared with M. testaceum KU313, and S. aquatilis KU408, and MgSO4 treatments ( and ). Volatiles from M. testaceum KU313 significantly (p < .05) reduced the germination of A. fumigatus AF8 only compared with untreated and negative controls ( and ).
3.4. In vivo antifungal activity of bacterial volatiles against Aspergillus and Penicillium on I-plates
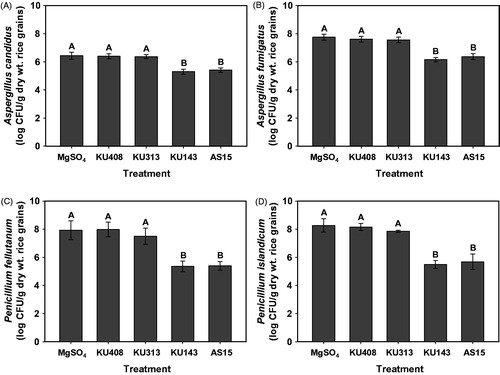
When the antifungal activity of bacterial volatiles was evaluated against the populations of the tested fungi using unhulled rice grains, volatiles produced by B. megaterium KU143 and P. protegens AS15 significantly (p < .05) inhibited the fungal populations, compared with M. testaceum KU313, S. aquatilis KU408 (bacterial negative control), and MgSO4 (untreated control) treatments, regardless of the tested fungus (Supplementary Figure 2; ). Volatiles from M. testaceum failed to reduce the populations of the tested fungi, compared with negative and untreated controls ().
Figure 3. Antifungal activities of the volatiles produced by antagonistic bacterial strains, Microbacterium testaceum KU313, Bacillus megaterium KU143, and Pseudomonas protegens AS15, and negative control strain Sphingomonas aquatilis KU408 against populations of (A) Aspergillus candidus AC317, (B) Aspergillus fumigatus AF8, (C) Penicillium fellutanum KU53, and (D) Penicillium islandicum KU101, 7 d after treatment in unhulled rice grains on the I-plates. Bacterial strains or 10 mM MgSO4 solution (untreated control) were smeared on one side (nutrient agar) of the I-plates and the other side (rice grains) was inoculated with a conidial suspension of the tested fungi, 24 h after bacterial treatment. Fungal populations in rice grains were assessed on DG18 after 4 d of incubation at 28 °C. Different capital letters on the vertical bars indicate significant (p < .05) differences between treatments according to the least significant difference test. CFU: colony-forming unit.
3.5. Volatile compounds produced by B. megaterium KU143 and P. protegens AS15
GC-MS analysis of volatiles produced by B. megaterium KU143 and P. protegens AS15 revealed 12 and 17 distinct peaks, respectively (). The 12 peaks from B. megaterium KU143 identified using the best match in the Chemstation library were: heptane [peak number (PN) 1], 2,5-dimethyltetrahydrofuran (PN 2), methylcyclohexane (PN 3), 3-hexanone (PN 4), 2-hexanone (PN 5), trimethyl [4-(1,1,3,3,-tetramethylbutyl) phenoxy] silane (PN 6), 3-hexene-2,5-diol (PN 7), 2-bromohexane (PN 8), 2,4,4-trimethyl-1-pentanol (PN 9), 2,6-dimethylpiperazine (PN 10), 2,2-dimethyl-3-hexanol (PN 11), and 5-methyl-2-phenyl-1H-indole (PN 12). Similarly, the 17 peaks from P. protegens AS15 were identified as: heptane (PN 1), dimethyl disulfide (PN 2), 2,2,3-trimethylpentane (PN 3), 4-methylheptane (PN 4), 3-hexanone (PN 5), 2-hexanone (PN 6), 2,4-dimethyl-1-heptene (PN 7), 3-methylcyclopentanol (PN 8), 3,3-dimethyloctane (PN 9), 2,6-dimethylnonane (PN 10), 1-(2-methylbutyl)-1-(1-methylpropyl)-cyclopropane (PN 11), 2-butyl-1-octanol (PN 12), undecane (PN 13), 1,3-di-tert-butylbenzene (PN 14), 4-trifluoroacetoxyhexadecane (PN 15), 2-isopropyl-5-methyl-1-heptanol (PN 16), and isotridecanol (PN 17). However, no distinct volatile compounds were detected in the TSB control ().
Figure 4. Gas chromatographs of the volatiles produced by (A) Bacillus megaterium KU143 and (B) Pseudomonas protegens AS15. Antagonistic bacterial strains were cultured in tryptic soy broth (TSB) at 28 °C for 24 h. Volatile compounds from B. megaterium KU143 were identified as: heptane [peak number (PN) 1], 2,5-dimethyltetrahydrofuran (PN 2), methylcyclohexane (PN 3), 3-hexanone (PN 4), 2-hexanone (PN 5), trimethyl [4-(1,1,3,3,-tetramethylbutyl) phenoxy] silane (PN 6), 3-hexene-2,5-diol (PN 7), 2-bromohexane (PN 8), 2,4,4-trimethyl-1-pentanol (PN 9), 2,6-dimethylpiperazine (PN 10), 2,2-dimethyl-3-hexanol (PN 11), and 5-methyl-2-phenyl-1 H-indole (PN 12). Volatile compounds from P. protegens AS15 were identified as: heptane (PN 1), dimethyl disulfide (PN 2), 2,2,3-trimethylpentane (PN 3), 4-methylheptane (PN 4), 3-hexanone (PN 5), 2-hexanone (PN 6), 2,4-dimethyl-1-heptene (PN 7), 3-methylcyclopentanol (PN 8), 3,3-dimethyloctane (PN 9), 2,6-dimethylnonane (PN 10), 1-(2-methylbutyl)-1-(1-methylpropyl)-cyclopropane (PN 11), 2-butyl-1-octanol (PN 12), undecane (PN 13), 1,3-di-tert-butylbenzene (PN 14), 4-trifluoroacetoxyhexadecane (PN 15), 2-isopropyl-5-methyl-1-heptanol (PN 16), and isotridecanol (PN 17). However, no distinct volatile compounds were detected in the TSB control.
![Figure 4. Gas chromatographs of the volatiles produced by (A) Bacillus megaterium KU143 and (B) Pseudomonas protegens AS15. Antagonistic bacterial strains were cultured in tryptic soy broth (TSB) at 28 °C for 24 h. Volatile compounds from B. megaterium KU143 were identified as: heptane [peak number (PN) 1], 2,5-dimethyltetrahydrofuran (PN 2), methylcyclohexane (PN 3), 3-hexanone (PN 4), 2-hexanone (PN 5), trimethyl [4-(1,1,3,3,-tetramethylbutyl) phenoxy] silane (PN 6), 3-hexene-2,5-diol (PN 7), 2-bromohexane (PN 8), 2,4,4-trimethyl-1-pentanol (PN 9), 2,6-dimethylpiperazine (PN 10), 2,2-dimethyl-3-hexanol (PN 11), and 5-methyl-2-phenyl-1 H-indole (PN 12). Volatile compounds from P. protegens AS15 were identified as: heptane (PN 1), dimethyl disulfide (PN 2), 2,2,3-trimethylpentane (PN 3), 4-methylheptane (PN 4), 3-hexanone (PN 5), 2-hexanone (PN 6), 2,4-dimethyl-1-heptene (PN 7), 3-methylcyclopentanol (PN 8), 3,3-dimethyloctane (PN 9), 2,6-dimethylnonane (PN 10), 1-(2-methylbutyl)-1-(1-methylpropyl)-cyclopropane (PN 11), 2-butyl-1-octanol (PN 12), undecane (PN 13), 1,3-di-tert-butylbenzene (PN 14), 4-trifluoroacetoxyhexadecane (PN 15), 2-isopropyl-5-methyl-1-heptanol (PN 16), and isotridecanol (PN 17). However, no distinct volatile compounds were detected in the TSB control.](/cms/asset/79617019-24c6-4471-b627-3a71584de10d/tmyb_a_1454015_f0004_c.jpg)
4. Discussion
Biological control of plant pathogens or deleterious storage fungi with microbial antagonists or metabolites is an environmentally sound alternative to conventional chemical control, especially at the postharvest stage, when the use of agricultural chemicals is potentially undesirable for animal or human health and development of fungicide resistance [Citation39]. The investigation of biocontrol or antimicrobial characteristics of antagonistic microbes is often important for understanding biocontrol mechanisms to get better control efficacy of the antagonists and for isolating biological compounds that are effective against deleterious grain fungi [Citation40]. In our previous studies [Citation20,Citation34], we selected three antagonistic bacterial strains, B. megaterium KU143, M. testaceum KU313, and P. protegens AS15, from a total of 460 strains isolated from rice grains, against A. flavus. The selected strains exhibited significant in vitro and in vivo antifungal activity against A. flavus. These strains were efficient colonizers of the surface of rice grains and good competitors of A. flavus of the grains, as shown in our previous study [Citation20]. Furthermore, the strains exhibited an inhibitory effect against aflatoxin production in rice grains. Among these strains, P. protegens AS15 could degrade aflatoxin B1 and utilize the toxin for bacterial growth in nutrient-deficient conditions. In this study, the selected antagonistic strains were examined if they could show a broad spectrum of antifungal or biocontrol activity against A. candidus, A. fumigatus, P. fellutanum, and P. islandicum, which are deleterious grain fungi predominant in stored rice. Consequently, we found that these strains inhibited the growth and development of all the tested fungi on media and rice grains.
The investigation of biocontrol activity of the strains in this study revealed that they could produce extracellular secondary metabolites with antifungal activity against the tested fungi from stored rice grains. These strains inhibited mycelial growth of the fungi in dual-culture assays. The cell-free culture filtrates also suppressed conidial germination, germ tube elongation, and mycelial growth of the fungi. Previously, viable cells and supernatants of Lactobacillus spp. have been shown to inhibit the growth of A. flavus and to reduce germination rates and germ-tube elongation of fungal conidia [Citation41]. Our results from culture filtrates of the antagonistic strains in this study were consistent with previous observations by Mannaa et al. [Citation20], which showed that the culture filtrates significantly inhibited the growth of A. flavus in rice grains. Reddy et al. [Citation42] also showed that the culture filtrates of several biocontrol agents were inhibitory against A. flavus and aflatoxin production in rice grains, while filtrates from Pseudomonas fluorescens and Rhodococcus erythropolis produced the most suppressive activities against A. flavus growth and aflatoxin production. Similarly, in this study, culture filtrates from P. protegens AS15 exhibited the most effective antifungal activity of the tested bacterial strains against the growth of all the tested fungi.
One of the potential biocontrol methods is bio-rational approach, which uses antifungal compounds produced by antagonistic microbes, particularly when the direct application of the biocontrol agents is not feasible due to practical limitations [Citation43]. For example, biocontrol species Bacillus subtilis has been shown to produce antifungal peptides (peptidolipid iturins), such as bacillomycin and mycosubtilin. These compounds can inhibit not only spore germination of mycotoxigenic Aspergillus, Fusarium, and Penicillium, but also the fungal contamination of stored grains, such as corn, peanuts, and cottonseeds [Citation43–45]. Several other compounds from Bacillus pumilus are also known to have inhibitory activities against Mucoraceae and Aspergillus, including A. flavus and A. fumigatus [Citation46,Citation47]. Dikin et al. [Citation48] reported that the supernatant of a M. testaceum strain contained aminopyrrole, phenazine, phenylpyrrole, and pyrrolnitrin, which could inhibit the growth of Schizophyllum commune. Meanwhile, Zhang et al. [Citation49] observed that cell-free culture filtrates from a strain of B. subtilis reduced the growth and spore germination of A. flavus, and possessed bacillomycin-like active compounds, which showed complete inhibition of A. flavus in peanuts [Citation49]. Thus, further investigation is needed for purification and characterization of the antifungal compounds produced by the bacterial strains tested in this study.
Volatile compounds from antagonistic bacteria are one of the important traits for their biocontrol or antifungal activity [Citation50]. For example, Fernando et al. [Citation51] attempted to identify bacterial volatiles from Pseudomonas aurantiaca, Pseudomonas chlororaphis, Pseudomonas corrugata, and Pseudomonas fluorescens. They presented several compounds, such as benzothiazole, cyclohexanol, n-decanal, dimethyl trisulfide, 2-ethyl-1-hexanol, and nonanal, as having inhibitory activities against ascospore germination and mycelial growth of Sclerotinia sclerotiorum. Other studies [Citation30,Citation31] demonstrated that 2,4-di-tert-butylphenol produced by Flavobacterium johnsoniae inhibited mycelial growth, sporulation, and spore germination of Colletotrichum acutatum and Phytophthora capsici. Volatiles from Bacillus amyloliquefaciens are also known to have antifungal activity, inhibiting the growth and spore germination of Fusarium oxysporum f. sp. cubense [Citation52]. In our previous study [Citation21], volatiles from B. megaterium KU143 and P. protegens AS15 inhibited mycelial growth, sporulation, conidial germination, and aflatoxin production by A. flavus on media and rice grains. The biocontrol activity of the volatiles from these two strains was also observed in this study against the growth and development of A. candidus, A. fumigatus, P. fellutanum, and P. islandicum. However, M. testaceum failed to produce significant antifungal volatiles against the tested fungi, except for A. fumigatus in medium tests.
In the GC-MS analysis of bacterial volatiles, 12 and 17 volatile compounds produced by B. megaterium KU143 and P. protegens AS15, respectively, were identified. Among these compounds, 5-methyl-2-phenyl-1H-indole from B. megaterium KU143 and 2-butyl 1-octanal, dimethyl disulfide, 2-isopropyl-5-methyl-1-heptanol, and 4-trifluoroacetoxyhexadecane from P. protegens AS15 were reported as having antifungal or antimicrobial activities [Citation53–59]. Derivatives and related compounds of 5-methyl-2-phenyl-1H-indole were previously found to possess biological activity against not only bacteria such as Pseudomonas aeruginosa and Klebsiella pneumoniae but also fungi such as Aspergillus niger and Aspergillus oryzae [Citation54,Citation57]. Wang et al. [Citation59] identified volatiles from biocontrol species Streptomyces albidoflavus, in which inhibited Aspergillus spp. and Fusarium moniliforme. Among the volatiles, dimethyl disulfide completely inhibited the mycelial growth and sporulation of F. moniliforme. Similarly, Li et al. [Citation53] observed that volatiles from Streptomyces globisporus had antifungal activity against Penicillium italicum on citrus fruits. When the identified compounds were tested for antifungal activity, it was found that dimethyl disulfide inhibited mycelial growth, sporulation, and conidial germination of P. italicum. Additionally, 2-butyl 1-octanal was identified from the biocontrol agent Trichoderma harzianum [Citation58]. Antimicrobial compound 4-trifluoroacetoxyhexadecane was identified from secondary metabolites of the leaves and bark of Naringi crenulate [Citation55]. Previously, 2-isopropyl-5-methyl-1-heptanol was reported as one of the prevailing compounds from the leaves of Feronia elephantum, an Indian tree that has been used for medical purposes [Citation56].
Taken together, the present results show that the tested bacterial strains, B. megaterium KU143, M. testaceum KU313 and P. protegens AS15, possess a broad spectrum of antifungal activities against Aspergillus and Penicillium spp., which are predominant in stored rice grains. These strains produced extracellular secondary metabolites with inhibitory activity against mycelial growth, conidial germination, and germ-tube elongation of the tested fungi. In addition, B. megaterium KU143 and P. protegens AS15 could produce volatiles with antifungal activity against the tested fungi. Among the identified volatile compounds, the antifungal compound, 5-methyl-2-phenyl-1H-indole, was produced by strain KU143, whereas the antimicrobial compounds, 2-butyl 1-octanal, dimethyl disulfide, 2-isopropyl-5-methyl-1-heptanol, and 4-trifluoroacetoxyhexadecane, were produced by strain AS15. Therefore, the bacterial strains in this study, especially B. megaterium KU143 and P. protegens AS15, may be potential biocontrol agents against Aspergillus and Penicillium species during rice grain storage.
Supplemental Material
Download MS Power Point (2.3 MB)Supplemental Material
Download MS Power Point (1.5 MB)Acknowledgments
This work was supported by a Korea University Grant. M. Mannaa was supported by the Korean Government Scholarship Program (KGSP) during his Ph.D. study at Korea University, Seoul, Korea.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Magan N, Hope R, Cairns V, et al. Post-harvest fungal ecology: impact of fungal growth and mycotoxin accumulation in stored grain. Eur J Plant Pathol. 2003;109:723–730.
- Oh JY, Jee SN, Nam Y, et al. Populations of fungi and bacteria associated with samples of stored rice in Korea. Mycobiology. 2007;35:36–38.
- Mew TW, Gonzales P. A handbook of rice seedborne fungi. Los Baños: International Rice Research Institute; 2002.
- Mannaa M, Kim KD. Influence of temperature and water activity on deleterious fungi and mycotoxin production during grain storage. Mycobiology. 2017;45:240–254.
- Park JW, Choi SY, Hwang HJ, et al. Fungal mycoflora and mycotoxins in Korean polished rice destined for humans. Int J Food Microbiol. 2005;103:305–314.
- Mannaa M, Kim KD. Microbe-mediated control of mycotoxigenic grain fungi in stored rice with focus on aflatoxin biodegradation and biosynthesis inhibition. Mycobiology. 2016;44:67–78.
- Scheidegger KA, Payne GA. Unlocking the secrets behind secondary metabolism: a review of Aspergillus flavus from pathogenicity to functional genomics. J Toxicol Toxin Rev. 2003;22:423–459.
- Oh JY, Kim EN, Ryoo MI, et al. Morphological and molecular identification of Penicillium islandicum isolate KU101 from stored rice. Plant Pathol J. 2008;24:469–473.
- Oh JY, Sang MK, Lee HJ, et al. First detection of Penicillium fellutanum from stored rice in Korea. Res Plant Dis. 2011;17:216–221.
- Oh JY, Sang MK, Oh JE, et al. Microbial population, aflatoxin contamination and predominant Aspergillus species in Korean stored rice. Plant Pathol J. 2010;26:121–129.
- Boller RA, Schroeder HW. Influence of Aspergillus candidus on production of aflatoxin in rice by Aspergillus parasiticus. Phytopathology. 1974;64:121–123.
- Krysińska-Traczyk E, Dutkiewicz J. Aspergillus candidus: a respiratory hazard associated with grain dust. Ann Agric Environ Med. 2000;7:101–109.
- Takahashi C, Sekita S, Yoshihira K, et al. The structures of toxic metabolites of Aspergillus candidus. II. The compound B (xanthoascin), a hepato- and cardio-toxic xanthocillin analog. Chem Pharm Bull. 1976;24:2317–2321.
- Takahashi C, Yoshihira K, Natori S, et al. The structures of toxic metabolites of Aspergillus candidus. I. The compounds A and E, cytotoxic p-terphenyls. Chem Pharm Bull. 1976;24:613–620.
- Abad A, Fernández-Molina JV, Bikandi J, et al. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol. 2010;27:155–182.
- Panaccione DG, Coyle CM. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl Environ Microbiol. 2005;71:3106–3111.
- Marumo S. Islanditoxin, a toxic metabolite produced by Penicillium islandicum sopp: part III. Bull Agric Chem Soc Jpn. 1959;23:428–437.
- Kozlovsky AG, Vinokurova NG, Adanin VM, et al. New diketopiperazine alkaloids from Penicillium fellutanum. J Nat Prod. 2000;63:698–700.
- Vinokurova NG, Boichenko LV, Arinbasarov MU. Production of alkaloids by fungi of the genus Penicillium grown on wheat grain. Appl Biochem Microbiol. 2003;39:403–406.
- Mannaa M, Oh JY, Kim KD. Microbe-mediated control of Aspergillus flavus in stored rice grains with a focus on aflatoxin inhibition and biodegradation. Ann Appl Biol. 2017;171:376–392.
- Mannaa M, Oh JY, Kim KD. Biocontrol activity of volatile-producing Bacillus megaterium and Pseudomonas protegens against Aspergillus flavus and aflatoxin production on stored rice grains. Mycobiology. 2017;45:213–219.
- Mannaa M, Kim KD. Control strategies for deleterious grain fungi and mycotoxin production from preharvest to postharvest stages of cereal crops: a review. Life Sci Nat Resour Res. 2017;25:13–27.
- Muir WE, Wallace HAH. Effects of treating damp grain with formaldehyde to prevent storage deterioration. Can J Plant Sci. 1972;52:375–379.
- Bothast RJ, Lancaster EB, Hesseltine CW. Ammonia kills spoilage molds in corn. J Dairy Sci. 1973;56:241–245.
- Sauer DB, Burroughs R. Efficacy of various chemicals as grain mold inhibitors. Trans ASAE. 1974;17:557–559.
- Dunkel F, Lung PZ, Chuan L, et al. Insect and fungal response to sorbic acid-treated wheat during storage in south China. J Econ Entomol. 1982;75:1083–1088.
- White DG, Toman J, Burnette DC, et al. The effect of postharvest fungicide application on storage fungi of corn during ambient air drying and storage. Plant Dis. 1993;77:562–568.
- White DG, Toman J. Effects of postharvest oil and fungicide application on storage fungi in corn following high-temperature grain drying. Plant Dis. 1994;78:38–43.
- Gerhardson B. Biological substitutes for pesticides. Trends Biotechnol. 2002;20:338–343.
- Sang MK, Kim JD, Kim BS, et al. Root treatment with rhizobacteria antagonistic to Phytophthora blight affects anthracnose occurrence, ripening, and yield of pepper fruit in the plastic house and field. Phytopathology. 2011;101:666–678.
- Sang MK, Kim KD. The volatile-producing Flavobacterium johnsoniae strain GSE09 shows biocontrol activity against Phytophthora capsici in pepper. J Appl Microbiol. 2012;113:383–398.
- Hua SST, Beck JJ, Sarreal SBL, et al. The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 2014;30:71–78.
- Wilson MC, Mori T, Rückert C, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62.
- Lee SY, Oh JY, Ryoo MI, et al. Biological control of the rice storage fungi Aspergillus and Penicillium species by antagonistic bacteria originated from rice. Plant Pathol J. 2007;23:328.
- Gu Q, Han N, Liu J, et al. Expression of Helicobacter pylori urease subunit B gene in transgenic rice. Biotechnol Lett. 2006;28:1661–1666.
- Hocking AD, Pitt JI. Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl Environ Microbiol. 1980;39:488–492.
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675.
- Levene H. Robust tests for equality of variances. In: Olkin, I, Ghurye, SG, Hoeffeling, W, Madow, WG, Mann, HB, editors. Contributions to probability and statistics: essays in honor of Harold Hotelling. Stanford (CA): Stanford University Press; 1960.
- Sharma RR, Singh D, Singh R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: a review. Biol Control. 2009;50:205–221.
- Wisniewski ME, Wilson CL. Biological control of postharvest diseases of fruit and vegetables: recent advances. HortScience. 1992;27:94–98.
- Gourama H, Bullerman LB. Inhibition of growth and aflatoxin production of Aspergillus flavus by Lactobacillus species. J Food Prot. 1995;58:1249–1256.
- Reddy KRN, Reddy CS, Muralidharan K. Potential of botanicals and biocontrol agents on growth and aflatoxin production by Aspergillus flavus infecting rice grains. Food Control. 2009;20:173–178.
- Klich MA, Lax AR, Bland JM. Inhibition of some mycotoxigenic fungi by iturin A, a peptidolipid produced by Bacillus subtilis. Mycopathologia. 1991;116:77–80.
- Gueldner RC, Reilly CC, Pusey PL, et al. Isolation and identification of iturins as antifungal peptides in biological control of peach brown rot with Bacillus subtilis. J Agric Food Chem. 1988;36:366–370.
- Klich MA, Arthur KS, Lax AR, et al. Iturin A: a potential new fungicide for stored grains. Mycopathologia. 1994;127:123–127.
- Munimbazi C, Bullerman LB. Isolation and partial characterization of antifungal metabolites of Bacillus pumilus. J Appl Microbiol. 1998;84:959–968.
- Bottone EJ, Peluso RW. Production by Bacillus pumilus (MSH) of an antifungal compound that is active against Mucoraceae and Aspergillus species: preliminary report. J Med Microbiol. 2003;52:69–74.
- Dikin A, Sijam K, Kadir J, et al. Mode of action of antimicrobial substances from Burkholderia multivorans and Microbacterium testaceum against Schizophyllum commune Fr. Int J Agric Biol. 2007;9:311–314.
- Zhang T, Shi ZQ, Hu LB, et al. Antifungal compounds from Bacillus subtilis B-FS06 inhibiting the growth of Aspergillus flavus. World J Microbiol Biotechnol. 2008;24:783–788.
- Xu CK, Mo MH, Zhang LM, et al. Soil volatile fungistasis and volatile fungistatic compounds. Soil Biol Biochem. 2004;36:1997–2004.
- Fernando WD, Ramarathnam R, Krishnamoorthy AS, et al. Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol Biochem. 2005;37:955–964.
- Yuan J, Raza W, Shen Q, et al. Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl Environ Microbiol. 2012;78:5942–5944.
- Li Q, Ning P, Zheng L, et al. Fumigant activity of volatiles of Streptomyces globisporus JK-1 against Penicillium italicum on Citrus microcarpa. Postharvest Biol Technol. 2010;58:57–165.
- Rao RM, Reddy GN, Sreeramulu J. Synthesis of some new pyrazolo-pyrazole derivatives containing indoles with antimicrobial activity. Der Pharma Chem. 2011;3:301–309.
- Sarada K, Margret RJ, Mohan VR. GC-MS Determination of bioactive components of Naringi crenulata (Roxb) Nicolson. Int J Chem Tech Res. 2011;3:1548–1555.
- Muthulakshmi A, Margret R, Mohan VR. GC-MS analysis of bioactive components of Feronia elephantum Correa (Rutaceae). J Appl Pharm Sci. 2012;2:69–74.
- Raghunath SA, Manjunatha Y, Rayappa K. Synthesis, antimicrobial, and antioxidant activities of some new indole analogues containing pyrimidine and fused pyrimidine systems. Med Chem Res. 2012;21:3809–3817.
- Siddiquee S, Cheong BE, Taslima K, et al. Separation and identification of volatile compounds from liquid cultures of Trichoderma harzianum by GC-MS using three different capillary columns. J Chromatogr Sci. 2012;50:358–367.
- Wang C, Wang Z, Qiao X, et al. Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1. FEMS Microbiol Lett. 2013;341:45–51.
