Abstract
Mon1 is a guanine nucleotide exchange factor subunit that activates the Ypt7 Rab GTPase and is essential for vacuole trafficking and autophagy in eukaryotic organisms. Here, we identified and characterized the function of Mon1, an ortholog of Saccharomyces cerevisiae Mon1, in a human fungal pathogen, Cryptococcus neoformans. Mutation in mon1 resulted in hypersensitivity to thermal stress. The mon1 deletion mutant exhibited increased sensitivity to cell wall and endoplasmic reticulum stress. However, the mon1 deletion mutant showed more resistance to the antifungal agent fluconazole. In vivo studies demonstrated that compared to the wild-type strain, the mon1 deletion mutant attenuated virulence in the Galleria mellonella insect model. Moreover, the mon1 deletion mutant was avirulent in the murine inhalation model. These results demonstrate that Mon1 plays a crucial role in stress survival and pathogenicity in C. neoformans.
Keywords:
1. Introduction
Vacuoles, the fungal equivalent of lysosomes, are key organelles essential for the vesicular transport of molecules between the different membrane organelles in fungi [Citation1–3]. These organelles contain various intrinsic vacuolar hydrolases that degrade cellular components and recycle cytoplasmic organelles [Citation4]. Vacuolar hydrolases are transported from donor organelles (vacuoles) to acceptor organelles via selective or non-selective pathways [Citation5–7]. Intracellular membrane fusion, including vacuole fusion, is a key step in these pathways that involves multiple membrane proteins such as the soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) complex, homotypic fusion and vacuolar protein sorting (C-Vps/HOPS) complex, and Rab GTPase Ypt7p [Citation8–12].
Mon1 is a subunit of the Mon1-Ccz1 hetero-complex that acts as a guanine nucleotide exchange factor (GEF) in Saccharomyces cerevisiae [Citation13–15]. This heterodimeric complex activates Ypt7p, thereby regulating endosomal maturation and vacuole fusion in autophagosomes [Citation16–19]. Recently, a detailed molecular mechanism underlying the Mon1-Ccz1 hetero-complex was discovered via structural and biochemical analyses [Citation20]. In many fungi, Mon1 orthologs play a role in vacuole targeting (Cvt) and autophagy pathways in the cytoplasm [Citation1,Citation20–23]. Moreover, Mon1 also plays an essential role in fungal development and pathogenicity in two plant pathogens, Fusarium graminearum and Magnaporthe oryzae [Citation21,Citation22,Citation24]. However, the pathogenic role of Mon1 in human pathogenic fungi remains unclear.
Cryptococcus neoformans is a basidiomycetous yeast that is used as a model organism for studying fungal pathogenesis and antifungal drug discovery [Citation25,Citation26]. This ubiquitous fungus is widespread in the environment and can be inhaled by humans in the form of spores or desiccated yeast cells [Citation27,Citation28]. The inhaled fungal pathogen colonizes in the lung and results in pulmonary infectious diseases or meningoencephalitis once it crosses the blood-brain-barrier (BBB) in immunocompromised hosts such as organ transplant and AIDS patients [Citation29–32]. Recently, various comprehensive analyses have been conducted that have provided insights into the virulence mechanisms of C. neoformans [Citation33–38].
Several studies demonstrated the importance of the vacuolar trafficking pathway in C. neoformans pathogenesis [Citation39]. Deletion of genes encoding proteins associated with vacuole sorting reduced or eliminated virulence [Citation34,Citation40–42]. Although the function of several vacuole-sorting proteins has been studied, the role of Mon1 in the virulence of C. neoformans has not yet been elucidated. In this study, we investigated the roles of the Mon1 ortholog in C. neoformans (Cnmon1) and found that the deletion of mon1 (Cnmon1Δ) caused growth defects at high temperatures and hypersensitivity to dithiothreitol (DTT; an ER stress inducer) and sodium dodecyl sulfate (SDS; a membrane destabilizing agent). Moreover, loss of Cnmon1 led to significant defects in fungal pathogenicity in vivo, suggesting its role in the response against environmental stresses and virulence in C. neoformans.
2. Material and methods
2.1 Fungal strains and culture conditions
Cryptococcus neoformans strains used in this study are listed in . For general cultures, fungal strains were grown in yeast extract-peptone-dextrose (YPD; Difco, Sparks, MD, USA) broth or agar supplemented with relevant antibiotics and incubated at 30 °C. To test thermotolerance, 5 µL of cells cultured in YPD broth overnight were serially diluted 10-fold and spotted onto YPD agar. The cells were then incubated at different temperatures (30, 37, 38, and 39 °C). To examine stress tolerance, each strain was serially diluted 10-fold and spotted onto YPD agar supplemented with the following compounds: dithiothreitol (DTT; Sigma), sodium dodecyl sulfate (SDS; Fisher, Fair Lawn, NJ, USA), and fluconazole (Sigma). The strains were incubated at 30 °C and the plates were photographed from 48 or 72 hours post-treatment.
Table 1. Cryptococcus neoformans strains used in this study.
2.2 Generation of mon1 mutant strains
The oligonucleotides used in this study are listed in . To generate mon1 deletion mutants (Cnmon1Δ), Cnmon1 deletion cassettes were constructed using double-joint PCR as described previously [Citation43]. The 5′- and 3′-flanking regions of CnMON1 were amplified using primer pairs JOHE41125–JOHE41127 and JOHE41126–JOHE41128, respectively, with the C. neoformans serotype A H99 [Citation44,Citation45] genomic DNA as the template (). The selectable marker NEO [Citation46] was amplified with the primer pair JOHE40706–JOHE40707 using pJAF1. The final deletion cassettes were generated using the primer pair JOHE41129–JOHE41130 and the 5′- and 3′-flanking regions and markers as templates. The mon1 deletion cassettes were purified using the QIAquick Gel Extraction kit (Qiagen) and combined with gold microcarrier beads (Bio-Rad). The gold bead/DNA particles were then introduced into H99 employing the biolistic transformation method. Multiple stable transformants were isolated in independent experiments, selected on YPD agar containing G418, and then confirmed for the 5′- and 3′-junctions via diagnostic PCR followed by restriction enzyme digestion.
Table 2. Oligonucleotides used in this study.
To generate the complementary Cnmon1 strain, the CnMON1 region, including its predicted promoter, was amplified using the primers JOHE41306 and JOHE41307. Next, the PCR product was digested with NotI and cloned into pHP1 [Citation47]. The resulting plasmid pHSP2 was then introduced into the recipient Cnmon1Δ strain (HP55). Multiple transformants were selected on YPD agar containing hygromycin (Sigma) and confirmed using PCR analysis.
2.3 The insect-based virulence assay
For the insect-based virulence assay, Galleria mellonella was purchased from Vanderhorst Inc. (St. Marys, OH, USA) and infected as previously described [Citation48]. Briefly, the wild-type (WT) and mutant fungal strains were grown overnight in YPD broth at 30 °C, washed thrice, and resuspended in PBS. Cell density was quantified using a hemocytometer and cell suspensions were prepared at a density of 2 × 106 cells/mL. Groups containing 12 G. mellonella larvae were injected with 4 μL of the cell suspension (8 × 104 cells/larvae) from the second to the last prolegs of each larva using a syringe (PB600-1; Hamilton). For non-infection controls, a larval group was injected with PBS only. The larvae were maintained at 37 °C and monitored daily for 15 days. Larvae that pupated during the experiments were censored. Survival curves were analyzed using the Prism 5 software (GraphPad, San Diego, CA, USA) and statistically analyzed using the log-rank (Mantel-Cox) test.
2.4 Mouse virulence assay
For the survival test, groups containing 10 female A/J mice (16–20 g; Jackson Labs or NCI/Charles River Laboratories) per strain were used. The fungal strains were grown overnight in YPD broth at 30 °C. Cultured cells were collected, washed twice with PBS, and the final concentration was adjusted to 1 × 107 CFU/mL. Mice were anesthetized with pentobarbital (Lundbeck Inc. Deerfield, IL, USA) and inoculated at a density of 5 × 105 CFU in 50 μL via intranasal inhalation as previously described [Citation49]. Survival was monitored daily, and survival curves were generated based on the Kaplan-Meier method using the Prism 5 software (GraphPad). Statistical significance (p values) was assessed using the log-rank test.
2.5 Ethics statement
Animal care and all the experiments were conducted in accordance with the ethical guidelines of the Institutional Animal Care and Use Committee (IACUC) of Duke University Medical Center (DUMC). Studies on mice were conducted at the Division of Laboratory Animal Resources (DLAR) of DUMC. The animals were handled according to the guidelines defined by the United States Animal Welfare Act and in full compliance with the DUMC IACUC. The DUMC IACUC approved all the vertebrate studies under the protocol number A245-13-09.
3. Results
3.1 Identification of CnMon1 in C. neoformans
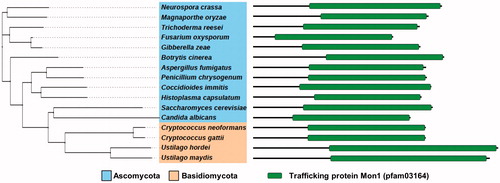
To identify the gene encoding the C. neoformans Mon1 protein (CnMon1), the S. cerevisiae Mon1 (ScMon1) protein sequence was blasted against the C. neoformans serotype A H99 genome database, where CNAG_00971 (XP_012049217) was identified as the most similar protein (31% identity). This gene encodes a 620-amino acid protein featuring a domain related to the CnMon1 trafficking protein on the C-terminus region. Mon1 is highly conserved in all eukaryotic organisms, including fungal species such as ascomycetes and basidiomycetes (). To further study the function of CnMon1 in C. neoformans, multiple Cnmon1Δ mutants and complementary strains were generated from independent experiments.
Figure 1. Phylogenetic tree depicting Mon1 in various fungal species. Sequences from Neurospora crassa OR74A (XP_959164.3), Magnaporthe oryzae 70-15 (XP_003710676.1), Trichoderma reesei QM6a (XP_006963764.1), Fusarium oxysporum Fo47 (EWZ45038.1), Gibberella zeae PH-1 (XP_011326784.1), Botrytis cinerea T4 (CCD56988.1), Aspergillus fumigatus AF293 (Q4WHL1.2), Penicillium chrysogenum (KZN85230.1), Coccidioides immitis RS (XP_001247287.2), Histoplasma capsulatum H88 (EGC46448.1), Candida albicans SC5314 (KHC78337.1), Saccharomyces cerevisiae S288c (NP_011391), Cryptococcus neoformans H99 (XP_012049217.1), Cryptococcus gattii Ru294 (KIR57394.1), Ustilago hordei (CCF52623.1), and Ustilago maydis 521 (XP_011389404.1) were compared and a phylogenetic tree of the Mon1 orthologs was generated via the MEGA 5 software (http://www.megasoftware.net/) using the alignment data from ClustalW2. The tree results were submitted to iTOL (http://itol.embl.de/) to generate the figure (left). Domain architecture of the Mon1 orthologs in fungi (right).
3.2 Mutation in Cnmon1 confers stress sensitivity
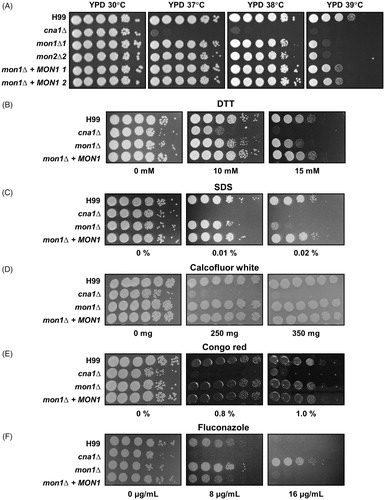
Since the Scmon1Δ mutant S. cerevisiae strain was hypersensitive to both brefeldin A and monensin (compounds known to affect intracellular transport) [Citation50], we tested the sensitivity of the Cnmon1Δ mutant against brefeldin A and monensin. Unlike the Scmon1Δ mutant, the Cnmon1Δ mutant strain was not sensitive to brefeldin A and monensin (Figure S1). Next, phenotypic analysis was conducted under various stress conditions using multiple Cnmon1Δ mutants and complementary strains. The Cnmon1Δ mutant exhibited growth defects at a high temperature of 39 °C (). Deletion of CnMON1 also caused increased sensitivity towards the ER stress inducer DTT (). As shown in , the Cnmon1Δ mutant was more sensitive to SDS than the WT and complemented strains. However, this mutant exhibited growth characteristics similar to those of the WT and could be stained with Congo red and calcofluor-white (). Interestingly, the Cnmon1Δ mutant also showed an increased resistance to fluconazole (). Importantly, normal sensitivity to temperature, SDS, DTT, and fluconazole was restored by reintroducing the WT CnMON1 gene. Overall, these results indicated that Mon1 is essential for appropriate stress survival.
Figure 2. Phenotypes of the Cnmon1Δ mutant following exposure to various stresses. Spot dilution assays with WT (H99), Cncna1Δ (KK1), Cnmon1Δ (HP55 and HP56), and Cnmon1 + CnMON1 (HPC3 and HPC4). Cells were incubated overnight, diluted 10-fold, and plated on YPD agar. Plated cells were incubated for 2 days at 30, 37, 38, and 39 °C. (A) Serially diluted cells were also plated on YPD agar without or with (B) dithiothreitol (DTT), (C) sodium dodecylsulfate (SDS), (D) calcofluor white, (E) congo red, and (F) fluconazole at the indicated concentrations. Results shown are representative of two independent experiments.
3.3 CnMon1 is required for fungal pathogenicity
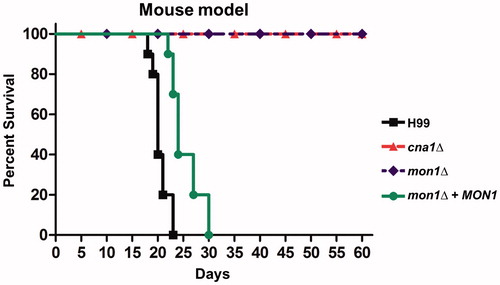
Since Mon1 is crucial for the virulence of plant pathogenic fungi [Citation21,Citation22], we hypothesized that CnMon1 is also involved in C. neoformans virulence. To demonstrate the role of CnMon1 in fungal infection, 2 in vivo models (insect- and murine inhalation-based) were employed. In the Galleria mellonella model, 2 independent Cnmon1Δ mutants were used, which were observed to attenuate virulence when compared to the virulence exhibited by the WT strain (). We also found that the Cnmon1Δ mutant was avirulent up to 60 days post-infection in the murine model (). Importantly, the virulence of the Cnmon1Δ strain was restored upon reintroducing the CnMON1 WT allele. Overall, these results demonstrated that Mon1 is essential for fungal virulence in both the insect and murine models.
Figure 3. Virulence of the Cnmon1Δ mutant in the insect larvae model. Each Galleria larva (12 per group) was infected with approximately 80,000 WT (H99), Cncna1Δ (KK1), or Cnmon1Δ (HP55 and HP56) cells. The infected larvae were maintained at 37 °C and monitored daily for 12 days.
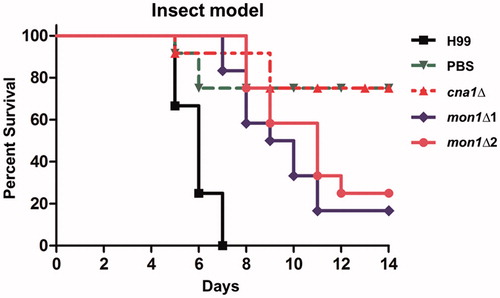
Figure 4. Virulence of the Cnmon1Δ mutant in the murine inhalation model. Cnmon1Δ mutant exhibits avirulence when compared to the WT and Cnmon1 + CnMON1 complement strains. WT (H99), Cncna1Δ (KK1), Cnmon1Δ (HP55), and Cnmon1 + CnMON1 (HPC3) cells were grown overnight in YPD broth at 30 °C and washed with PBS. Cells (5 × 105) were inoculated into A/J mice via intranasal instillation. Animal survival was monitored for 60 days post-infection.
4. Discussion
Mon1 is a highly conserved protein that plays a crucial role in membrane trafficking in all eukaryotic organisms ranging from yeast to mammals. The Mon1/SAND-1 protein activates Rab7 during endosome-controlling membrane fusion in the mammalian system [Citation51,Citation52]. In Drosophila, Dmon1 (the Drosophila ortholog of Mon1/SAND-1) recruits Rab7 to the maturing endosomes, thereby facilitating phagosome maturation and the trans-synaptic mechanism [Citation53–55]. In plants, Mon1 is conserved in Rab7 activation, vacuolar traffic control, and plant growth [Citation56]. In yeast, ScMon1 is essential for vacuole fusion, autophagy, and the CVT pathway [Citation15]. In filamentous fungi, the Mon1 ortholog proteins are essential for proper vacuole formation, autophagy, and asexual spore formation [Citation21,Citation22]. Importantly, Mon1 is a key factor for fungal virulence, which is demonstrated by the inability of mon1 deletion mutants in 2 plant pathogenic fungi to infect plants [Citation21,Citation22]. Although the virulent functions of Mon1 have been studied in plant pathogenic fungi, the pathogenic role of the Mon1 ortholog in animal/human pathogenic fungi has not yet been investigated. In this study, we demonstrated that Cnmon1 deletion results in attenuated virulence in the insect model and avirulence in the murine model (), suggesting that CnMon1 is required for fungal pathogenicity in the opportunistic human pathogenic fungus, C. neoformans. These results indicate that the pathogenic role of Mon1 is conserved in plant and human pathogenic fungi.
Previous studies demonstrated that several genes involved in vacuole trafficking played important roles in the regulation of C. neoformans growth and virulence at high temperatures [Citation34,Citation39–42]. For example, deletion of vps15, which encodes a serine/threonine protein kinase required for the CVT pathway, results in reduced fungal virulence, growth defects at high temperatures, and hypersensitivity to multiple stresses [Citation34]. Similar to the vps15 or vps23 null mutants, the Cnmon1Δ mutant was also highly susceptible to multiple stresses and demonstrated growth defects and reduced virulence. These results strongly show vacuolar sorting to play a crucial role in the fungal pathogenicity and stress response of C. neoformans.
Owing to its role in vacuole fusion, ScMon1 is required for autophagy, an intracellular process in S. cerevisiae that is essential for the degradation of cytoplasmic components [Citation16,Citation57,Citation58]. Autophagy has been reported to be essential in some pathogenic fungi [Citation40]. For example, the ATG8 knockdown mutant strains of C. neoformans showed attenuated virulence in 2 mouse models [Citation40]. Our preliminary data also show that atg8 or atg15 deletion results in attenuated virulence (data not shown). Although we did not test the importance of CnMon1 in the autophagy pathway in C. neoformans, we speculate that CnMon1 could regulate autophagosome formation, thereby regulating fungal virulence. The relationship between CnMon1 and autophagy in C. neoformans needs to be thoroughly examined. Alternatively, autophagy also affects stress granules and/or P-bodies in yeast [Citation59]. Loss-of-function of components involved in the autophagy process causes an increased accumulation of stress granules and P-bodies in the cytosol [Citation59]. For example, Cnmon1 deletion increases stress granules and P-bodies. We examined the role of Mon1 in P-body formation and found that it does not affect the formation of P-bodies in C. neoformans (data not shown).
In yeast, the Scmon1Δ mutant was shown to be hypersensitive to brefeldin A and monensin (inhibitors of intracellular protein transport), suggesting that the Cnmon1Δ mutant could also be sensitive to these compounds [Citation50]. Thus, we examined the sensitivity of the Cnmon1Δ mutant strain to these 2 compounds and found that it was not sensitive to brefeldin A and monensin (Figure S1). These results propose that Mon1 probably plays a different role in C. neoformans and yeast. To further test the role of CnMon1 in endocytosis, we examined the uptake of FM4-64, a fluorescent endocytic marker, in WT and mutants. However, we did not observe any differences in the WT and Cnmon1Δ mutant strains. Overall, these results propose that CnMon1 may not be a key factor for intracellular protein transport.
In summary, we identified CnMon1 and demonstrated its crucial role in fungal growth at high temperatures, sensitivity against several stresses, and fungal virulence in C. neoformans. The cellular and biochemical roles of CnMon1, which includes vacuolar fusion, autophagy, endocytosis, and GEF, will be the subject of future studies in C. neoformans.
Supplemental Material
Download TIFF Image (1.7 MB)Acknowledgments
The authors are grateful to Joseph Heitman for his helpful suggestions and critical reading of the manuscript. We also thank Ci Fu for the discussions and critical reading of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Rodrigues ML, Nosanchuk JD, Schrank A, et al. Vesicular transport systems in fungi. Future Microbiol. 2011;6:1371–1381.
- Jahn R, Scheller RH. SNAREs-engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643.
- Nickel W, Brugger B, Wieland FT. Vesicular transport: the core machinery of COPI recruitment and budding. J Cell Sci. 2002;115:3235–3240.
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292.
- Yamasaki A, Noda NN. Structural biology of the Cvt pathway. J Mol Biol. 2017;429:531–542.
- Teter SA, Klionsky DJ. Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation. Semin Cell Dev Biol. 2000;11:173–179.
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366.
- Sato K, Wickner W. Functional reconstitution of ypt7p GTPase and a purified vacuole SNARE complex. Science. 1998;281:700–702.
- Wickner W. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136.
- Hickey CM, Stroupe C, Wickner W. The major role of the Rab Ypt7p in vacuole fusion is supporting HOPS membrane association. J Biol Chem. 2009;284:16118–16125.
- Schimmoller F, Riezman H. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J Cell Sci. 1993;106:823–830.
- Wang T, Ming Z, Xiaochun W, et al. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal. 2011;23:516–521.
- Wang CW, Stromhaug PE, Shima J, et al. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J Biol Chem. 2002;277:47917–47927.
- Wang CW, Stromhaug PE, Kauffman EJ, et al. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163:973–985.
- Cabrera M, Engelbrecht-Vandre S, Ungermann C. Function of the Mon1-Ccz1 complex on endosomes. Small GTPases. 2014;5:1–3.
- Meiling-Wesse K, Barth H, Voss C, et al. Yeast Mon1p/Aut12p functions in vacuolar fusion of autophagosomes and cvt-vesicles. FEBS Lett. 2002;530:174–180.
- Nordmann M, Cabrera M, Perz A, et al. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–1659.
- Cabrera M, Nordmann M, Perz A, et al. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J Cell Sci. 2014;127:1043–1051.
- Cabrera M, Ungermann C. Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J Biol Chem. 2013;288:28704–28712.
- Kiontke S, Langemeyer L, Kuhlee A, et al. Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat Commun. 2017;8:14034.
- Gao HM, Liu XG, Shi HB, et al. MoMon1 is required for vacuolar assembly, conidiogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae. Res Microbiol. 2013;164:300–309.
- Li Y, Li B, Liu L, et al. FgMon1, a guanine nucleotide exchange factor of FgRab7, is important for vacuole fusion, autophagy and plant infection in Fusarium graminearum. Sci Rep. 2015;5:18101.
- Polupanov AS, Nazarko VY, Sibirny AA. CCZ1, MON1 and YPT7 genes are involved in pexophagy, the Cvt pathway and non-specific macroautophagy in the methylotrophic yeast Pichia pastoris. Cell Biol Int. 2011;35:311–319.
- Liu XH, Gao HM, Xu F, et al. Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy. 2012;8:1415–1425.
- Loftus BJ, Fung E, Roncaglia P, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324.
- Bouklas T, Fries BC. Cryptococcus neoformans constitutes an ideal model organism to unravel the contribution of cellular aging to the virulence of chronic infections. Curr Opin Microbiol. 2013;16:391–397.
- Idnurm A, Bahn YS, Nielsen K, et al. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Micro. 2005;3:753–764.
- Kwon-Chung KJ, Fraser JA, Doering TL, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4:a019760.
- Williamson PR, Jarvis JN, Panackal AA, et al. Cryptococcal meningitis: epidemiology, immunology, diagnosis and therapy. Nat Rev Neurol. 2017;13:13–24.
- Perfect JR, Bicanic T. Cryptococcosis diagnosis and treatment: What do we know now. Fungal Genet Biol. 2015;78:49–54.
- Kidd SE, Hagen F, Tscharke RL, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci USA. 2004;101:17258–17263.
- Park BJ, Wannemuehler KA, Marston BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530.
- Jung KW, Yang DH, Maeng S, et al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun. 2015;6:6757.
- Lee KT, So YS, Yang DH, et al. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat Commun. 2016;7:12766.
- Liu OW, Chun CD, Chow ED, et al. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188.
- Maier EJ, Haynes BC, Gish SR, et al. Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome Res. 2015;25:690–700.
- Gish SR, Maier EJ, Haynes BC, et al. Computational analysis reveals a key regulator of cryptococcal virulence and determinant of host response. mBio. 2016;7:e00313-16.
- Desjardins CA, Giamberardino C, Sykes SM, et al. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 2017;27:1207–1219.
- Godinho RM, Crestani J, Kmetzsch L, et al. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci Rep. 2014;4:6198.
- Hu G, Hacham M, Waterman SR, et al. PI3K signaling of autophagy is required for starvation tolerance and virulenceof Cryptococcus neoformans. J Clin Invest. 2008;118:1186–1197.
- Hu G, Caza M, Cadieux B, et al. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect Immun. 2013;81:292–302.
- Liu X, Hu G, Panepinto J, et al. Role of a VPS41 homologue in starvation response, intracellular survival and virulence of Cryptococcus neoformans. Mol Microbiol. 2006;61:1132–1146.
- Yu JH, Hamari Z, Han KH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981.
- Perfect JR, Toffaletti DL, Rude TH. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61:4446–4451.
- Janbon G, Ormerod KL, Paulet D, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014;10:e1004261.
- Fraser JA, Subaran RL, Nichols CB, et al. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2:1036–1045.
- Park H-S, Chow EW, Fu C, et al. Calcineurin targets involved in stress survival and fungal virulence. PLoS Pathog. 2016;12:e1005873.
- Mylonakis E, Moreno R, El Khoury JB, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73:3842–3850.
- Cox GM, Mukherjee J, Cole GT, et al. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448.
- Muren E, Oyen M, Barmark G, et al. Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast. 2001;18:163–172.
- Yasuda S, Morishita S, Fujita A, et al. Mon1-Ccz1 activates Rab7 only on late endosomes and dissociates from the lysosome in mammalian cells. J Cell Sci. 2016;129:329–340.
- Gerondopoulos A, Langemeyer L, Liang JR, et al. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012;22:2135–2139.
- Deivasigamani S, Basargekar A, Shweta K, et al. Presynaptic regulatory system acts transsynaptically via Mon1 to regulate glutamate receptor levels in Drosophila. Genetics. 2015;201:651–664.
- Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782.
- Yousefian J, Troost T, Grawe F, et al. Dmon1 controls recruitment of Rab7 to maturing endosomes in Drosophila. J Cell Sci. 2013;126:1583–1594.
- Cui Y, Zhao Q, Gao C, et al. Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell. 2014;26:2080–2097.
- Hegedus K, Takats S, Boda A, et al. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell. 2016;27:3132–3142.
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361.
- Buchan JR, Kolaitis RM, Taylor JP, et al. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474.
- Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101:177–194.
- Kojima K, Bahn YS, Heitman J. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology. 2006;152:591–604.
