Abstract
Black rot disease in orchids is caused by the water mold Phytophthora palmivora. To gain better biocontrol performance, several factors affecting growth and antifungal substance production by Pseudomonas aeruginosa RS1 were verified. These factors include type and pH of media, temperature, and time for antifungal production. The results showed that the best conditions for P. aeruginosa RS1 to produce the active compounds was cultivating the bacteria in Luria-Bertani medium at pH 7.0 for 21 h at 37 °C. The culture filtrate was subjected to stepwise ammonium sulfate precipitation. The precipitated proteins from the 40% to 80% fraction showed antifungal activity and were further purified by column chromatography. The eluted proteins from fractions 9–10 and 33–34 had the highest antifungal activity at about 75% and 82% inhibition, respectively. SDS-PAGE revealed that the 9–10 fraction contained mixed proteins with molecular weights of 54 kDa, 32 kDa, and 20 kDa, while the 33–34 fraction contained mixed proteins with molecular weights of 40 kDa, 32 kDa, and 29 kDa. Each band of the proteins was analyzed by LC/MS to identify the protein. The result from Spectrum Modeler indicated that these proteins were closed similarly to three groups of the following proteins; catalase, chitin binding protein, and protease. Morphological study under scanning electron microscopy demonstrated that the partially purified proteins from P. aeruginosa RS1 caused abnormal growth and hypha elongation in P. palmivora. The bacteria and/or these proteins may be useful for controlling black rot disease caused by P. palmivora in orchid orchards.
1. Introduction
Synthetic chemical fungicide application is the most popular method used to reduce the incidence of plant diseases caused by fungi and molds. However, using chemical agents has several negative impacts such as the chemical residues that remain in environment, the reduction of beneficial soil microorganisms, the physiological adaption of pathogens to resist the chemical fungicide, and the direct exposure to chemical mixtures on human health [Citation1,Citation2]. Owing to recent concerns regarding the impact of chemical fungicides on the environment and human health, natural bioactive compounds as well as biological control agents are being used more often instead of agrochemicals [Citation3]. In addition, the biological control method was reported to be an eco-friendly and safe approach to control plant pathogens, thus is now used as an alternative method to prevent and/or protect plants from diseases [Citation4]. Various beneficial microorganisms have been used as biocontrol agents against fungal and mold plant pathogens [Citation5]. Good examples of microorganisms that are able to antagonize the growth of phytopathogenic fungi and mold include many species of bacteria in the genera Streptomyces, Bacillus, and Pseudomonas [Citation6–8].
Numerous bacterial biocontrol agents have been manufactured using Pseudomonas spp., some of which have become commercially available [Citation9,Citation10]. Pseudomonas spp. can produce many kinds of extracellular biofungicides. For example, Pseudomonas fluorescens produces volatile 1-undecene to inhibit Phytophthora infestans [Citation11,Citation12], Pseudomonas aeruginosa SS14 produces biosurfactant against Fusarium verticillioides [Citation13], P. fluorescens produces a cyclic lipopeptide that suppresses Gaeumannomyces graminis var. tritici and Rhizoctonia solani [Citation14], and P. aeruginosa produces pafungin to inhibit Fusarium oxysporum [Citation15]. Several Pseudomonas spp. are also reported to produce antifungal proteins as biocontrol substances. For example, two bifunctional chitinases/lysozymes from P. aeruginosa K-187 have shown antifungal activity against 36 strains of fungi [Citation9], and protease from P. aeruginosa has demonstrated antifungal activity by inhibiting spore germination and hyphal elongation of Fusarium solani [Citation16].
Orchidaceae is not only the largest flowering plant family but also produces the greatest floral diversity of all Angiosperms [Citation17]. Orchids are found in several countries worldwide such as Netherlands, Thailand, Taiwan, Singapore, and New Zealand [Citation18]. In Thailand, the orchid export industry generated around US$50 million in 2001 [Citation19]. However, the orchid industry suffers financially from poor crop yields due to the problem of plant diseases. Most orchid diseases are acquired through infection from bacteria [Citation20], viruses [Citation21], and fungi [Citation22]. Thus, controlling disease is an important practice for maintaining the quality of orchids.
Fungal and mold pathogens cause several diseases in orchids, such as Curvularia lunata, causes rust spot disease on the flowers [Citation23], C. eragrostidis causes rusty flower disease [Citation24], Fusarium moniliforme causes column blight disease, Fusarium solani causes leaf yellowing and root and collar rot diseases [Citation25], Fusarium oxysporum causes wilt disease, Sclerotium rolfsii causes collar rot disease, Phoma exigua causes flower spot disease, Colletotrichum gloeosporioides causes anthracnose disease [Citation26], and Phytophthora palmivora causes black rot disease [Citation23]. P. palmivora is considered a water mold that causes black rot disease in wide variety of orchids including Aerides, Ascocenda, Brassavola, Dendrobium, Gongora, Maxillaria, Miltonia, Oncidium, Paphiopedilum, Phalaenopsis, Rhynchostylis, and Schomburgkia [Citation27]. After infection, orchids initially display small black lesions on the root or basal portion of the pseudobulbs, afterwards the black lesions expand to completely cover the pseudobulbs and leaves [Citation27]. The mold spreads and infects other parts of the orchid until the orchid dies.
In this research, the natural antagonistic bacteria that inhibit the growth of P. palmivora were selected. The antifungal proteins were purified and then identified by LC-mass spectrometry.
2. Materials and methods
2.1. Microorganisms and culture media
P. aeruginosa RS1 (GenBank accession number MF344551) from the culture collection of Department of Microbiology, Faculty of Science, Chulalongkorn University under Thailand Bioresource Research Center TBRC as MSCU 0860 which was found to inhibit the growth of P. palmivora, was maintained on a nutrient agar (NA) plate (0.5% peptone, 0.3% beef extract and 1.5% agar) at 37 °C.
Phytophthora palmivora (DOAC2057) which was obtained from the Department of Agriculture (Bangkok, Thailand) was cultured on a V8 agar plate (5% V8 vegetable juice (original) and 1.5% agar) at 26 °C.
Inhibition of P. palmivora by P. aeruginosa RS1 was performed by inoculating an agar plug of P. palmivora at the center of the V8 agar plate, and dropping the suspended bacteria next to the fungal plug. The plate was incubated at 26 °C for 2 days. The vegetative growth of P. palmivora was measured by comparing it with the growth of the mold growing without bacteria. The percentage inhibition was calculated using formula IP = (C – T/C) * 100; where IP = inhibitory percentage, C = average diameter of P. palmivora from the control, and T = average diameter from the test plate [Citation28]. Mycelia at the periphery of the colony from both conditions were observed under light microscope.
2.2. Optimization conditions for producing antifungal substances from P. aeruginosa RS1
P. aeruginosa RS1 was cultured in nutrient broth (NB) at 37 °C until the optical density (OD) at 660 nm reached about 0.5. A 1% inoculum of the culture was added to nutrient broth (NB), Luria-Bertani (LB) broth, and tryptic soy broth (TSB); all at pH 6. The culture was incubated at 37 °C for 18 h with shaking at 200 rpm. The culture was then centrifuged at 4 °C, 5870 × g for 15 min to obtain a supernatant. The supernatant was tested for antifungal activity as described below. The medium that showed the highest antifungal activity was selected for pH optimization, which included pH 6, 7, 8, and 9. The bacteria were cultured in the optimum medium and pH at different temperatures; 30 °C, 37 °C, and 40 °C. After the optimum temperature was obtained, the optimum cultivation time was determined.
2.3. Purification of the antifungal proteins
P. aeruginosa RS1 was cultured under optimum conditions and the supernatant was obtained. The antifungal proteins were then precipitated by ammonium sulfate precipitation followed by anion exchange chromatography. Briefly, the proteins in the P. aeruginosa RS1 supernatant were precipitated by the ammonium sulfate concentration in two steps; 0–40% and 40–80%. The protein pellet was dissolved with the minimum volume of 50 mM Tris-HCl buffer at pH 7.5 and dialyzed three times (5 h each) using a dialysis bag with the cutoff at 3500 (SnakeSkin Dialysis Tubing, Thermo Fisher Scientific, Rockford, IL) against 50 mM Tris-HCl buffer at pH 7.5. The final step of dialysis was performed in the same buffer which contained 30% of glycerol. The crude protein from the ammonium sulfate precipitation step was applied to a DEAE-cellulose column that pre-equilibrated with 50 mM Tris-HCl buffer at pH 7.5. The unbound protein fractions were collected and the bound protein fractions were eluted with a linear gradient of 0.0–1.0 M NaCl in the same buffer. All fractions were monitored at 280 nm for the presence of proteins. Two eluted fractions were combined to decrease the sample numbers during the antifungal activity assay. Antifungal activity was calculated as an Arbitrary Unit (AU) defined as the highest dilution of the protein that yielded a clear inhibition zone against P. palmivora.
2.4. Electrophoresis and LC mass spectrometry
The protein from ammonium sulfate precipitation and the protein fractions from the DEAE-cellulose column were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using the method of Laemmli and Favre [Citation29]. A page ruler prestained protein ladder (10-170 kDa) (Thermo Fisher Scientific, Rockford, IL) was used as molecular weight marker. Identification of purified proteins was accomplished by liquid chromatography equipped with Q ExactiveTM Plus hybrid quadrupole-orbitrap mass spectrometry (LC/MS) (Thermo Fisher Scientific, Rockford, IL) at the Proteomics Core Facility, Faculty of Medicine, Chulalongkorn University (Bangkok, Thailand). Data analysis was performed by using an X! TANDEM Spectrum Modeler (http://www.thegpm.org) against the Global Proteome Machine Database. Searches were run with trypsin as the cleaving enzyme with a maximum of one missed cleavage, carbamidomethyl cysteine as a fixed modification, and oxidation of methionine as the potential variable modification.
2.5. Antifungal activity assay
The supernatant which was collected from each condition was filtered through a 0.45 µm membrane filter to remove cells. The antifungal activity was inspected by pour plate method [Citation30] using a 1:9 ratio of supernatant to V8 media for all experiments, except the protein fractions from ammonium sulfate precipitation, which used a 1:1 ratio. The agar plug with a 6-mm diameter of P. palmivora was placed at the center of each plate, and the plate was incubated at 26 °C for 48 h. The control plate contained the fungus growing on the media with only the V8 agar. The vegetative growth of the fungus was measured, and the percentage of inhibition was calculated by using formula provided above. The experiments were replicated four times.
2.6. Microscopy
The effect of purified protein to P. palmivora was examined under optical microscope and scanning electron microscope (SEM). The result was compared with the control P. palmivora growing without bacterial protein.
3. Results
3.1. Antagonistic activity of P. aeruginosa RS1 against P. Palmivora
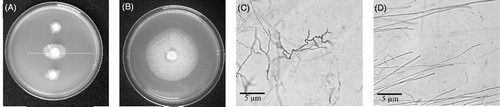
Pseudomonas aeruginosa RS1 was shown to inhibit the growth of P. palmivora on a V8 agar plate as demonstrated in comparing to the control in . The inhibition efficiency was around 50%. Observation under microscope revealed malformed hyphae of P. palmivora growing with P. aeruginosa RS1, while a normal appearance with smooth elongate hyphae was observed in P. palmivora growing alone (, respectively).
3.2. Optimization conditions for P. aeruginosa RS1 to produce antifungal substances
Pseudomonas aeruginosa RS1 was cultured in different media (LB, NB and TSB) and the culture filtrate to medium ratio of 1:9 was used to determine the antifungal activity on P. palmivora. The result showed that the highest inhibition activity was obtained from cultivation in LB, follow by NB and TSB with the percentage inhibition at 53.33 ± 2.72%, 29.03 ± 2.63%, and 13.33 ± 3.85%, respectively (). Bacteria were cultured in LB medium with pH levels at 6, 7, 8, and 9. The result revealed that LB medium at pH 7 demonstrated 52.63 ± 2.15% inhibition, which was better than at pH 6, pH 8, and pH 9 (48.57 ± 2.33%, 32.35 ± 2.40%, and 22.58 ± 2.63% inhibition, respectively) (). After bacteria were cultured in LB medium at pH 7 at different temperatures; 30 °C, 37 °C, or 40 °C; it was found that the best inhibition activity was observed at 37 °C with the inhibition percentage at 54.84 ± 3.72% (). A time course experiment was performed at this optimum condition, which demonstrated that the growth of P. aeruginosa RS1 entered a logarithmic phase at 3–6 h, and a late logarithmic phase at 6–15 h, after which the growth gradually reduced and entered a stationary phase at about 18 h (). The culture filtrate started to show antifungal activity from 9 h of growth, and reached the highest activity at 21 h, which yielded an inhibition percentage of about 52.94 ± 1.60% ().
Figure 2. Inhibition percentage against P. palmivora and absorbance at OD660 nm of P. aeruginosa RS1 in LB medium (pH 7) at 37 °C, at time points measured every 3 h, 24 h.
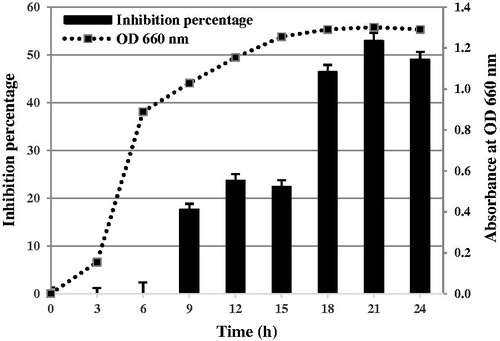
Table 1. Percentage of inhibition by culture filtrate from P. aeruginosa RS1 at different variable factors.
3.3. Purification and electrophoresis of the antifungal proteins
The antifungal protein from P. aeruginosa RS1 was purified by ammonium sulfate precipitation and DEAE-cellulose chromatography. The crude protein from ammonium sulfate precipitation at 40–80% inhibited the growth of P. palmivora up to 100% (). The protein from this fraction was further purified by DEAE-cellulose column chromatography, and the chromatogram is shown in . Several protein peaks were obtained. Every protein fraction peak showed antifungal activity ranging from 27.94–82.35% (data not shown). The protein from fractions 9–10 and 33–34 exhibited the highest antifungal activity at about 75 ± 2.08% and 82.35 ± 2.08% inhibition, respectively (). The 9–10 fraction was eluted before adding the gradient of NaCl, while the 33–34 fraction was eluted during about 0.7–0.9 M NaCl ().
Figure 3. Chromatogram from DEAE-cellulose column chromatography indicates the absorbance at OD280 nm of the proteins eluting from the column using gradient concentrations of NaCl.
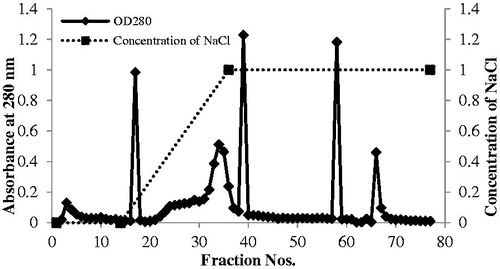
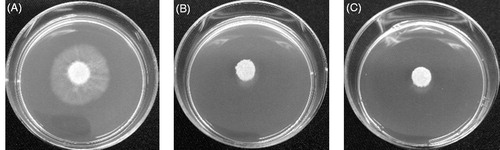
Figure 4. Inhibition of P. palmivora by eluted proteins from DEAE-cellulose column chromatography. P. palmivora grew on a V8 agar plate: control without protein (A), with protein fraction 9–10 (B), and with protein fraction 33–34.
As shown in , the culture filtrate, 40–80% ammonium sulfate fraction, DEAE fraction 9–10, and DEAE fraction 33–34 yielded total activity of 8000, 384, 8.4, and 16 AU, respectively. The ion exchange chromatography resulted in 11.25 and 1.67 fold increases in purification of the proteins from the 9–10 and 33–34 fractions, respectively. These fractions revealed the specific activity of 250 AU/mg and 37.04 AU/mg, respectively. SDS-PAGE confirmed the presence of proteins from these fractions. Fewer protein bands appeared in the DEAE-cellulose column chromatography step compared with those in 40–80% ammonium sulfate precipitation step (). Some bands were concentrated after the DEAE-cellulose column chromatography step because those bands did not appear in the ammonium sulfate fraction. The results also demonstrated that the proteins in DEAE fraction 9–10 and DEAE fraction 33–34 were partially purified because several polypeptide bands were observed. At least three bands appeared in DEAE fraction 9–10 with molecular weights of about 54 kDa, 32 kDa, and 20 kDa, respectively (, #1–3); whereas at least three bands appeared in DEAE fraction 33–34 with molecular weights of about 40 kDa, 32 kDa, and 29 kDa, respectively (, #4–6).
Figure 5. SDS-PAGE of partial purification of the antifungal proteins from the ammonium sulfate precipitation and anion exchange chromatography steps. The labels 9–10 and 33–34 refer to the proteins from DEAE fractions 9–10 and 33–34, respectively.
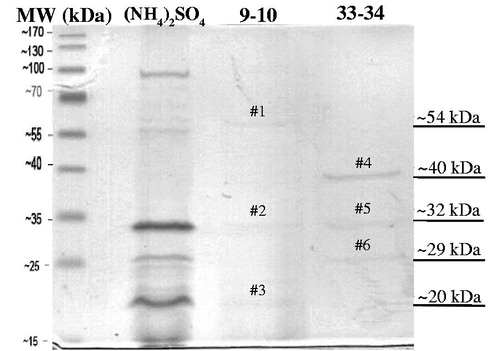
Table 2. Summary for purification of the antifungal protein from P. aeruginosa RS1.
3.4. Identification of antifungal proteins with LC mass spectrometry analysis
All six protein bands (#1–6) were excised from the gel and subjected to protein identification by LC/MS. The data obtained were compared with the Global Proteome Machine Database, and the results of each protein band are reported in . Protein band #1–3 which was obtained from fraction 9–10 showed high similarity to catalase, protease, and protease IV, with a protein coverage of 65%, 53%, and 40%, respectively. Protein band #4–6 from fraction 33–34 showed high similarity to chitin binding domain, protease, and protease with protein coverage of 86%, 53%, and 33%, respectively. These matched proteins belonged to the proteins from Pseudomonas aeruginosa LESB58.
Table 3. Identification of each protein band from anion exchange chromatography fractions 9–10 and 33–34.
3.5. Microscopy
Morphology of P. palmivora after incubation with purified antifungal protein for 2 days was observed under SEM. The result revealed that the hyphae of the control P. palmivora without the purified protein had straight and smooth hyphae (). However, P. palmivora grown with both the purified protein from fractions 9–10 and 33–34 had abnormal growth showing short branched, rugous, and swollen hyphal tips (, respectively).
4. Discussion
In this research, to obtain high antifungal activity against P. palmivora by P. aeruginosa RS1, cultivation conditions for the bacteria including type of medium, pH, temperature, and time-course were optimized. The antifungal proteins were purified from supernatant of the bacteria by ammonium sulfate precipitation, followed by DEAE-column chromatography, and identified by LC/MS. This work demonstrates that P. aeruginosa RS1 is capable of producing several anti-P. palmivora proteins that may be useful for controlling black rot disease caused by P. palmivora in orchid orchards.
The results revealed that varying the type of culture medium was highly influential to the antifungal activity, while the variation of pH and temperature showed less affect. In our study, LB was found to be the best medium for producing the antifungal substance as compared with TSB and NB at 4 and 1.8 fold, respectively. The difference in inhibition efficiency also indicated that media composition has an effect on anti-P. palmivora metabolite production. Although generally, TSB is the richest nutrient composition medium when compared with NB and LB media, and can well support the growth of several bacteria, TSB and NB may interfere with antifungal production as previously observed [Citation31,Citation32]. The original conditions, pH 7 and 37 °C, seemed to be optimal and demonstrated the highest percentage of inhibition when compared with other tested conditions. Thus, the optimal condition for producing an antifungal agent by P. aeruginosa RS1 in this study was to culture the bacteria in LB medium at pH 7 and at 37 °C for 21 h. The bioactive antifungal compounds were non-growth associated products since the activity was appeared in the late logarithmic phase of P. aeruginosa RS1, which are similar to most metabolites produced by bacterial biocontrol agents.
The purification of antifungal proteins in this research consisted of two steps including ammonium sulfate precipitation and DEAE ion exchange chromatography. The 40–80% ammonium sulfate saturation contained antifungal proteins. The proteins in this fraction were purified further by DEAE-cellulose column chromatography. Several pools of eluted proteins retained antifungal activity, which indicated that there were several types of antifungal proteins produced by P. aeruginosa RS1. The highest activity was obtained from fractions 9–10 and 33–34 with the inhibition of 75.00 ± 2.08% and 82.35 ± 2.08%, respectively. The proteins in fraction 9–10 were eluted before starting the gradient of NaCl, while the proteins in fraction 33–34 were eluted at about 0.7–0.9 M gradient of NaCl. These results indicated that the proteins in fraction 9–10 have no negative charge with which to bind a positive ion onto DEAE, whereas the proteins in fraction 33–34 have a strong negative charge [Citation33–35]. SDS-PAGE indicated that there were three apparent protein bands from each pooled fraction. Bands #1–3 were from fraction 9–10 and bands #4–6 were from fraction 33–34. LC/MS revealed that these proteins were closed similarly to catalase, protease and chitin binding proteins.
Catalase is an enzyme that is produced by variety of organisms exposed to oxygen. It catalyzes the decomposition of hydrogen peroxide to water and oxygen [Citation36]. Catalase was reported as antifungal substance, for example, the Bacillus subtilis strain LEV-006 showed antifungal activity against Sclerotinia sclerotiorum, Rhizoctonia solani, Alternaria brassicae, and Leptosphaeria maculans. One antifungal protein was found to be similar to vegetative catalase 1; however, expression of catalase in Escherichia coli showed catalase activity but no antifungal activity [Citation37]. Thus, the antifungal mechanism of catalase is still unknown and needs to be clarified.
Proteases can be found in animals, plants, fungi, bacteria, archaea, and viruses. They are also called peptidases or proteinases, and are any enzyme that breaks down proteins into smaller polypeptides or amino acids by hydrolysis of peptide bonds. The different classes of protease illustrate the same reaction by completely different catalytic mechanisms [Citation38]. In antifungal applications, alkaline antifungal protease was discovered from B. subtilis strain N7. The enzyme completely inhibited spore and hyphae growth of Fusarium oxysporum f. sp. cucumerinum by hydrolysis and loss of integrity of the fungal cell wall [Citation39]. Based on SEM, it appeared that the action of the partial purified proteins occurred during hypha development or cell wall synthesis. In case of Phytophthora, cell wall compositions were composed of 80–90% glucan, 5–10% protein, and 1–2% lipids [Citation40]. In addition, cell-wall-associated proteins were also identified in this oomycete mold, and these proteins were proposed to be important for pathogenicity [Citation41]. The antifungal proteins of the protein bands #2, #3, #5, and #6 may individually or in combination act on the proteins on the cell wall of P. palmivora.
The secretion of several chitinolytic enzymes and chitin-binding proteins has been found in the extracellular environment of several species of Serratia, Bacillus, Pseudomonas, and Vibrio [Citation42]. The functions of chitinolytic enzymes and chitin-binding proteins are to degrade chitin on the cell wall of fungi. It was reported that proteins that consisted of only chitin-binding domains revealed fungal growth inhibition [Citation43]. In addition, the chitin-binding protein of P. aeruginosa strain PAO25 has been reported to secrete chitin-binding proteins and have antifungal activity against R. solani and F. oxysporum [Citation43]. Although it was reported that the cell wall of oomycete molds does not contain chitin, recent research has found that these water molds could temporarily produce chitin at the hyphal tips before converting it to cellulose [Citation44]. Thus, the chitin binding domain of protein band #4 could act on chitin at this stage. It is also possible that the chitinolytic activity alone may not directly affect the growth of P. palmivora and requires a synergistic action of other bioactive compounds to be effective [Citation45,Citation46].
In this study, it was demonstrated that P. aeruginosa RS1 produced several proteins that can inhibit the growth of P. palmivora. Although the mode of action of each purified protein needs to be verified, these mixed proteins can also be used to obtain synergistic antifungal actions. Recent study reported that P. aeruginosa strain JO and strain JO7 produced β-1,3-glucanase, cellulase, protease, and chitinase. These production of several wall lytic enzymes attributed to high antifungal efficiency against F. oxysporum and Alternaria solani [Citation47].
Several P. aeruginosa strains were found to produce antifungal substances against both human and plant pathogenic fungi. For instance, several derivatives of phenazine were produced by P. aeruginosa, which had antifungal activity towards human pathogenic fungi such as Candida albicans [Citation48]. Pseudomonas aeruginosa K-187 produced pafungin that affected F. oxysporum, causing swelling and lysis of the fungal hyphae, which subsequently inhibited damping-off disease in alfalfa [Citation15]. Generally, due to public concern regarding human health and environment, P. aeruginosa has not been widely accepted as a safe biocontrol agent in agriculture. However, non-pathogenic P. aeruginosa strains ATCC 15442 with noninvasive and noncytotoxic properties has been reported [Citation49]. Therefore, genome sequencing as well as these properties of P. aeruginosa RS1 would suggest further for its public concern issue. Nevertheless, its cell-free metabolites would be an alternative approach in agriculture to inhibit the growth of P. palmivora, the causal pathogen of orchids.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Mauffret A, Baran N, Joulian C. Effect of pesticides and metabolites on groundwater bacterial community. Sci Total Environ. 2017;576:879–887.
- Aktar MW, Sengupta D, Chowdhury A. Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip Toxicol. 2009;2:1–12.
- Martínez-Medina A, Del Mar Alguacil M, Pascual JA, et al. Phytohormone profiles induced by Trichoderma isolates correspond with their biocontrol and plant growth-promoting activity on melon plants. J Chem Ecol. 2014;40:804–815.
- DiTomaso JM, Van Steenwyk RA, Nowierski RM, et al. Addressing the needs for improving classical biological control programs in the USA. Biol Control. 2017;106:35–39.
- Velivelli SL, De Vos P, Kromann P, et al. Biological control agents: from field to market, problems, and challenges. Trends Biotechnol. 2014;32:493–496.
- Aksoy HM, Kaya Y, Ozturk M, et al. Pseudomonas putida – induced response in phenolic profile of tomato seedlings (Solanum lycopersicum L.) infected by Clavibacter michiganensis subsp michiganensis. Biol Control. 2017;105:6–12.
- Law JW, Ser HL, Khan TM, et al. The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front Microbiol. 2017;8:3.
- Torres MJ, Pérez Brandan C, Sabaté DC, et al. Biological activity of the lipopeptide-producing Bacillus amyloliquefaciens PGPBacCA1 on common bean Phaseolus vulgaris L. pathogens. Biol Control. 2017;105:93–99.
- Wang SL, Yieh TC, Shih IL. Production of antifungal compounds by Pseudomonas aeruginosa K-187 using shrimp and crab shell powder as a carbon source. Enzyme Microb Technol. 1999;25:142–148.
- Botelho GR, Mendonça-Hagler LC. Fluorescent pseudomonads associated with the rhizosphere of crops - an overview. Braz J Microbiol. 2006;37:401–416.
- Guyer A, De Vrieze M, Bonisch D, et al. The anti-Phytophthora effect of selected potato-associated Pseudomonas strains: from the laboratory to the field. Front Microbiol. 2015;6:1309.
- Hunziker L, Bonisch D, Groenhagen U, et al. Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl Environ Microbiol. 2015;81:821–830.
- Borah SN, Goswami D, Sarma HK, et al. Rhamnolipid biosurfactant against Fusarium verticillioides to control stalk and ear rot disease of maize. Front Microbiol. 2016;7:1505.
- Yang MM, Wen SS, Mavrodi DV, et al. Biological control of wheat root diseases by the CLP-producing strain Pseudomonas fluorescens HC1-07. Phytopathology. 2014;104:248–256.
- Wanga SL, Yieh TC, Shih IL. Purification and characterization of a new antifungal compound produced by Pseudomonas aeruginosa K-187 in a shrimp and crab shell powder medium. Enzyme Microb Technol. 1999;25:439–446.
- Yen YH, Li PL, Wang CL, et al. An antifungal protease produced by Pseudomonas aeruginosa M-1001 with shrimp and crab shell powder as a carbon source. Enzyme Microb Technol. 2006;39:311–317.
- Hsiao YY, Pan ZJ, Hsu CC, et al. Research on orchid biology and biotechnology. Plant Cell Physiol. 2011;52:1467–1486.
- De LC, Pathak P, Rao AN, et al. 2 Global Orchid Industry. In: De LC, editor. Commercial Orchids. Berlin: De Gruyter Open; 2014. p. 13–19.
- Pizano M. International market trends - tropical flowers. Proceeding of the V International Symposium on New Floricultural Crops; 2003 Aug 26-30; Paranà. International Society for Horticultural Science (ISHS), Leuven; 2003. p. 79–86.
- Moon H, Park HJ, Jeong A, et al. Isolation and identification of Burkholderia gladioli on Cymbidium orchids in Korea. Biotechnol Biotechnol Equip. 2016;31:280–288.
- Sudha DR, Rani GU. Detection, diagnosis of orchid virus and inactivation of cymbidium mosaic virus (CYMV) on plants. Int J Plant Sci. 2016;11:302–306.
- Li J, Wang R, Wang Z, et al. The phylogenetic relationship and non-specific symbiotic habit of mycorrhiza fungi from a terrestrial orchid (Cymbidium). Nord J Bot. 2016;34:343–348.
- Maketon C, Tongjib Y, Patipong T, et al. Greenhouse evaluations of harpin protein and microbial fungicides in controlling Curvularia lunata, Fusarium moniliforme, and Phythopthora palmivora, major causes of orchid diseases in Thailand. Life Sci J. 2015;12:125–132.
- Středa T, Krédl Z, Pokorný R, et al. Effect of wetting period on infection of orchid flowers by Alternaria alternata and Curvularia eragrostidis. N Z J Crop Hortic Sci. 2013;41:1–8.
- Laurence MH, Howard C, Summerell BA, et al. Identification of Fusarium solani f. sp. phalaenopsis in Australia. Australasian Plant Dis Notes. 2016;11:3.
- Meera T, Louis V, Beena S. Diseases of Phalaenopsis: symptoms, etiology and management. Int J Agric Res Innov Technol. 2016;5:296–300.
- Cating R, Palmateer A, Stiles C, et al. Black rot of orchids caused by Phytophthora cactorum and Phytophthora palmivora in Florida. Plant Health Progr. 2010 [cited June 14]. June 14. DOI:10.1094/PHP-2010-0614-01-DG
- Dooh JPN, Ambang Z, Bekolo N, et al. Effect of extracts of Thevetia peruviana on the development of Phytophthora megakarya causal agent of black pod disease of cocoa. J App Bioscience. 2014;77:6564–6574.
- Laemmli UK, Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973;80:575–599.
- Behdani M, Pooyan M, Abbasi S. Evaluation of antifungal activity of some medicinal plants essential oils against Botrytis cinerea, causal agent of postharvest apple rot, in vitro. Intl J Agri Crop Sci. 2012;4:1012–1016.
- Valentine N, Wunschel S, Wunschel D, et al. Effect of culture conditions on microorganism identification by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry. Appl Environ Microbiol. 2005;71:58–64.
- Lee HA, Kim JH. Isolation of Bacillus amyloliquefaciens strains with antifungal activities from Meju. Prev Nutr Food Sci. 2012;17:64–70.
- Cummins PM, Dowling O, O'Connor BF. Ion-exchange chromatography: basic principles and application to the partial purification of soluble mammalian prolyl oligopeptidase. Methods Mol Biol. 2011;681:215–228.
- Hegedüs N, Marx F. Antifungal proteins: more than antimicrobials? Fungal Biol Rev. 2013;26:132–145.
- Ouedraogo JP, Hagen S, Spielvogel A, et al. Survival strategies of yeast and filamentous fungi against the antifungal protein AFP. J Biol Chem. 2011;286:13859–13868.
- Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208.
- Hou X, Boyetchko SM, Brkic M, et al. Characterization of the anti-fungal activity of a Bacillus spp. associated with sclerotia from Sclerotinia sclerotiorum. Appl Microbiol Biotechnol. 2006;72:644–653.
- Oda K. New families of carboxyl peptidases: serine-carboxyl peptidases and glutamic peptidases. J Biochem. 2012;151:13–25.
- Luo Y, Sun L, Zhu Z, et al. Identification and characterization of an anti-fungi Fusarium oxysporum f. sp. cucumerium protease from the Bacillus subtilis strain N7. J Microbiol. 2013;51:359–366.
- Tokunaga J, Bartnicki-Garcia S. Structure and differentiation of the cell wall of Phytophthora palmivora: cysts, hyphae and sporangia. Archiv Fur Mikrobiologie. 1971;79:293–310.
- Meijer HJ, van de Vondervoort PJ, Yin QY, et al. Identification of cell wall-associated proteins from Phytophthora ramorum. Mol Plant-Microbe Interact. 2006;19:1348–1358.
- Folders J, Algra J, Roelofs MS, et al. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J Bacteriol. 2001;183:7044–7052.
- Folders J, Tommassen J, van Loon LC, et al. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J Bacteriol. 2000;182:1257–1263.
- Guerriero G, Avino M, Zhou Q, et al. Chitin synthases from Saprolegnia are involved in tip growth and represent a potential target for anti-oomycete drugs. PLoS Pathog. 2010;6:e1001070.
- Kim YC, Jung H, Kim KY, et al. An effective biocontrol bioformulation against Phytophthora blight of pepper using growth mixtures of combined chitinolytic bacteria under different field conditions. Eur J Plant Pathol. 2008;120:373–382.
- Arora NK, Kim MJ, Kang SC, et al. Role of chitinase and beta-1,3-glucanase activities produced by a fluorescent pseudomonad and in vitro inhibition of Phytophthora capsici and Rhizoctonia solani. Can J Microbiol. 2007;53:207–212.
- Paramanandham P, Rajkumari J, Pattnaik S, et al. Biocontrol potential against Fusarium oxysporum f. sp. lycopersici and Alternaria solani and tomato plant growth due to plant growth-promoting rhizobacteria. Int J Veg Sci. 2017;23:294–303.
- Morales DK, Jacobs NJ, Rajamani S, et al. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol Microbiol. 2010;78:1379–1392.
- Wang Y, Li C, Gao C, et al. Genome sequence of the nonpathogenic Pseudomonas aeruginosa strain ATCC 15442. Genome Announc. 2014;2:e00421-14.