Abstract
A new species belonging to the genus Alternaria was isolated from the necrotic leaf spots of Brassica rapa subsp. pekinensis in Yuseong district, Daejeon, Korea. It is an occasional isolate, not an etiological agent, which is morphologically similar to A. broccoli-italicae, but differs in conidial size and conidiophore shape. Phylogenetic analysis using the sequence datasets of the internal transcribed spacer (ITS) region of the rDNA, glyceraldehyde-3-phosphate dehydrogenase (gpd), and plasma membrane ATPase genes showed that it is distantly related to A. broccoli-italicae and closely related to Alternaria species in the section Pseudoalternaria, which belonged to a clade basal to the section Infectoriae. Morphologically, the species is unique because it produces solitary conidia or conidial chains (two units), unlike the four members in the section Pseudoalternaria that produce conidia as short branched chains. It exhibits weak pathogenicity in the host plant. This report includes the description and illustration of A. brassicifolii as a new species.
The genus Brassica is known to include important agricultural and horticultural crops. In Asia, especially in Korea, Brassica rapa L. subsp. pekinensis (Lour.) Hanelt is one of the most popular leafy vegetables used to prepare kimchi, a traditional Korean food. Three species of Alternaria, namely, A. brassicae (Berk) Sacc, A. brassicicola (Schwein) Wiltshire, and A. japonica Yoshii have been isolated from Brassica rapa in Korea [Citation1]. Alternaria leaf spots caused by these three species is the most common and destructive fungal disease occurring in many cruciferous plants worldwide [Citation2,Citation3]. Other Alternaria species are also reported from Brassica plants such as A. brassicinae Simmons, A. broccoli-italicae Simmons, A. ethzedia Simmons, and A. nepalensis Simmons [Citation4].
Alternaria was originally described by Nees (1816), with A. tenuis Nees being the type species. Since then, approximately 280 Alternaria species have been reported as plant pathogens and saprophytes, resulting in poor crop yield and spoilage during storage [Citation4,Citation5]. The taxonomy of Alternaria species is mainly based on the shape, size, and septation of the conidia, as well as sporulation patterns [Citation1,Citation4,Citation6,Citation7]. Since the 21st century, molecular approaches, especially sequence analyses, have been popularly adopted to identify the Alternaria species [Citation8–10]. Both morphological and molecular phylogenetic analyses work in a complementary manner for the classification of this species [Citation11–13]. Recently, Alternaria has been classified into 27 sections, and 14 other genera have been synonymized [Citation14,Citation15].
Leaves showing necrotic spots on Brassica rapa L. subsp. pekinensis (Lour.) Hanelt (Brassicaceae) were collected from Yuseong-gu district, Daejeon, Korea, in June 2011. The samples were processed for sporulation, and Alternaria isolates were obtained using the methods described by Deng et al. [Citation12]. Pure cultures were deposited in the Culture Collection center of the Chungnam National University (CNU) in Daejeon, and the ex-type of the species (CNU 111118) was stored in the Korean Agricultural Culture Collection (KACC), Suwon, Korea.
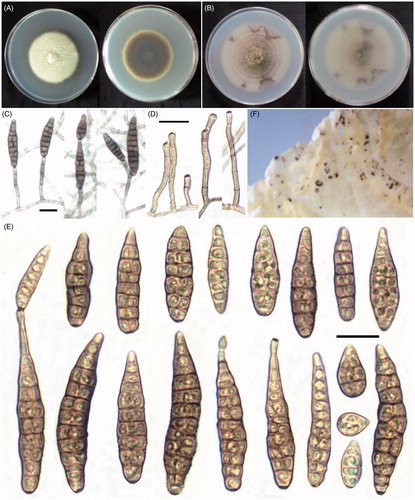
To determine colony characteristics, the isolates were cultured on potato dextrose agar (PDA; Difco, Montreal, Canada) for 7 days at 25 °C under dark condition. To observe sporulation patterns and conidial morphology, the isolates were transferred to potato carrot agar (PCA: 20 g white potato, 20 g carrot, and 20 g agar in 1 L) for 7 days at 22 °C under alternating light and dark conditions (8/16 light/dark) [Citation4]. Sporulation patterns and conidia (50) were digitally photographed () and measured using an OLYMPUS BX50 light microscope (OLYMPUS, Tokyo, Japan) equipped with an Artcam 300MI digital camera (ARTRAY, Tokyo, Japan).
Taxonomy
Alternaria brassicifolii S. H. Yu and J. X. Deng, sp. nov. ().
Mycobank: MB 824688.
Description: Colonies on PDA at 7 days, 52–57 mm diameter in size, cottony, olivaceous buffer (); on PCA at 7 days, 65–68 mm diameter in size, velvety, pale gray with conspicuous concentric rings, presenting sectors at the edge (). Conidia solitary with a large proportion, some formed chains by means of an apical secondary conidium on the PCA medium (). Conidiophores simple, erect or bent, smooth, terminally or laterally from hyphae, septate (1–9), usually only one pigmented terminal conidiogenous site, sometimes with one more lateral conidiogenous locus, 15–100 × 2.5–4.5 μm (). Juvenile conidia subcylindric, bluntly rounded base and apex, or ovoid with tapered apex containing 1–2 septa, measuring 19–35 × 7–12 μm. Matured conidia long elliptical or obclavate, beakless tapering gradually to a rounded or conical apex, sometimes with a short apical secondary conidiophore, straight or slightly curved, up to 40–80 (∼87) × 9–17 μm, dilute tan in color, smoothly or evenly verruculose in the conidial wall with 5–10 transverse septa and 1–2 longitudinal or oblique septum in the transverse segments (). Its teleomorph stage was not observed.
Etymology: Brassicifolii, refers to the genus of the host plant (Brassica) and the leaf (folium) from it was collected.
Type: Korea, Daejeon, Yuseong-gu, from the leaves of Brassica rapa L. subsp. pekinensis (Lour.) Hanelt, June 2011, by S. H. Yu and J. X. Deng, cultures: CNU 111118 and CNU 111116.
The morphological characteristics of A. brassicifolii were different from those of any other Alternaria species described, except A. broccoli-italicae, which produces solitary and long-ovoid conidia, and reported, for the first time, from Brassica oleracea L. var. italica Plenck (Brassicaceae) [Citation4]. However, A. broccoli-italicae grows slowly on the PCA medium (2–3 cm diameter) in 5–7 days and produce smaller conidia (35–60 × 8–12 μm) with respect to the Alternaria species described in this study. The most obvious difference between these two species is found in the conidiophores. The conidiophores of A. broccoli-italicae frequently extended into twisted, branching elements that are conidiogenous at several successive loci on the PCA medium [Citation4]. This type of conidiophore was not observed among the isolates of A. brassicifolii.
To perform molecular analysis, genomic DNA of the isolate CNU 111118 and the ex-type strain E.G.S. 40-134 of A. broccoli-italicae was extracted using a method described by Park et al. [Citation16] with some modifications. PCR amplification of ITS, gpd, and ATPase genes was performed using the primer pairs of ITS1/ITS4 [Citation17], gpd1/gpd2 [Citation18], and ATPDF1/ATPDR1 [19], respectively. PCR was performed as described by Lawrence et al. [Citation19]. The products were purified using a Wizard PCR prep kit (Promega, Madison, WI) and sequenced by a commercial sequencing service provider (Macrogen, Daejeon, Korea). Each gene sequence was deposited in GenBank and assigned an accession number ().
Table 1. NCBI GenBank accession numbers of the Alternaria isolates used in the study.
The sequences obtained in this work and the sequences of related species from the sections of Alternaria phylogeny [Citation14,Citation19,Citation20] were used for phylogenetic analysis. Sequence alignment was generated with the Clustal X program [Citation21] and manually adjusted. The three gene sequences were assembled in a single sequence resulting in a 2460 characters. Maximum likelihood analysis was performed using the GTRCAT model in RAxML program [Citation22]. Branch support measures were calculated with 1000 bootstrap replicates. The resulting phylogenetic tree () was constructed with Mega v5.05 [Citation23]. In the phylogram, A. brassicifolii fell into a clade (100% bootstrap values) comparing species of the sections Infectoriae and Pseudoalternaria. Moreover, the species was closely related to four members in the section Pseudoalternaria (A. arrhenatheria, A. kordkuyana, A. parvicaespitosa, and A. rosea) and they all gathered in a clade (81% bootstrap values) basal to the section Infectoriae. Meanwhile, A. brassicifolii, which is phylogenetically distant to A. broccoli-italicae, fell into the section Infectoria. Morphologically, the species in the section Pseudoalternaria commonly produced conidia in short branched chains; however, A. brassicifolii is unique and mostly produces solitary conidia, at times with simple chains of up to two conidia (). The molecular phylogenetic data available and the unique morphology of this fungus further confirms that it is a new species belonging to Alternaria, proposed to be named as A. brassicifolii sp. nov.
Figure 2. Maximum likelihood tree obtained from the combined datasets of ITS, gpd, and ATPase gene sequences of Alternaria brassicifolii and other related species. Bootstrap values (>60%) calculated for 1000 replicates are shown above the branches. The bar indicates the number of substitutions per position. The sections were referenced from previously published reports [Citation14,Citation19].
![Figure 2. Maximum likelihood tree obtained from the combined datasets of ITS, gpd, and ATPase gene sequences of Alternaria brassicifolii and other related species. Bootstrap values (>60%) calculated for 1000 replicates are shown above the branches. The bar indicates the number of substitutions per position. The sections were referenced from previously published reports [Citation14,Citation19].](/cms/asset/b5d50be7-c891-41b2-bb97-8aad3f70d421/tmyb_a_1468054_f0002_b.jpg)
Table 2. Comparison of the conidial morphology of Alternaria brassicifolii sp. nov. with Pseudoalternaria species on the PCA medium.
The pathogenicity of A. brassicifolii sp. nov. was evaluated. Spore suspension (106 spores/mL) was sprayed on the detached leaves of Brassica rapa L. subsp. pekinensis (Lour.) Hanelt and kept in a clean moistened box at 25 °C for 5 days. Small necrotic spots (1–2 mm) were observed on the leaves, which were used to isolate and identify the fungus involved. The test was repeated thrice and showed similar results. No symptoms were observed in control samples. These results indicate that A. brassicifolii exhibits weak pathogenicity in the host, which may reduce its market value.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Yu SH. Alternaria species and allied genera. Incheon: National Institute of Biological Resources Ministry of Environment; 2015.
- Nowicki M, Nowakowska M, Niezgoda A, et al. Alternaria black spot of crucifers: symptoms, importance of disease, and perspectives of resistance breeding. Veg Crops Res Bull. 2012;76:5–19.
- Kumar D, Maurya N, Bharati YK, et al. Alternaria blight of oilseed Brassicas: a comprehensive review. Afr J Microbiol Res. 2014;8:2816–2829.
- Simmons EG. Alternaria: an identification manual. CBS Biodiversity Series 6. Utrecht: Centraalbureau voor Schimmelcultures; 2007.
- Andersen B, Krøger E, Roberts RG. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol Res. 2001;105:291–299.
- Zhang TY. Flora fungorum sinicorum, vol. 16 Alternaria. Beijing: Science Press; 2003.
- Nishikawa J, Nakashima C. Taxonomic characterization and experimental host ranges of four newly recorded species of Alternaria from Japan. J Phytopathol. 2013;161:604–616.
- Pryor BM, Bigelow DM. Molecular characterization of Embellisia and Nimbya species and their relationship to Alternaria, Ulocladium and Stemphylium. Mycologia. 2003;95:1141–1154.
- Hong SG, Cramer RA, Lawrence CB, et al. Alt a 1 allergen homologs from Alternaria and related taxa: analysis of phylogenetic content and secondary structure. Fungal Genet Biol. 2005;42:119–129.
- Park MS, Romanoski CE, Pryor BM. A re-examination of the phylogenetic relationship between the causal agents of carrot black rot, Alternaria radicina and A. carotiincultae. Mycologia. 2008;100:511–527.
- Tóth B, Csosz M, Szabo-Hever A, et al. Alternaria hungarica sp. nov., a minor foliar pathogen of wheat in Hungary. Mycologia. 2011;103:94–100.
- Deng JX, Cho HS, Paul NC, et al. A novel species belonging to Alternaria isolated from Peucedanum japonicum in Korea. Mycobiology. 2014;42:12–16.
- Woudenberg J, Truter M, Groenewald J, et al. Large-spored Alternaria pathogens in section Porri disentangled. Stud Mycol. 2014;79:1–47.
- Woudenberg JH, Groenewald JZ, Binder M, et al. Alternaria redefined. Stud Mycol. 2013;75:171–212.
- Lawrence DP, Rotondo F, Gannibal PB. Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol Prog. 2016;15:3.
- Park MS, Seo GS, Bae KS, et al. Characterization of Trichoderma spp. associated with green mold of Oyster mushroom by PCR-RFLP and sequence analysis of ITS regions of rDNA. Plant Pathol J. 2005;21:229–236.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322.
- Berbee ML, Pirseyedi M, Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977.
- Lawrence DP, Gannibal PB, Dugan FM, et al. Characterization of Alternaria isolates from the infectoria species-group and a new taxon from Arrhenatherum, Pseudoalternaria arrhenatheria sp. nov. Mycol Progress. 2014;13:257–276.
- Poursafar A, Ghosta Y, Orina AS, et al. Taxonomic study on Alternaria sections Infectoriae and Pseudoalternaria associated with black (sooty) head mold of wheat and barley in Iran. Mycol Progress. 2018;17:343–356.
- Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882.
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771.
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Bio Evol. 2011;28:2731–2739.
- Gannibal PB, Lawrence DP. Distribution of Alternaria species among sections. 3. Sections Infectoriae and Pseudoalternaria. Mycotaxon. 2016;131:781–790.